Systemic Blood Proteome Patterns Reflect Disease Phenotypes in Neovascular Age-Related Macular Degeneration
Abstract
1. Introduction
2. Results
2.1. Study Population and Baseline Clinical Characteristics
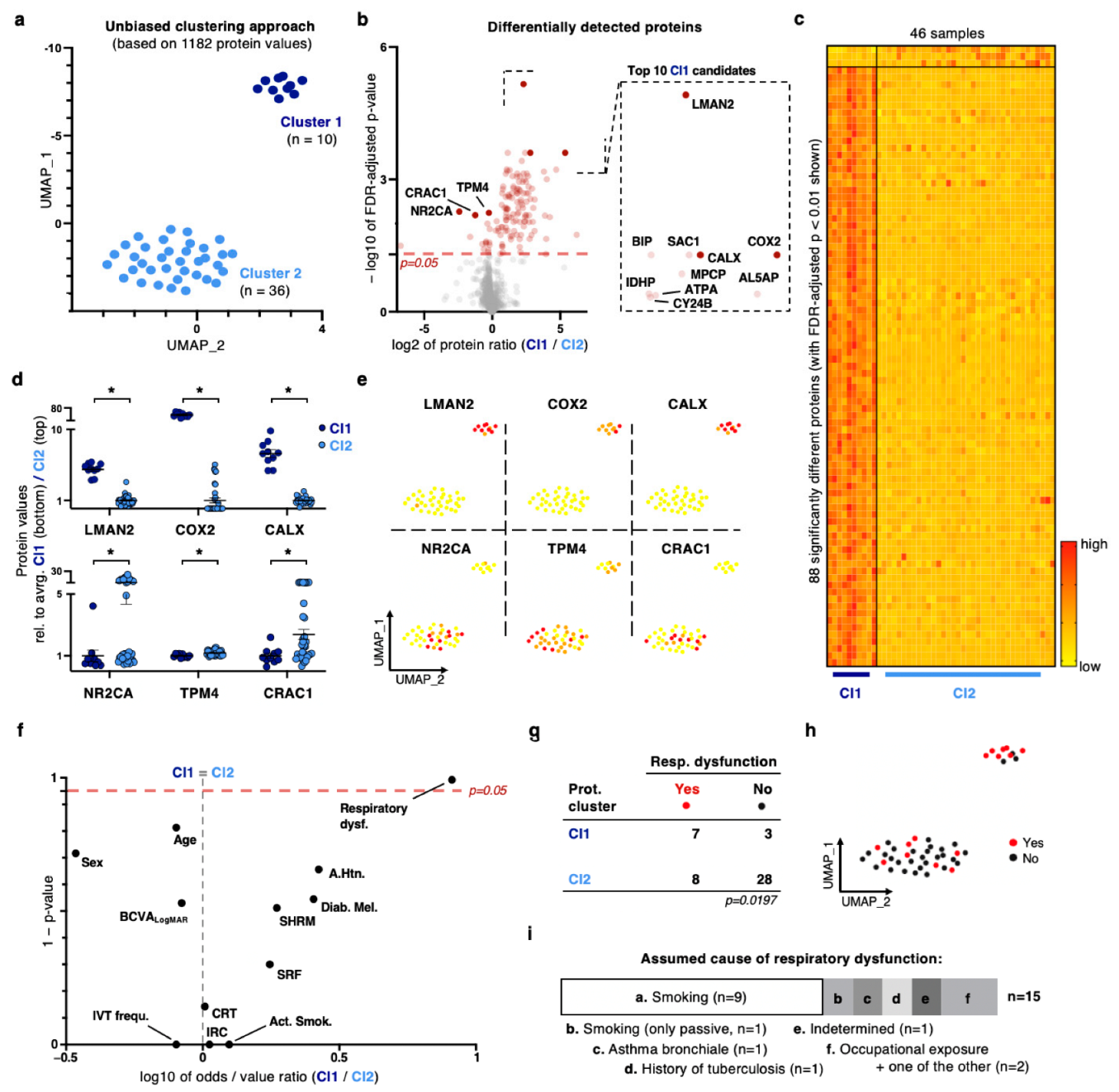
2.2. Unbiased Dimensionality Reduction and Clustering Reveal a Distinct Patient Sub-Cohort with a Strong Proteomic Signature for Oxidative Stress Response and Respiratory Dysfunction as an Underlying Health Condition
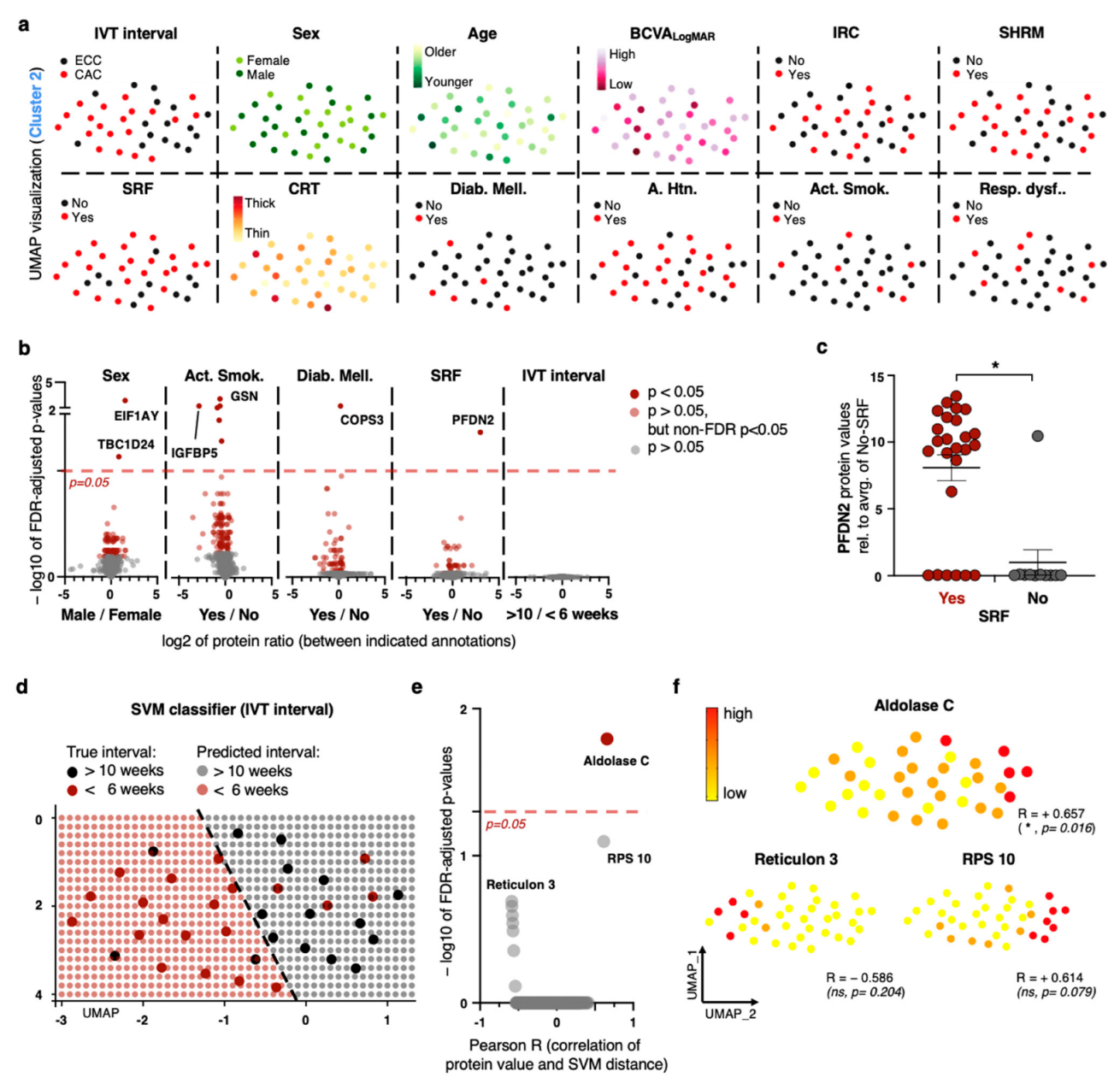
2.3. Exploiting Meta-Feature Annotations on a Patient Level Yields a Limited Number of Biomarker Discoveries
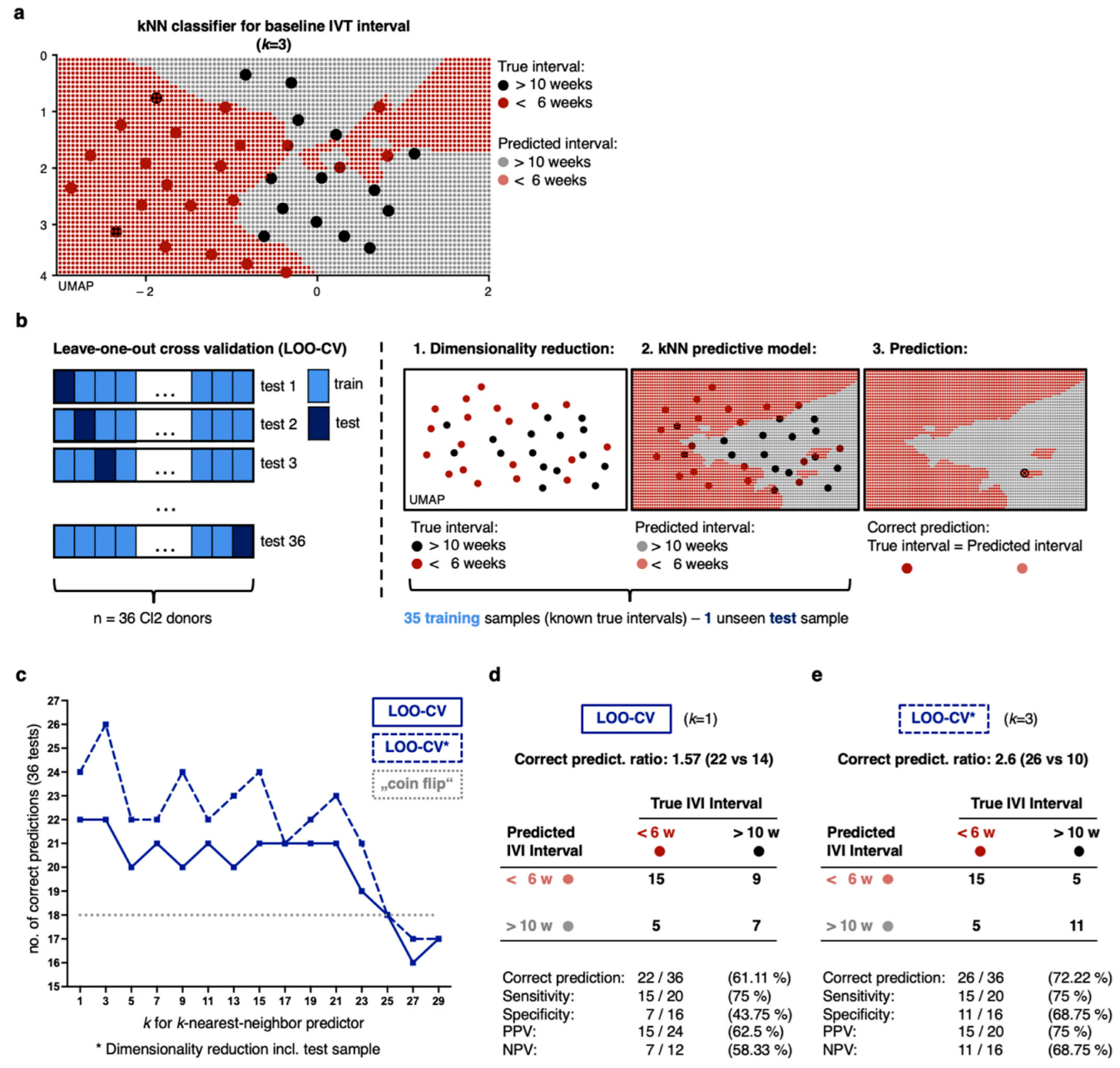
2.4. Non-Linear Classification Model Detects the Level of CNV Activity Based on Similarities in a High Number of Proteomic Dimensions
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. Study Protocol and Subject Recruitment
4.3. Clinical Examination and Meta-Feature Logging
4.4. Sample Collection and Mass Spectrometry Analysis
4.5. Statistical Approach and Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| anti-VEGF | anti-vascular endothelial growth factor |
| BCVA | best-corrected visual acuity |
| CAC | chronically active CNV |
| CNV | choroidal neovascularization |
| CRT | central retinal thickness |
| ECC | effectively controlled CNV |
| ETDRS | The Early Treatment Diabetic Retinopathy Study Group |
| FAF | fundus autofluorescence |
| FA | fluorescein angiography |
| IRC | intraretinal cysts |
| IVT | intravitreal treatment |
| kNN | k-nearest neighbors |
| LC-MS | liquid chromatography with tandem mass spectrometry |
| LOO-CV | leave-one-out cross validation |
| ML | machine learning |
| nAMD | neovascular age-related macular degeneration |
| OCT | optical coherence tomography |
| SHRM | subretinal hyperreflective material |
| SRF | subretinal fluid |
| UMAP | Uniform Manifold Approximation and Projection |
References
- Fleckenstein, M.; Keenan, T.D.; Guymer, R.H.; Chakravarthy, U.; Schmitz-Valckenberg, S.; Klaver, C.C.; Wong, W.T.; Chew, E.Y. Age-related macular degeneration. Nat. Rev. Dis. Prim. 2021, 7, 31. [Google Scholar] [CrossRef]
- Chong, V. Ranibizumab for the treatment of wet AMD: A summary of real-world studies. Eye 2016, 30, 270–286. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, P.J.; Brown, D.M.; Heier, J.S.; Boyer, D.S.; Kaiser, P.K.; Chung, C.Y.; Kim, R.Y. Ranibizumab for Neovascular Age-Related Macular Degeneration. N. Engl. J. Med. 2006, 355, 1419–1431. [Google Scholar] [CrossRef] [PubMed]
- Mullard, A. FDA approves bispecific antibody for two eye diseases. Nat. Rev. Drug Discov. 2022, 21, 170. [Google Scholar] [CrossRef]
- Heier, J.S.; Khanani, A.M.; Ruiz, C.Q.; Basu, K.; Ferrone, P.J.; Brittain, C.; Figueroa, M.S.; Lin, H.; Holz, F.G.; Patel, V.; et al. Efficacy, durability, and safety of intravitreal faricimab up to every 16 weeks for neovascular age-related macular degeneration (TENAYA and LUCERNE): Two randomised, double-masked, phase 3, non-inferiority trials. Lancet 2022, 399, 729–740. [Google Scholar] [CrossRef]
- Li, M.; Dolz-Marco, R.; Messinger, J.D.; Ferrara, D.; Freund, K.B.; Curcio, C.A. Neurodegeneration, gliosis, and resolution of haemor-rhage in neovascular age-related macular degeneration, a clinicopathologic correlation. Eye 2021, 35, 548–558. [Google Scholar] [CrossRef]
- Kauppinen, A.; Paterno, J.J.; Blasiak, J.; Salminen, A.; Kaarniranta, K. Inflammation and its role in age-related macular degeneration. Cell. Mol. Life Sci. 2016, 73, 1765–1786. [Google Scholar] [CrossRef]
- Ferrara, N. Molecular and biological properties of vascular endothelial growth factor. J. Mol. Med. 1999, 77, 527–543. [Google Scholar] [CrossRef]
- Apte, R.S.; Chen, D.S.; Ferrara, N. VEGF in Signaling and Disease: Beyond Discovery and Development. Cell 2019, 176, 1248–1264. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.W.; Le Couter, J.; Strauss, E.C.; Ferrara, N. Vascular endothelial growth factor A in intraocular vascular disease. Ophthalmology 2013, 120, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, N.; Gerber, H.-P.; LeCouter, J. The biology of VEGF and its receptors. Nat. Med. 2003, 9, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Lamb, J.R.; Jennings, L.L.; Gudmundsdottir, V.; Gudnason, V.; Emilsson, V. It’s in Our Blood: A Glimpse of Personalized Medicine. Trends Mol. Med. 2021, 27, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Emilsson, V.; Ilkov, M.; Lamb, J.R.; Finkel, N.; Gudmundsson, E.F.; Pitts, R.; Gudnason, V. Co-regulatory networks of human serum proteins link genetics to disease. Science 2018, 36, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Suhre, K.; McCarthy, M.I.; Schwenk, J.M. Genetics meets proteomics: Perspectives for large population-based studies. Nat. Rev. Genet. 2020, 22, 19–37. [Google Scholar] [CrossRef]
- García-Quintanilla, L.; Rodríguez-Martínez, L.; Bandín-Vilar, E.; Gil-Martínez, M.; González-Barcia, M.; Mondelo-García, C.; Fernández-Ferreiro, A.; Mateos, J. Recent Advances in Proteomics-Based Approaches to Studying Age-Related Macular Degeneration: A Systematic Review. Int. J. Mol. Sci. 2022, 23, 14759. [Google Scholar] [CrossRef]
- Yuan, X.; Gu, X.; Crabb, J.S.; Yue, X.; Shadrach, K.; Hollyfield, J.G.; Crabb, J.W. Quantitative proteomics: Comparison of the macular bruch membrane/choroid complex from age-related macular degeneration and normal eyes. Mol. Cell. Proteom. 2010, 9, 1031–1046. [Google Scholar] [CrossRef]
- Joo, J.-H.; Kim, H.; Shin, J.-H.; Moon, S.W. Aqueous humor cytokine levels through microarray analysis and a sub-analysis based on optical coherence tomography in wet age-related macular degeneration patients. BMC Ophthalmol. 2021, 21, 399. [Google Scholar] [CrossRef]
- Rinsky, B.; Beykin, G.; Grunin, M.; Amer, R.; Khateb, S.; Tiosano, L.; Almeida, D.; Hagbi-Levi, S.; Elbaz-Hayoun, S.; Chowers, I. Analysis of the aqueous humor proteome in patients with age-related macular degeneration. Investig. Opthalmol. Vis. Sci. 2021, 62, 18. [Google Scholar] [CrossRef]
- Erie, J.C.; Good, J.A.; Butz, J.A.; Hodge, D.O.; Pulido, J.S. Urinary Cadmium and Age-related Macular Degeneration. Am. J. Ophthalmol. 2007, 144, 414–418.e1. [Google Scholar] [CrossRef]
- Sivaprasad, S.; Chong, N.V.; Bailey, T.A. Serum elastin-derived peptides in age-related macular degeneration. Investig. Opthalmology Vis. Sci. 2005, 46, 3046–3051. [Google Scholar] [CrossRef]
- Kim, H.J.; Ahn, S.J.; Woo, S.J.; Hong, H.K.; Suh, E.J.; Ahn, J.; Park, J.H.; Ryoo, N.K.; Lee, E.J.; Kim, K.W.; et al. Proteomics-based identification and validation of novel plasma biomarkers phospholipid transfer protein and mannan-binding lectin serine protease-1 in age-related macular degeneration. Sci. Rep. 2016, 6, 32548. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhu, M.; Geng-Spyropoulos, M.; Shardell, M.; Gonzalez-Freire, M.; Gudnason, V.; Eiriksdottir, E.G.; Schaumberg, D.; Van Eyk, J.E.; Ferrucci, L.; et al. A novel, multiplexed targeted mass spectrometry assay for quantification of complement factor H (CFH) variants and CFH-related proteins 1–5 in human plasma. Proteomics 2017, 17, 1600237. [Google Scholar] [CrossRef] [PubMed]
- Krishnakumar, S.; Sivagurunathan, S.; Selvan, L.N.; Khan, A.; Parameswaran, S.; Bhattacharjee, H.; Gogoi, K.; Gowda, H.; Prasad, T.K.; Pandey, A.; et al. Proteomics-based approach for differentiation of age-related macular degeneration sub-types. Indian J. Ophthalmol. 2021, 69, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Emilsson, V.; Gudmundsson, E.F.; Jonmundsson, T.; Jonsson, B.G.; Twarog, M.; Gudmundsdottir, V.; Li, Z.; Finkel, N.; Poor, S.; Liu, X.; et al. A proteogenomic signature of age-related macular degeneration in blood. Nat. Commun. 2022, 13, 3401. [Google Scholar] [CrossRef]
- Senabouth, A.; Daniszewski, M.; Lidgerwood, G.E.; Liang, H.H.; Hernández, D.; Mirzaei, M.; Keenan, S.N.; Zhang, R.; Han, X.; Neavin, D.; et al. Transcriptomic and proteomic retinal pigment epithelium signatures of age-related macular degeneration. Nat. Commun. 2022, 13, 4233. [Google Scholar] [CrossRef]
- Flores-Bellver, M.; Mighty, J.; Aparicio-Domingo, S.; Li, K.V.; Shi, C.; Zhou, J.; Cobb, H.; McGrath, P.; Michelis, G.; Lenhart, P.; et al. Extracellular vesicles released by human retinal pigment epithelium mediate increased polarised secretion of drusen proteins in response to AMD stressors. J. Extracell. Vesicles 2021, 10, e12165. [Google Scholar] [CrossRef]
- Cai, B.; Zhang, Z.; Sun, S.; Lin, T.; Ke, Y.; Li, Z.; Yang, J.; Li, X. A Pilot Application of an iTRAQ-Based Proteomics Screen Estimates the Effects of Cigarette Smokers’ Serum on RPE Cells with AMD High-Risk Alleles. Transl. Vis. Sci. Technol. 2022, 11, 15. [Google Scholar] [CrossRef]
- Koss, M.J.; Hoffmann, J.; Nguyen, N.; Pfister, M.; Mischak, H.; Mullen, W.; Husi, H.; Rejdak, R.; Koch, F.; Jankowski, J.; et al. Proteomics of Vitreous Humor of Patients with Exudative Age-Related Macular Degeneration. PLoS ONE 2014, 9, e96895. [Google Scholar] [CrossRef]
- Nobl, M.; Reich, M.; Dacheva, I.; Siwy, J.; Mullen, W.; Schanstra, J.P.; Choi, C.Y.; Kopitz, J.; Kretz, F.T.; Auffarth, G.U.; et al. Proteomics of vitreous in neovascular age-related macular degeneration. Exp. Eye Res. 2016, 146, 107–117. [Google Scholar] [CrossRef]
- Harrell, C.R.; Markovic, B.S.; Fellabaum, C.; Arsenijevic, A.; Djonov, V.; Volarevic, V. Molecular mechanisms underlying therapeutic potential of pericytes. J. Biomed. Sci. 2018, 25, 21. [Google Scholar] [CrossRef]
- Schori, C.; Trachsel, C.; Grossmann, J.; Zygoula, I.; Barthelmes, D.; Grimm, C. the proteomic landscape in the vitreous of patients with age-related and diabetic retinal disease. Investig. Opthalmol. Vis. Sci. 2018, 59, AMD31–AMD40. [Google Scholar] [CrossRef]
- Winiarczyk, M.; Kaarniranta, K.; Winiarczyk, S.; Adaszek, S.; Winiarczyk, D.; Mackiewicz, J. Tear film proteome in age-related macular degeneration. Graefe’s Arch. Clin. Exp. Ophthalmol. 2018, 256, 1127–1139. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.; Liew, G.; Gopinath, B.; Wong, T.Y. Age-related macular degeneration. Lancet 2018, 392, 1147–1159. [Google Scholar] [CrossRef] [PubMed]
- Kowal-Bielecka, O.; Chwieśko-Minarowska, S.; Bernatowicz, P.L.; Allanore, Y.; Radstake, T.; Matucci-Cerinic, M.; Broen, J.; Hesselstrand, R.; Krasowska, D.; Riemekasten, G.; et al. The arachidonate 5-lipoxygenase activating protein gene polymorphism is associated with the risk of scleroderma-related interstitial lung disease: A multicentre European Scleroderma Trials and Research group (EUSTAR) study. Rheumatology 2017, 56, 844–852. [Google Scholar] [CrossRef]
- Nakada, E.M.; Sun, R.; Fujii, U.; Martin, J.G. The Impact of Endoplasmic Reticulum-Associated Protein Modifications, Folding and Degradation on Lung Structure and Function. Front. Physiol. 2021, 12, 665622. [Google Scholar] [CrossRef]
- Choi, B.-H.; Kim, J.-S. Age-related decline in expression of calnexin. Exp. Mol. Med. 2004, 36, 499–503. [Google Scholar] [CrossRef] [PubMed]
- Lynch, A.M.; Wagner, B.D.; Weiss, S.J.; Wall, K.M.; Palestine, A.G.; Mathias, M.T.; Siringo, F.S.; Cathcart, J.N.; Patnaik, J.L.; Drolet, D.W.; et al. Proteomic profiles in advanced age-related macular degeneration using an aptamer-based proteomic technology. Transl. Vis. Sci. Technol. 2019, 8, 14. [Google Scholar] [CrossRef]
- Garcia, I.P.L.; Alfaro-Arnedo, E.; Canalejo, M.; Urtubia, A.; Roncero, A.; Ruiz-Martínez, C.; Hoeflich, A.; Höflich, C.; Gotera, C.; Barba, G.P.; et al. Insulin-Like Growth Factors and IGF-Binding Proteins levels in serum from smokers and patients with different grades of COPD, COPD and lung cancer and exacerbated COPD. Eur. Respir. J. 2021, 58, PA1952. [Google Scholar] [CrossRef]
- Zhang, Y.; Kent, J.; Olivier, M.; Ali, O.; Broeckel, U.; Abdou, R.; Dyer, T.; Comuzzie, A.; Curran, J.; Carless, M.; et al. QTL-based association analyses reveal novel genes influencing pleiotropy of metabolic syndrome (MetS). Obesity 2013, 21, 2099–2111. [Google Scholar] [CrossRef]
- Liang, J.; Xia, L.; Oyang, L.; Lin, J.; Tan, S.; Yi, P.; Han, Y.; Luo, X.; Wang, H.; Tang, L.; et al. The functions and mechanisms of prefoldin complex and prefoldin-subunits. Cell Biosci. 2020, 10, 87. [Google Scholar] [CrossRef]
- Huu, T.N.; Park, J.; Zhang, Y.; Park, I.; Yoon, H.J.; Woo, H.A.; Lee, S.-R. Redox Regulation of PTEN by Peroxiredoxins. Antioxidants 2021, 10, 302. [Google Scholar] [CrossRef]
- Noble, W.S. What is a support vector machine? Nat. Biotechnol. 2006, 24, 1565–1567. [Google Scholar] [CrossRef] [PubMed]
- Adamus, G. Impact of autoantibodies against glycolytic enzymes on pathogenicity of autoimmune retinopathy and other au-toimmune disorders. Front. Immunol. 2017, 8, 505. [Google Scholar] [CrossRef] [PubMed]
- Morohoshi, K.; Ohbayashi, M.; Patel, N.; Chong, V.; Bird, A.C.; Ono, S.J. Identification of anti-retinal antibodies in patients with age-related macular degeneration. Exp. Mol. Pathol. 2012, 93, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Lever, J.; Krzywinski, M.; Altman, N. Model selection and overfitting. Nat. Methods 2016, 13, 703–704. [Google Scholar] [CrossRef]
- Bian, Y.; Zheng, R.; Bayer, F.P.; Wong, C.; Chang, Y.-C.; Meng, C.; Zolg, D.P.; Reinecke, M.; Zecha, J.; Wiechmann, S.; et al. Robust, reproducible and quantitative analysis of thousands of proteomes by micro-flow LC–MS/MS. Nat. Commun. 2020, 11, 157. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J.; Saul, N.; Großberger, L. UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw. 2018, 3, 861. [Google Scholar] [CrossRef]
- Brischigliaro, M.; Zeviani, M. Cytochrome c oxidase deficiency. Biochim. Biophys. Acta Bioenerg. 2021, 1862, 148335. [Google Scholar] [CrossRef]
- Kraus, A.; Groenendyk, J.; Bedard, K.; Baldwin, T.A.; Krause, K.H.; Dubois-Dauphin, M.; Dyck, J.; Rosenbaum, E.E.; Korngut, L.; Colley, N.J.; et al. Calnexin deficiency leads to dysmye-lination. J. Biol. Chem. 2010, 285, 18928–18938. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Y.; Wu, Z.; Sun, P. Overexpression of TPM4 is associated with worse prognosis and immune infiltration in patients with glioma. BMC Neurol. 2023, 23, 17. [Google Scholar] [CrossRef]
- Jaeger, E.; members of the CORGI Consortium; Webb, E.; Howarth, K.; Carvajal-Carmona, L.; Rowan, A.; Broderick, P.; Walther, A.; Spain, S.; Pittman, A.; et al. Common genetic variants at the CRAC1 (HMPS) locus on chromosome 15q13.3 influence colorectal cancer risk. Nat. Genet. 2007, 40, 26–28. [Google Scholar] [CrossRef] [PubMed]
- Forman, H.J.; Zhang, H. Targeting oxidative stress in disease: Promise and limitations of antioxidant therapy. Nat. Rev. Drug Discov. 2021, 20, 689–709. [Google Scholar] [CrossRef] [PubMed]
- Chew, E.Y.; Clemons, T.E.; SanGiovanni, J.P.; Danis, R.; Ferris, F.L.; Elman, M. Lutein + zeaxanthin and omega-3 fatty acids for age-related macular degeneration: The Age-Related Eye Disease Study 2 (AREDS2) randomized clinical trial. JAMA 2013, 309, 2005–2015. [Google Scholar]
- Klein, R.; Klein, B.E.; Linton, K.L. Prevalence of age-related maculopathy. The Beaver Dam Eye Study. Ophthalmology 1992, 99, 933–943. [Google Scholar] [CrossRef] [PubMed]
- Moorthy, S.; Cheung, N.; Klein, R.; Shahar, E.; Wong, T.Y. Are lung disease and function related to age-related macular degeneration? Am. J. Ophthalmol. 2011, 151, 375–379. [Google Scholar] [CrossRef] [PubMed]
- De Fauw, J.; Ledsam, J.R.; Romera-Paredes, B.; Nikolov, S.; Tomasev, N.; Blackwell, S.; Ronneberger, O. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat. Med. 2018, 24, 1342–1350. [Google Scholar] [CrossRef]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M.; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902. [Google Scholar] [CrossRef]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.S.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018, 172, 1122–1131.e9. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Künzel, S.E.; Flesch, L.T.M.; Frentzel, D.P.; Knecht, V.A.; Rübsam, A.; Dreher, F.; Schütte, M.; Dubrac, A.; Lange, B.; Yaspo, M.-L.; et al. Systemic Blood Proteome Patterns Reflect Disease Phenotypes in Neovascular Age-Related Macular Degeneration. Int. J. Mol. Sci. 2023, 24, 10327. https://doi.org/10.3390/ijms241210327
Künzel SE, Flesch LTM, Frentzel DP, Knecht VA, Rübsam A, Dreher F, Schütte M, Dubrac A, Lange B, Yaspo M-L, et al. Systemic Blood Proteome Patterns Reflect Disease Phenotypes in Neovascular Age-Related Macular Degeneration. International Journal of Molecular Sciences. 2023; 24(12):10327. https://doi.org/10.3390/ijms241210327
Chicago/Turabian StyleKünzel, Steffen E., Leonie T. M. Flesch, Dominik P. Frentzel, Vitus A. Knecht, Anne Rübsam, Felix Dreher, Moritz Schütte, Alexandre Dubrac, Bodo Lange, Marie-Laure Yaspo, and et al. 2023. "Systemic Blood Proteome Patterns Reflect Disease Phenotypes in Neovascular Age-Related Macular Degeneration" International Journal of Molecular Sciences 24, no. 12: 10327. https://doi.org/10.3390/ijms241210327
APA StyleKünzel, S. E., Flesch, L. T. M., Frentzel, D. P., Knecht, V. A., Rübsam, A., Dreher, F., Schütte, M., Dubrac, A., Lange, B., Yaspo, M.-L., Lehrach, H., Joussen, A. M., & Zeitz, O. (2023). Systemic Blood Proteome Patterns Reflect Disease Phenotypes in Neovascular Age-Related Macular Degeneration. International Journal of Molecular Sciences, 24(12), 10327. https://doi.org/10.3390/ijms241210327






