Prolactin Regulates Ovine Ovarian Granulosa Cell Apoptosis by Affecting the Expression of MAPK12 Gene
Abstract
1. Introduction
2. Results
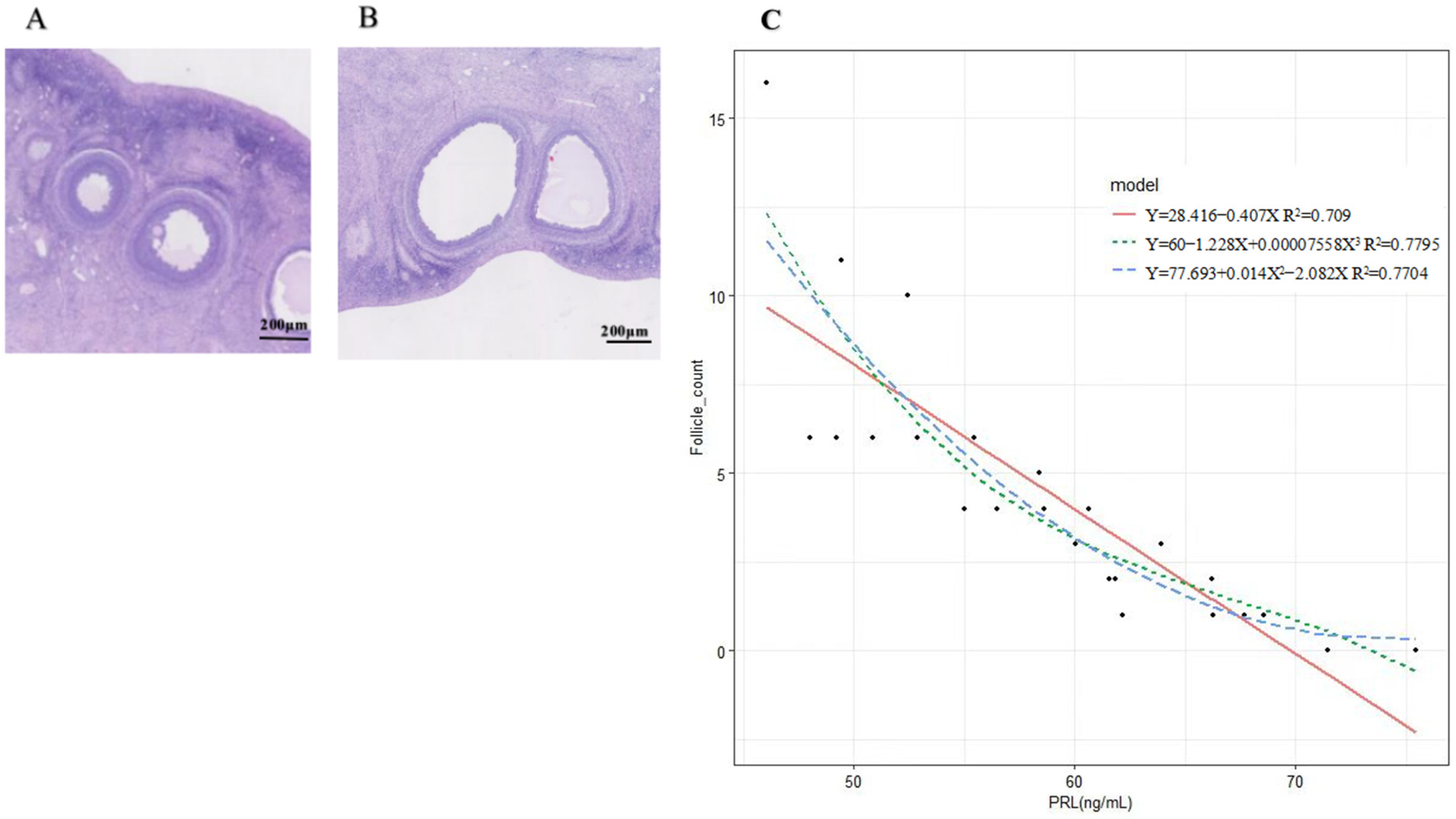
2.1. Regression Analysis
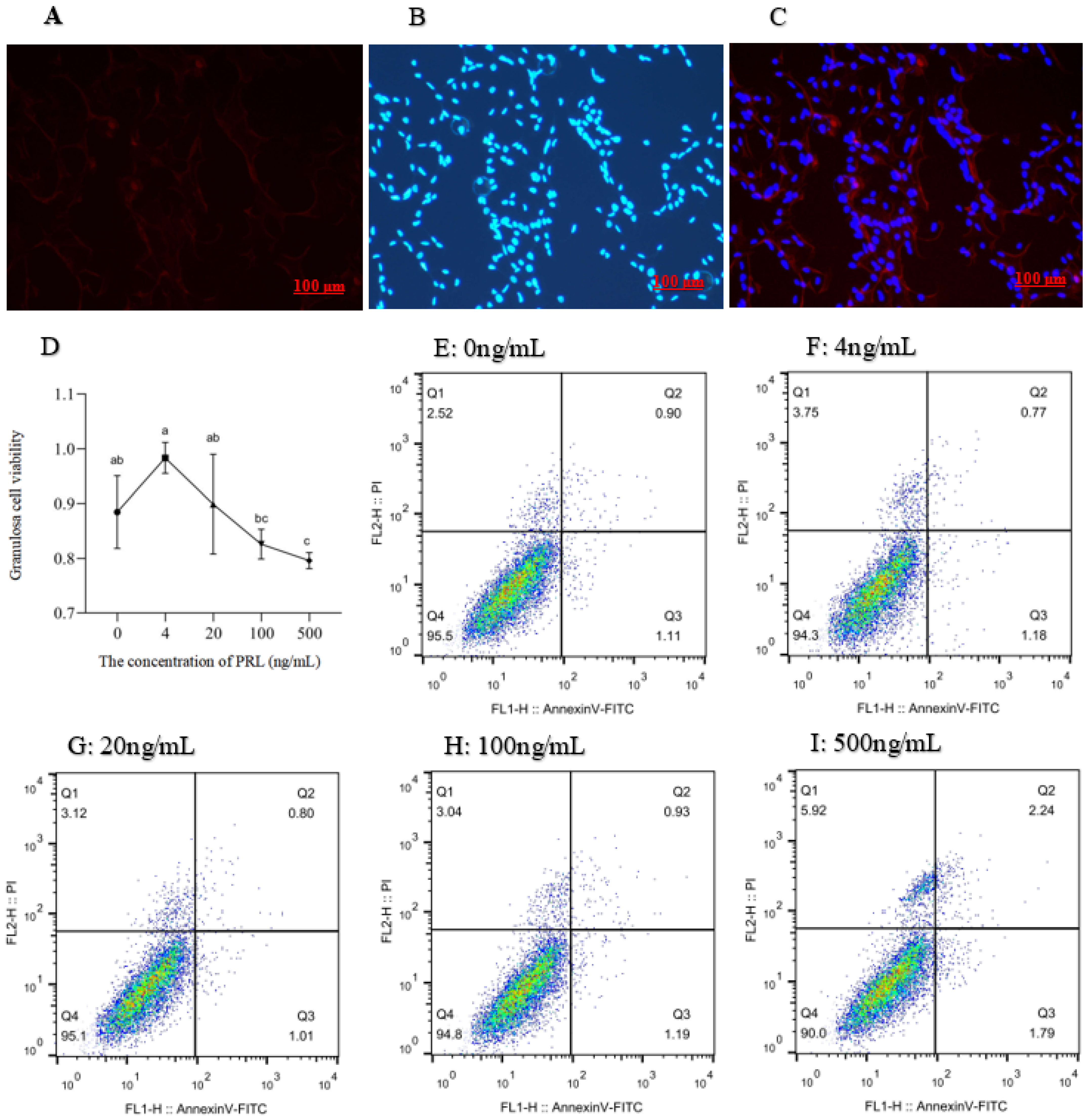
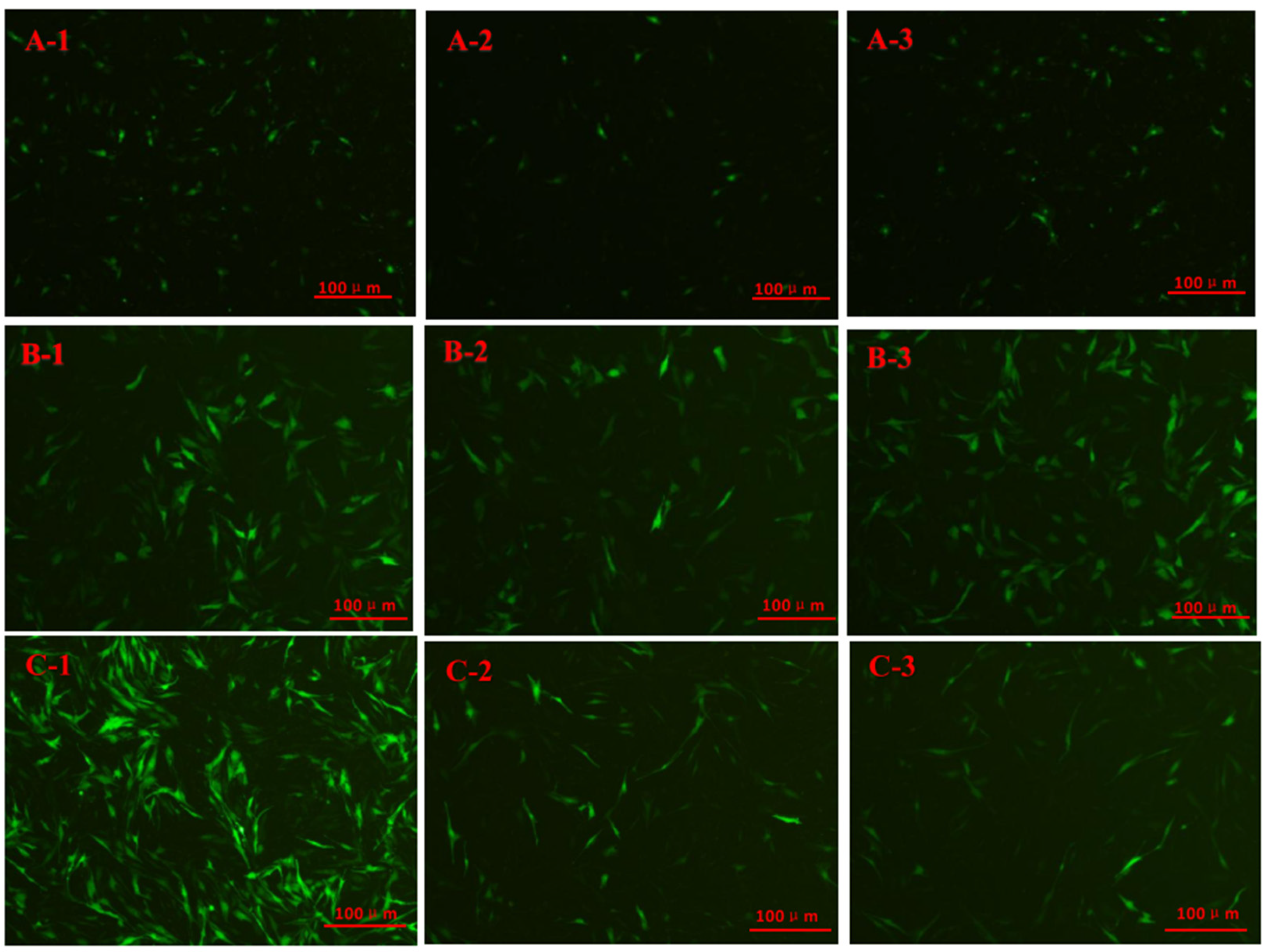
2.2. Immunofluorescence Staining
2.3. Cell Proliferation and Apoptosis Assay
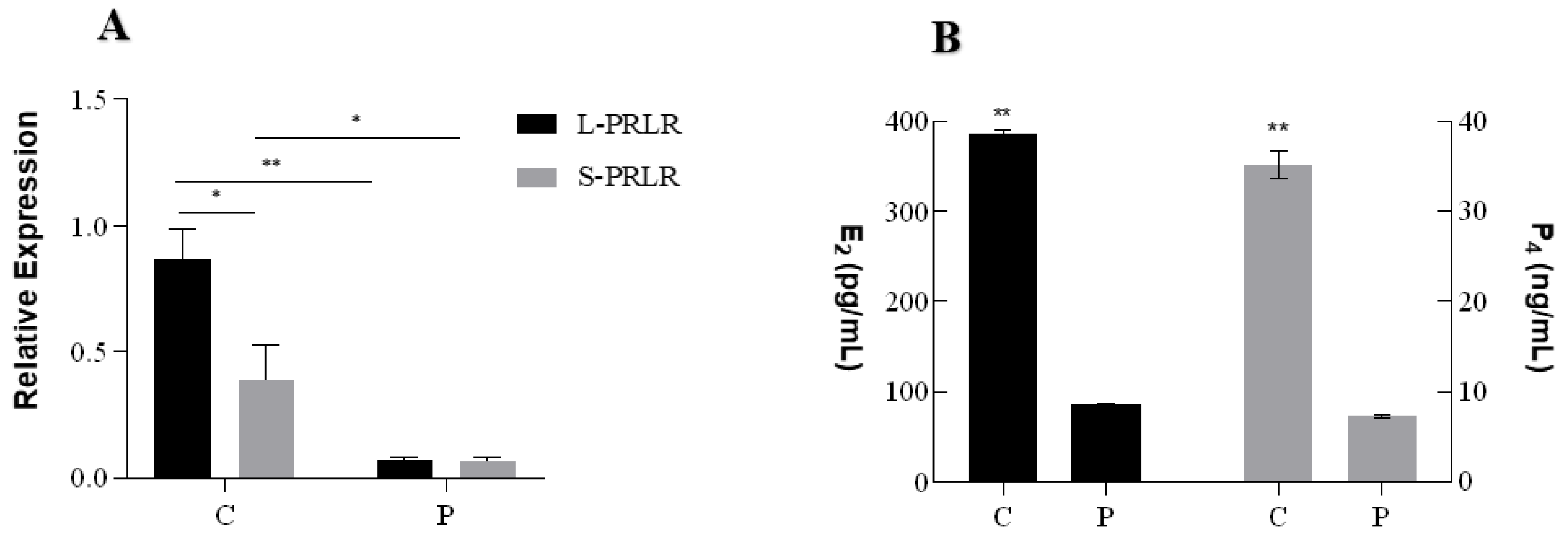
2.4. Effects of HPC on the Expression of L-PRLR and S-PRLR and the Secretion of Steroid Hormones (E2 and P4)
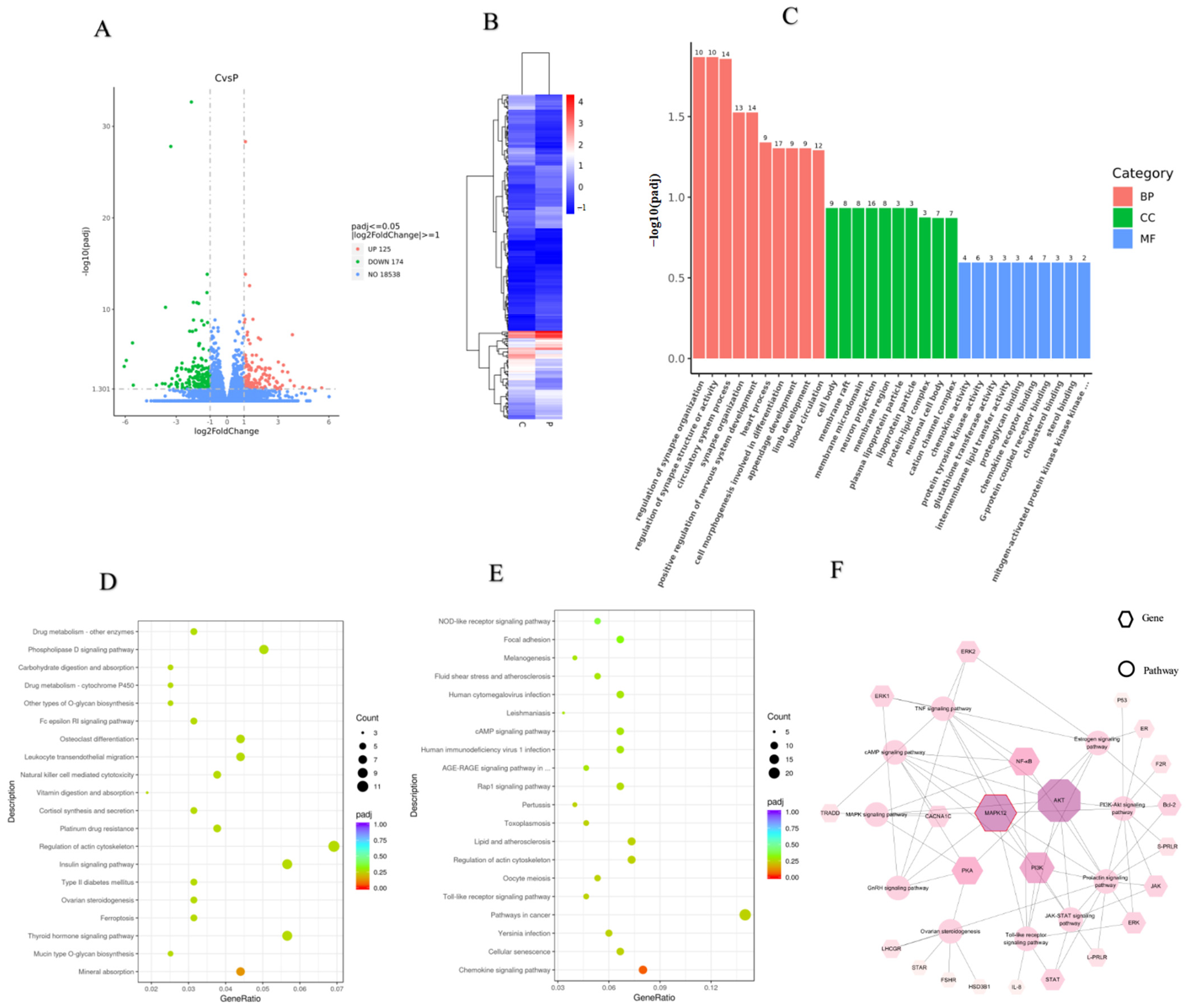
2.5. DEGs Analysis
2.6. GO and KEGG Pathway Enrichment Analysis
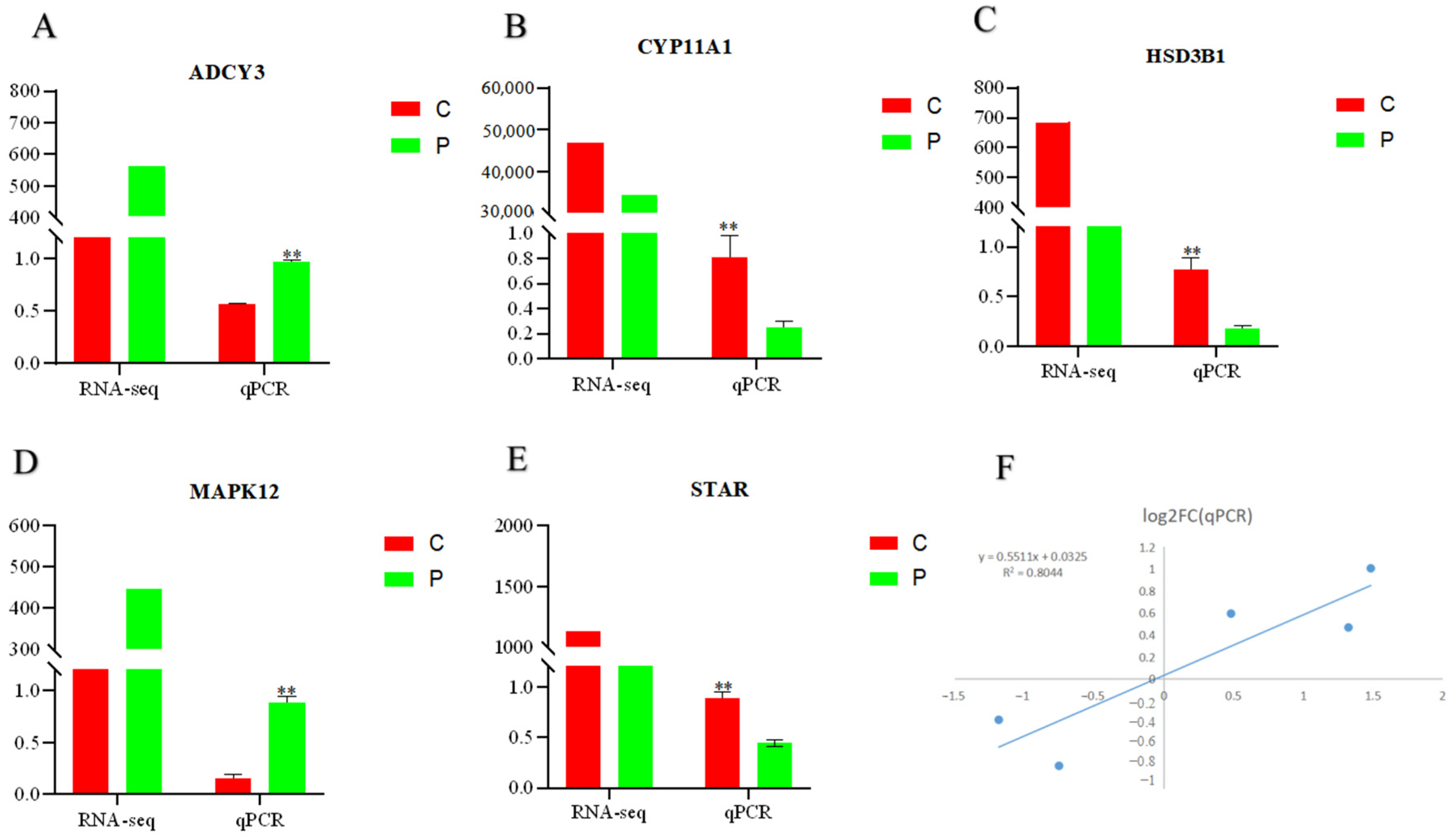
2.7. RT-qPCR Confirmation
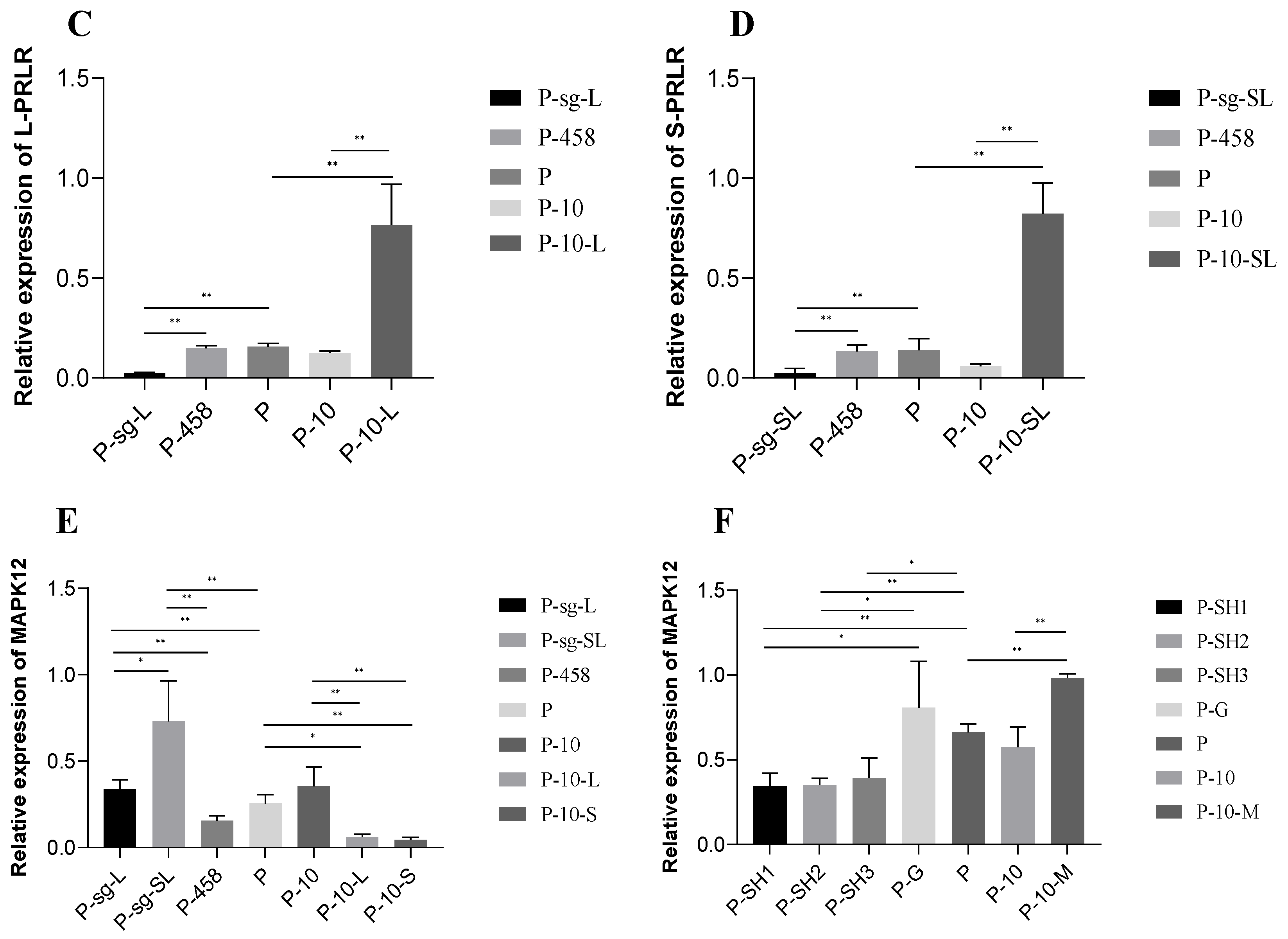
2.8. Expression of Related Genes after Knockdown and Overexpression of L-PRLR and S-PRLR
2.9. The Expression of Related Genes and the Secretion of Hormones in HPC GCs after Interference of MAPK12
2.9.1. The Expression of Apoptosis-Related Genes
2.9.2. The Secretion of Steroid Hormones (E2 and P4) and PRL
2.10. The Expression of Related Genes and the Secretion of Hormones in HPC GCs after Overexpression of MAPK12
2.10.1. The Expression of Apoptosis-Related Genes
2.10.2. The Secretion of Steroid Hormones (E2 and P4) and PRL
3. Discussion
3.1. The Effect of PRL on Proliferation and Apoptosis in GCs
3.2. The Effects of HPC on the Expression of L-PRLR and S-PRLR and the Secretion of Steroid Hormones (E2 and P4)
3.3. RNA-Seq Analysis
3.4. The Regulatory Mechanism of HPC on Apoptosis and Steroid Hormone (E2 and P4) Secretion in Sheep Ovarian GCs
4. Materials and Methods
4.1. Animals
4.2. Correlation between PRL and Follicle Counts
4.3. Experiment 1: PRL Model and Target Gene Identification
4.3.1. Ovaries GC Culture and Experimental Design
4.3.2. Cell Proliferation and Apoptosis Assay
4.3.3. P4 and E2 Analysis
4.3.4. The GC Model of PRL
4.3.5. RNA Extraction, Library Preparation, and Sequencing
4.3.6. Bioinformatics Analysis of RNA-Seq
4.3.7. Differentially Expressed Genes and Target Gene Identification
4.3.8. GCs of HPC Were Treated with CRISPR/Cas9 and Overexpression Techniques
4.4. Experiment 2: Functional Validation of MAPK12
4.4.1. Interference and Overexpression of MAPK12 in GCs with HPC
4.4.2. RNA Preparation and RT-qPCR
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kansaku, N.; Wakui, S.; Sasanami, T.; Ohkubo, T. Regulation of Prolactin Release at the End Stage of Chicken Embryogenesis. J. Poult. Sci. 2022, 59, 364–370. [Google Scholar] [CrossRef] [PubMed]
- Bole-Feysot, C.; Goffin, V.; Edery, M.; Binart, N.; Kelly, P.A. Prolactin (PRL) and its receptor: Actions, signal transduction pathways and phenotypes observed in PRL receptor knockout mice. Endocr. Rev. 1998, 19, 225–268. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.-C.; Hour, T.-C.; Liao, Y.-F.; Hung, Y.-C.; Liu, C.-C.; Chang, W.-H.; Kao, M.-C.; Tsay, G.J.; Hung, H.-C.; Liu, G.-Y. Increasing ornithine decarboxylase activity is another way of prolactin preventing methotrexate-induced apoptosis: Crosstalk between ODC and BCL-2. Apoptosis 2006, 11, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, E.; Otsuka, F.; Inagaki, K.; Miyoshi, T.; Yamanaka, R.; Tsukamoto, N.; Suzuki, J.; Ogura, T.; Makino, H. A Novel Antagonistic Effect of the Bone Morphogenetic Protein System on Prolactin Actions in Regulating Steroidogenesis by Granulosa Cells. Endocrinology 2010, 151, 5506–5518. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Zadworny, D. Effects of nonglycosylated and glycosylated prolactin on basal and gonadotropin-stimulated steroidogenesis in chicken ovarian follicles. Domest. Anim. Endocrinol. 2017, 61, 27–38. [Google Scholar] [CrossRef]
- Chen, Y.; Moutal, A.; Navratilova, E.; Kopruszinski, C.; Yue, X.; Ikegami, M.; Chow, M.; Kanazawa, I.; Bellampalli, S.S.; Xie, J.; et al. The prolactin receptor long isoform regulates nociceptor sensitization and opioid-induced hyperalgesia selectively in females. Sci. Transl. Med. 2020, 12, eaay7550. [Google Scholar] [CrossRef]
- Ramírez-De-Arellano, A.; Villegas-Pineda, J.C.; Hernández-Silva, C.D.; Pereira-Suárez, A.L. The Relevant Participation of Prolactin in the Genesis and Progression of Gynecological Cancers. Front. Endocrinol. 2021, 12, 747810. [Google Scholar] [CrossRef]
- Yang, R.; Duan, C.; Guo, Y.; Ma, Y.; Niu, N.; Zhang, Y.; Liu, Y. Sequence analysis and mRNA expression of prolactin receptor gene isoforms in different tissues of sheep during lactation and the post-weaning period. PeerJ 2021, 9, e11868. [Google Scholar] [CrossRef]
- Picazo, A.R.; Ruiz, J.P.G.; Moreno, J.S.; De Bulnes, A.G.; Muñoz, J.; Silván, G.; Lorenzo, P.L.; Illera, J.C. Cellular localization and changes in expression of prolactin receptor isoforms in sheep ovary throughout the estrous cycle. Reproduction 2004, 128, 545–553. [Google Scholar] [CrossRef]
- Bouilly, J.; Sonigo, C.; Auffret, J.; Gibori, G.; Binart, N. Prolactin signaling mechanisms in ovary. Mol. Cell. Endocrinol. 2012, 356, 80–87. [Google Scholar] [CrossRef]
- Grosdemouge, I.; Bachelot, A.; Lucas, A.; Baran, N.; Kelly, P.A.; Binart, N. Effects of deletion of the prolactin receptor on ovarian gene expression. Reprod. Biol. Endocrinol. 2003, 1, 12. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, M.; Ducheyne, K.D.; Deelen, C.; Beitsma, M.; Cristarella, S.; Quartuccio, M.; Stout, T.A.E.; De Ruijter-Villani, M. Advanced mare age impairs the ability of in vitro-matured oocytes to correctly align chromosomes on the metaphase plate. Equine Vet. J. 2019, 51, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Panzan, M.Q.; Soares Junior, J.M.; da Motta, E.L.A.; Haapalainen, E.F.; De Jesus Simoes, M.; Baptista, H.A.; Haidar, M.A.; Baracat, E.C. Metoclopramide-induced hyperprolactinaemia caused marked decline in pinopodes and pregnancy rates in mice. Hum. Reprod. 2006, 21, 2514–2520. [Google Scholar] [CrossRef]
- Adashi, E.Y.; Resnick, C.E. Prolactin as an inhibitor of granulosa cell luteinization: Implications for hyperprolactinemia-associated luteal phase dysfunction. Fertil. Steril. 1987, 48, 131–139. [Google Scholar] [CrossRef]
- Hoskova, K.; Bryant, N.K.; Chen, M.E.; Nachtigall, L.B.; Lippincott, M.F.; Balasubramanian, R.; Seminara, S.B. Kisspeptin Overcomes GnRH Neuronal Suppression Secondary to Hyperprolactinemia in Humans. J. Clin. Endocrinol. Metab. 2022, 107, e3515–e3525. [Google Scholar] [CrossRef] [PubMed]
- Leal Araujo, A.S.; Simoes, M.D.J.; Verna, C.; Simoes, R.S.; Soares Junior, J.M.; Baracat, E.C.; Teixeira Gomes, R.C. Influence of hyperprolactinemia on collagen fibers in the lacrimal gland of female mice. Clinics 2015, 70, 632–637. [Google Scholar] [CrossRef] [PubMed]
- Besnard, N.; Horne, E.A.L.; Whitehead, S.A. Prolactin and lipopolysaccharide treatment increased apoptosis and atresia in rat ovarian follicles. Acta Physiol. Scand. 2001, 172, 17–25. [Google Scholar] [CrossRef]
- Perks, C.M.; Newcomb, P.V.; Grohmann, M.; Wright, R.J.; Mason, H.D.; Holly, J.M.P. Prolactin acts as a potent survival factor against C2-ceramide-induced apoptosis in human granulosa cells. Hum. Reprod. 2003, 18, 2672–2677. [Google Scholar] [CrossRef]
- Zhang, Y. The effect of prolactin on cultured osteoblast of cow in vitro. Master’s Thesis, Huazhong Agricultural University, Wuhan, China, 2008. [Google Scholar]
- Zhang, H.; Wang, C.; Li, X.; Zhang, Y. Effects of pterostilbene on treating hyperprolactinemia and related mechanisms. Am. J. Transl. Res. 2016, 8, 3049–3055. [Google Scholar]
- Thompson, I.M.; Ozawa, M.; Bubolz, J.W.; Yang, Q.; Dahl, G.E. Bovine luteal prolactin receptor expression: Potential involvement in regulation of progesterone during the estrous cycle and pregnancy. J. Anim. Sci. 2011, 89, 1338–1346. [Google Scholar] [CrossRef]
- Clarke, D.L.; Linzer, D.I. Changes in prolactin receptor expression during pregnancy in the mouse ovary. Endocrinology 1993, 133, 224–232. [Google Scholar] [CrossRef]
- Palin, M.-F.; Caron, A.; Farmer, C. Effects of sustained hyperprolactinemia in late gestation on the mammary parenchymal tissue transcriptome of gilts. BMC Genom. 2023, 24, 1–16. [Google Scholar] [CrossRef]
- Hartwell, H.J.; Petrosky, K.Y.; Fox, J.G.; Horseman, N.D.; Rogers, A.B. Prolactin prevents hepatocellular carcinoma by restricting innate immune activation of c-Myc in mice. Proc. Natl. Acad. Sci. USA 2014, 111, 11455–11460. [Google Scholar] [CrossRef]
- Alkharusi, A.; AlMuslahi, A.; AlBalushi, N.; AlAjmi, R.; AlRawahi, S.; AlFarqani, A.; Norstedt, G.; Zadjali, F. Connections between prolactin and ovarian cancer. PLoS ONE 2021, 16, e0255701. [Google Scholar] [CrossRef]
- Goffin, V.; Bogorad, R.L.; Touraine, P. Identification of gain-of-function variants of the human prolactin receptor. In Methods in Enzymology 2010; Volume 484, Constitutive Activity in Receptors and Other Proteins, Part A; Conn, P.M., Ed.; Academic Press: Cambridge, MA, USA, 2010. [Google Scholar]
- Ma, K. Advantages of integrated Chinese and western medicine in diagnosis and treatment of anovulatory infertility due to kidney deficiency and blood stasis. China J. Chin. Mater. Med. 2021, 46, 2623–2628. [Google Scholar] [CrossRef]
- Prunet, P.; Sandra, O.; Le Rouzic, P.; Marchand, O.; Laudet, V. Molecular characterization of the prolactin receptor in two fish species, tilapia Oreochromis niloticus and rainbow trout, Oncorhynchus mykiss: A comparative approach. Can. J. Physiol. Pharm. 2000, 78, 1086–1096. [Google Scholar] [CrossRef]
- Cass, L.A.; Meinkoth, J.L. Differential effects of cyclic adenosine 3′,5′-monophosphate on p70 ribosomal S6 kinase. Endocrinology 1998, 139, 1991–1998. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Shimada, M.; Richards, J.S. The involvement of the Toll-like receptor family in ovulation. J. Assist. Reprod. Genet. 2008, 25, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Janelle, V.; Neault, M.; Lebel, M.; De Sousa, D.M.; Boulet, S.; Durrieu, L.; Carli, C.; Muzac, C.; Lemieux, S.; Labrecque, N.; et al. p16INK4a Regulates Cellular Senescence in PD-1-Expressing Human T Cells. Front. Immunol. 2021, 12, 698565. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Li, S.; Liu, J.; Wu, H.; Yao, G.; Sun, Y.; Chen, Z.J.; Li, W.; Du, Y. Up-regulated FHL2 inhibits ovulation through interacting with androgen receptor and ERK1/2 in polycystic ovary syndrome. Ebiomedicine 2020, 52, 102635. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Tong, C.; Chen, D.; Sun, Q. Roles of MAP kinase signaling pathway in oocyte meiosis. Chin. Sci. Bull. 2002, 47, 1157–1162. [Google Scholar] [CrossRef]
- He, Y.; Yang, Z.; Pi, J.; Cai, T.; Xia, Y.; Cao, X.; Liu, J. EGCG attenuates the neurotoxicity of methylglyoxal via regulating MAPK and the downstream signaling pathways and inhibiting advanced glycation end products formation. Food Chem. 2022, 384, 132358. [Google Scholar] [CrossRef] [PubMed]
- Hua, G.; George, J.W.; Clark, K.L.; Jonas, K.C.; Johnson, G.P.; Southekal, S.; Guda, C.; Hou, X.; Blum, H.R.; Eudy, J.; et al. Hypo-glycosylated hFSH drives ovarian follicular development more efficiently than fully-glycosylated hFSH: Enhanced transcription and PI3K and MAPK signaling. Hum. Reprod. 2021, 36, 1891–1906. [Google Scholar] [CrossRef] [PubMed]
- Simmons, A.J.; Banerjee, A.; Mckinley, E.T.; Scurrah, C.R.; Herring, C.A.; Gewin, L.S.; Masuzaki, R.; Karp, S.J.; Franklin, J.L.; Gerdes, M.J. Cytometry-based single-cell analysis of intact epithelial signaling reveals MAPK activation divergent from TNF-alpha-induced apoptosis in vivo. Mol. Syst. Biol. 2015, 11, 835, Erratum in Mol. Syst. Biol. 2016, 12, 881. [Google Scholar] [CrossRef]
- Yu, C.-P.; Pan, Y.-L.; Wang, X.-L.; Xin, R.; Li, H.-Q.; Lei, Y.-T.; Zhao, F.-F.; Zhang, D.; Zhou, X.-R.; Ma, W.-W.; et al. Stimulating the expression of sphingosine kinase 1 (SphK1) is beneficial to reduce acrylamide-induced nerve cell damage. Ecotoxicol. Environ. Saf. 2022, 237, 113511. [Google Scholar] [CrossRef]
- Seval, Y.; Cakmak, H.; Kayisli, U.A.; Arici, A. Estrogen-Mediated Regulation of p38 Mitogen-Activated Protein Kinase in Human Endometrium. J. Clin. Endocrinol. Metab. 2006, 91, 2349–2357. [Google Scholar] [CrossRef] [PubMed]
- Anerillas, C.; Abdelmohsen, K.; Gorospe, M. Regulation of senescence traits by MAPKs. Geroscience 2020, 42, 397–408. [Google Scholar] [CrossRef]
- Wang, Y.H.; Maurer, R.A. A role for the mitogen-activated protein kinase in mediating the ability of thyrotropin-releasing hormone to stimulate the prolactin promoter. Mol. Endocrinol. 1999, 13, 1094–1104. [Google Scholar] [CrossRef]
- Piccoletti, R.; Bendinelli, P.; Maroni, P. Signal transduction pathway of prolactin in rat liver. Mol. Cell. Endocrinol. 1997, 135, 169–177. [Google Scholar] [CrossRef]
- Cuadrado, A.; Nebreda, A.R. Mechanisms and functions of p38 MAPK signalling. Biochem. J. 2010, 429, 403–417. [Google Scholar] [CrossRef]
- Risco, A.; Cuenda, A. New Insights into the p38gamma and p38delta MAPK Pathways. J. Recept. Sig. Transd. 2012, 2012, 520289. [Google Scholar]
- Chen, H.; Wang, X.; Guo, F.; Li, P.; Peng, D.; He, J. Impact of p38 gamma mitogen-activated protein kinase (MAPK) on MDA-MB-231 breast cancer cells using metabolomic approach. Int. J. Biochem. Cell B 2019, 107, 6–13. [Google Scholar] [CrossRef]
- Deng, Z.; Fu, H.; Xiao, Y.; Zhang, B.; Sun, G.; Wei, Q.; Ai, B.; Hu, Q. Effects of selenium on lead-induced alterations in A beta production and Bcl-2 family proteins. Environ. Toxicol. Pharmacol. 2015, 39, 221–228. [Google Scholar] [CrossRef]
- Del Principe, M.I.; Dal Bo, M.; Bittolo, T.; Buccisano, F.; Rossi, F.M.; Zucchetto, A.; Rossi, D.; Bomben, R.; Maurillo, L.; Cefalo, M.; et al. Clinical significance of bax/bcl-2 ratio in chronic lymphocytic leukemia. Haematologica 2016, 101, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.F.; Jiang, C.C.; Kiejda, K.A.; Gillespie, S.; Zhang, X.D.; Hersey, P. Apoptosis Induction in Human Melanoma Cells by Inhibition of MEK Is Caspase-Independent and Mediated by the Bcl-2 Family Members PUMA, Bim, and Mcl-1. Clin. Cancer Res. 2007, 13, 4934–4942. [Google Scholar] [CrossRef]
- Chimento, A.; De Luca, A.; Avena, P.; De Amicis, F.; Casaburi, I.; Sirianni, R.; Pezzi, V. Estrogen Receptors-Mediated Apoptosis in Hormone-Dependent Cancers. Int. J. Mol. Sci. 2022, 23, 1242. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.-M.; Wang, J.Q. Synaptically Localized Mitogen-Activated Protein Kinases: Local Substrates and Regulation. Mol. Neurobiol. 2016, 53, 6309–6315. [Google Scholar] [CrossRef] [PubMed]
- Baumgarten, S.C.; Armouti, M.; Ko, C.; Stocco, C. IGF1R Expression in Ovarian Granulosa Cells Is Essential for Steroidogenesis, Follicle Survival, and Fertility in Female Mice. Endocrinology 2017, 158, 2309–2318. [Google Scholar] [CrossRef]
- Wells, J.A. Binding in the growth hormone receptor complex. Proc. Natl. Acad. Sci. USA 1996, 93, 1–6. [Google Scholar] [CrossRef]
- Begon, E.; Bernard, V. Prolactin and its receptor: From animal models to pituitary pathophysiology. Biol. Aujourdhui 2022, 216, 105–110. [Google Scholar] [CrossRef]
- Xia, Q.; Liu, Q.Y.; Wang, X.Y.; Hu, W.P.; Li, X.Y.; Pan, Z.Y.; Chu, M.X.; Di, R. Regulation of prolactin in sheep seasonal estrus. J. Anim. Ecol. 2017, 38, 1–4. (In Chinese) [Google Scholar]
- Ding, Q.-Y.; Zhang, Y.; Ma, L.; Chen, Y.-G.; Wu, J.-H.; Zhang, H.-F.; Wang, X. Inhibiting MAPK14 showed anti-prolactinoma effect. BMC Endocr. Disord. 2022, 20, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Milenkovic, L.; D’Angelo, G.; Kelly, P.A.; Weiner, R.I. Inhibition of gonadotropin hormone-releasing hormone release by prolactin from GT1 neuronal cell lines through prolactin receptors. Proc. Natl. Acad. Sci. USA 1994, 91, 1244–1247. [Google Scholar] [CrossRef]
- Hodson, D.J.; Henderson, H.L.; Townsend, J.; Tortonese, D.J. Photoperiodic Modulation of the Suppressive Actions of Prolactin and Dopamine on the Pituitary Gonadotropin Responses to Gonadotropin-Releasing Hormone in Sheep1. Biol. Reprod. 2012, 86, 122. [Google Scholar] [CrossRef]
- Song, Z.P.; Duan, C.H.; Li, Y.; Yue, S.C.; Guo, Y.X.; Liu, Y.Q.; Zhang, Y.J. Effects of P4 and FSH on follicular development and reproductive hormone secretion in growing ewes. Chin. J. Vet. Sci. 2022, 42, 1481–1488. (In Chinese) [Google Scholar]
- Wang, Z.; Shen, M.; Xue, P.; Divall, S.A.; Segars, J.; Wu, S. Female Offspring from Chronic Hyperandrogenemic Dams Exhibit Delayed Puberty and Impaired Ovarian Reserve. Endocrinology 2018, 159, 1242–1252. [Google Scholar] [CrossRef]
- Cao, X.; Wang, X.; Lu, L.; Li, X.; Di, R.; He, X.; Hu, W.; Zeng, X.; Liu, Q.; Chu, M. Expression and Functional Analysis of the BCL2-Associated Agonist of Cell Death (BAD) Gene in the Sheep Ovary During the Reproductive Cycle. Front. Endocrinol. 2018, 9, 512. [Google Scholar] [CrossRef]
- Osamura, R.Y.; Watanabe, K. Ultrastructural localization of prolactin in estrogen-induced prolactinoma of the rat pituitary. Experimental models for the human prolactinomas and the effects of bromocriptine. Acta Pathol. Jpn. 1986, 36, 1131–1137. [Google Scholar]
- Neradugomma, N.K.; Subramaniam, D.; Tawfik, O.W.; Goffin, V.; Kumar, T.R.; Jensen, R.A.; Anant, S. Prolactin signaling enhances colon cancer stemness by modulating Notch signaling in a Jak2-STAT3/ERK manner. Carcinogenesis 2014, 35, 795–806. [Google Scholar] [CrossRef] [PubMed]
- Lachmann, A.; Clarke, D.J.; Torre, D.; Xie, Z.; Ma’Ayan, A. Interoperable RNA-Seq analysis in the cloud. Biochim. Biophys. Acta (BBA)—Gene Regul. Mech. 2020, 1863, 194521. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Ren, E.; Chen, X.; Yu, S.; Xu, J.; Su, Y.; Zhu, W. Transcriptomic and metabolomic responses induced in the livers of growing pigs by a short-term intravenous infusion of sodium butyrate. Animal 2018, 12, 2318–2326. [Google Scholar] [CrossRef]

| Item | PRL Supplemented, ng/mL | SEM | p-Value | ||||
|---|---|---|---|---|---|---|---|
| 0 | 4 | 20 | 100 | 500 | |||
| Apoptotic rate, % | 1.60 ± 0.055 c | 1.93 ± 0.076 bc | 1.56 ± 0.399 c | 2.07 ± 0.055 b | 4.01 ± 0.220 a | 0.009 | 0.023 |
| Sample | Raw Reads | Clean Reads | Error Rate | Q20 | Q30 | GC pct |
|---|---|---|---|---|---|---|
| C1 | 47,452,474 | 46,017,302 | 0.03 | 97.28 | 92.75 | 51.02 |
| C2 | 50,424,042 | 49,060,726 | 0.03 | 97.76 | 93.72 | 52.09 |
| C3 | 45,485,418 | 44,286,592 | 0.03 | 97.58 | 93.3 | 51.09 |
| C4 | 46,667,212 | 45,237,462 | 0.03 | 97.83 | 93.94 | 52.97 |
| P1 | 43,582,446 | 42,289,948 | 0.03 | 97.69 | 93.62 | 52.82 |
| P2 | 46,354,432 | 45,081,360 | 0.03 | 97.8 | 93.86 | 52.74 |
| P3 | 48,253,124 | 46,923,820 | 0.03 | 97.45 | 93.04 | 52.50 |
| P4 | 45,705,810 | 44,655,822 | 0.03 | 97.64 | 93.4 | 52.45 |
| Genes | Gene ID | log2 Fold Change | p-Value |
|---|---|---|---|
| HSD3B1 | ENSOARG00000020402 | 1.32452 | 1.01 × 10−16 |
| STAR | ENSOARG00000001269 | 1.48462 | 4.89 × 10−8 |
| CDC20 | ENSOARG00000020542 | −1.51346 | 0.00037 |
| PLCE1 | ENSOARG00000003798 | 1.10614 | 1.35 × 10−7 |
| GRIA3 | ENSOARG00000013491 | 1.85560 | 0.00034 |
| IGSF10 | ENSOARG00000003977 | −3.29102 | 0.00053 |
| CACNAIC | ENSOARG00000013089 | 1.29897 | 0.00048 |
| RGS2 | ENSOARG00000009653 | 1.13720 | 3.20 × 10−7 |
| MAPK12 | ENSOARG00000019799 | −1.18458 | 6.80 × 10−16 |
| CTSK | ENSOARG00000020869 | −1.39051 | 0.00014 |
| NFATC4 | ENSOARG00000019084 | −2.02030 | 3.67 × 10−6 |
| Gene | Sequence 5′-3′ | Accession Number |
|---|---|---|
| L-PRLR-sgRNA1 | F:Caccg CAAATCCTCGCAGTCAGAAG R:Aaac CTTCTGACTGCGAGGATTTG c | O46561-1 |
| L-PRLR-sgRNA2 | F:Caccg CTTTGGAGGGGTGTGGCATC R:Aaac GATGCCACACCCCTCCAAAG c | O46561-1 |
| L-PRLR-sgRNA3 | F:Caccg TTTGCTGATGGAATTCATAG R:Aaac CTATGAATTCCATCAGCAAA c | O46561-1 |
| S-PRLR-sgRNA1 | F:Caccg CTTATTAAATGTCGGTCTCC R:Aaac GGAGACCGACATTTAATAAG c | O46561-2 |
| S-PRLR-sgRNA2 | F:Cacc GCGGTAAGTCAGTGTGTAAT R:Aaac ATTACACACTGACTTACCGC | O46561-2 |
| S-PRLR-sgRNA3 | F:Cacc GGAAACGTTCACCTGCTGGT R:Aaac ACCAGCAGGTGAACGTTTCC | O46561-2 |
| MAPK12-ShRNA1 | ACCGGCGTCATCCATAGGGACTTGACTCGAGTCAAGTCCCTATGGATGACGCTTTT | XM027968254.2 |
| MAPK12-ShRNA2 | ACCGGGACTGTGAGCTGAAGATTCTCTCGAGAGAATCTTCAGCTCACAGTCCTTTT | XM027968254.2 |
| MAPK12-ShRNA3 | ACCGGGAAGCGTGTCACATATAAAGCTCGAGCTTTATATGTGACACGCTTCCTTTT | XM027968254.2 |
| Gene | Sequence 5′-3′ | Size (bp) | Tm (°C) | Accession Number |
|---|---|---|---|---|
| StAR | F:ATTCAGGAGGCAAAGAGCAGC R:TCGGGTAAGGAAAATGGGTCA | 270 | 60 | XM015094520.2 |
| HSD13B1 | F:CAGTCTATGTTGGCAATGTGGC R:CGGTTGAAGCAGGGGTGGTAT | 283 | 60 | NM001135932.1 |
| CYP11A1 | F:GTTTCGCTTTGCCTTTGAGTC R:ACAGTTCTGGAGGGAGGTTGA | 120 | 60 | NM001093789.1 |
| ADCY3 | F:GCAACATCCAGGTGGTGGA R:TGGTCACATGTCCTTTCCCG | 282 | 60 | XM_027966562.2 |
| MAPK12 | F:GCAGGCAGACAGCGAGAT R:GGTCAGGACGGAGGCAAA | 307 | 62 | XM027968254.2 |
| Bax | F:TGCTCACTGCCTCACTCACC R:CCCAAGACCACTCCTCCCTA | 179 | 60 | XM027978592.1 |
| Bcl-2 | F:GATGACCGAGTACCTGAACCG R:GACAGCCAGGAGAAATCAAACA | 120 | 60 | XM012103831.3 |
| Caspase3 | F:GCTACAAGGTCCGTTATGCC R:GATGCTGCCGTATTCGTTCTC | 128 | 60 | XM015104559.2 |
| L-PRLR | F:CCCCTTGTTCTCTGCTAAACCC R: CTATCCGTCACCCGAGACACC | 129 | 60 | O46561-1 |
| S-PRLR | F:ACAGTAAGCGCCATCAACCA R:CTGGCTTGCATCGAATCTGC | 328 | 60 | O46561-2 |
| GAPDH | F:GGTCGGAGTGAACGGATTTG R:CTTGACTGTGCCGTGGAACTT | 222 | 60 | NM001190390.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, R.; Duan, C.; Zhang, S.; Liu, Y.; Zhang, Y. Prolactin Regulates Ovine Ovarian Granulosa Cell Apoptosis by Affecting the Expression of MAPK12 Gene. Int. J. Mol. Sci. 2023, 24, 10269. https://doi.org/10.3390/ijms241210269
Yang R, Duan C, Zhang S, Liu Y, Zhang Y. Prolactin Regulates Ovine Ovarian Granulosa Cell Apoptosis by Affecting the Expression of MAPK12 Gene. International Journal of Molecular Sciences. 2023; 24(12):10269. https://doi.org/10.3390/ijms241210269
Chicago/Turabian StyleYang, Ruochen, Chunhui Duan, Shuo Zhang, Yueqin Liu, and Yingjie Zhang. 2023. "Prolactin Regulates Ovine Ovarian Granulosa Cell Apoptosis by Affecting the Expression of MAPK12 Gene" International Journal of Molecular Sciences 24, no. 12: 10269. https://doi.org/10.3390/ijms241210269
APA StyleYang, R., Duan, C., Zhang, S., Liu, Y., & Zhang, Y. (2023). Prolactin Regulates Ovine Ovarian Granulosa Cell Apoptosis by Affecting the Expression of MAPK12 Gene. International Journal of Molecular Sciences, 24(12), 10269. https://doi.org/10.3390/ijms241210269





