Proteolytic Resistance Determines Albumin Nanoparticle Drug Delivery Properties and Increases Cathepsin B, D, and G Expression
Abstract
1. Introduction
2. Results
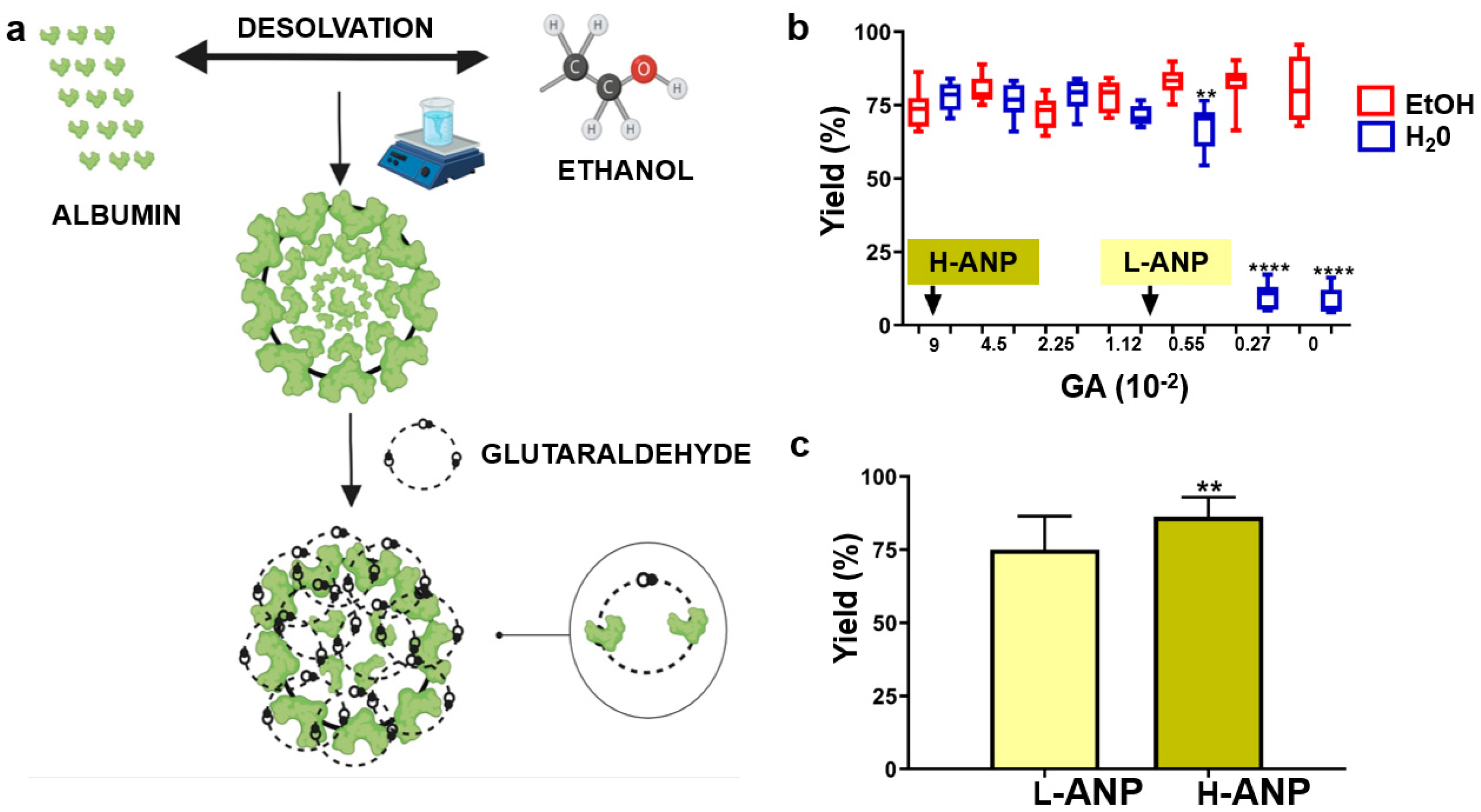
2.1. Determination of Cross-Linker Concentrations to Fabricate Lightly (L−ANP) and Heavily (H−ANP) Cross-Linked ANPs
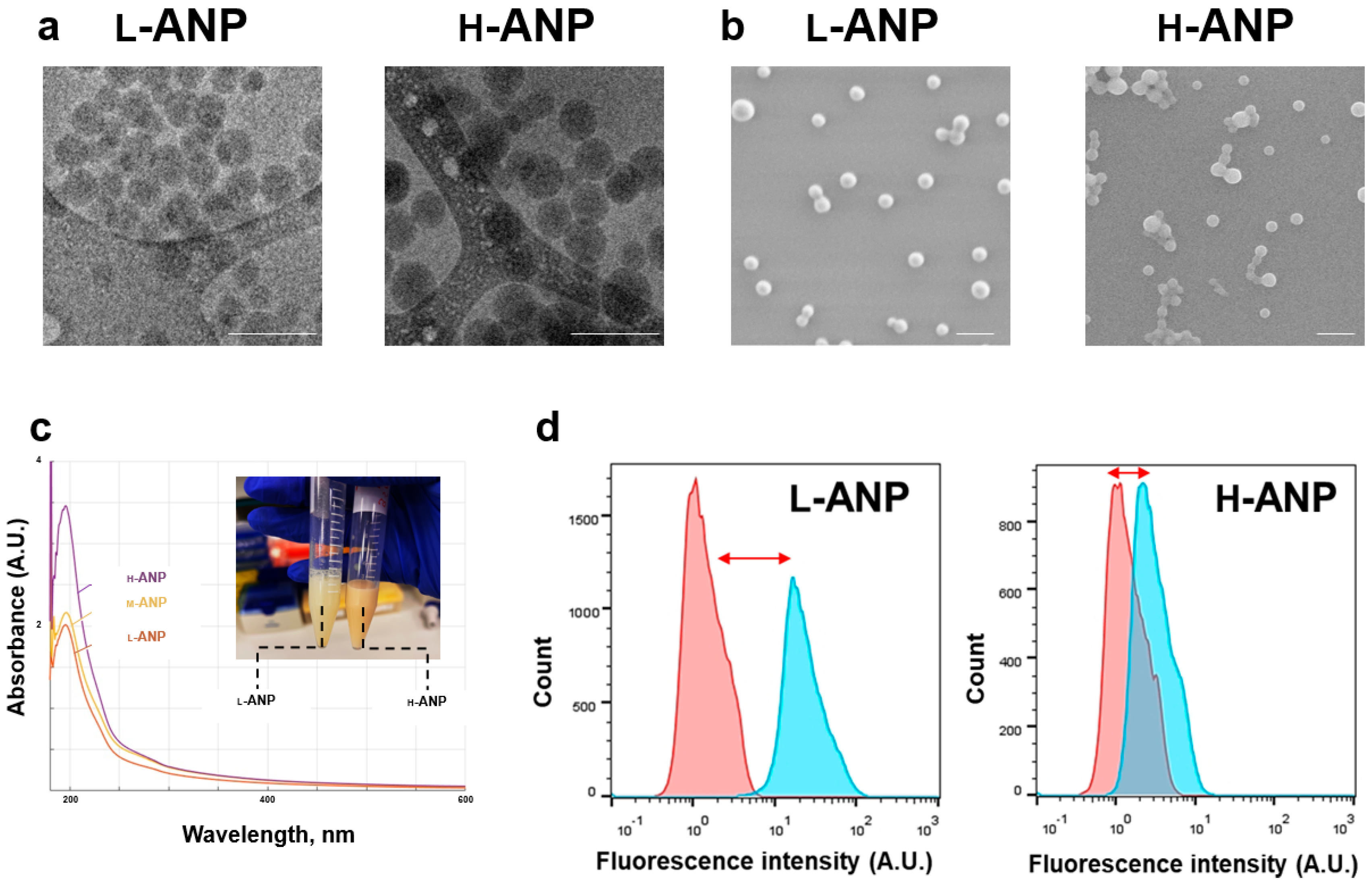
2.2. Analysis of Particle Physical and Surface Properties
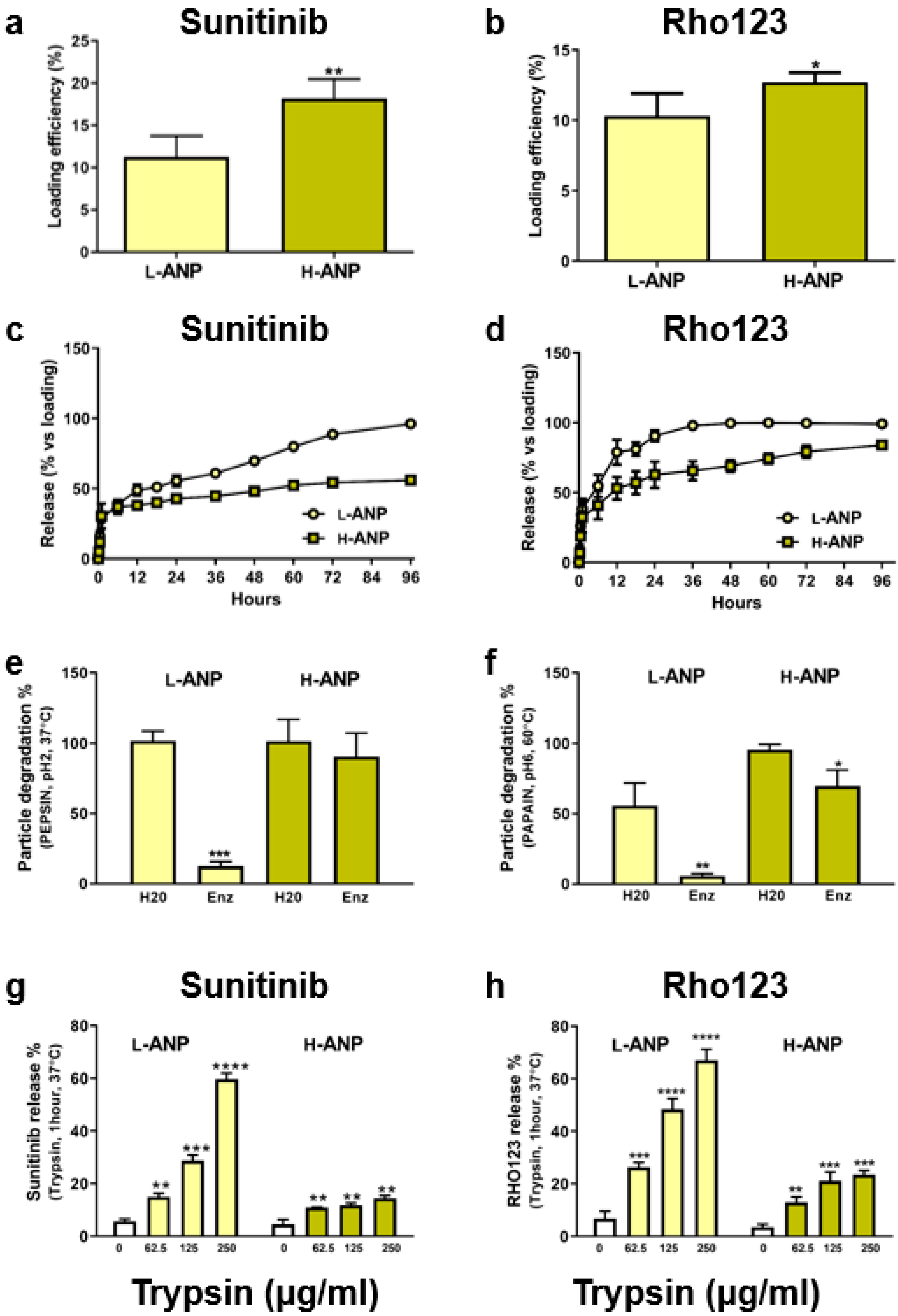
2.3. Particle Loading and Release in Normal and Proteolytic Conditions
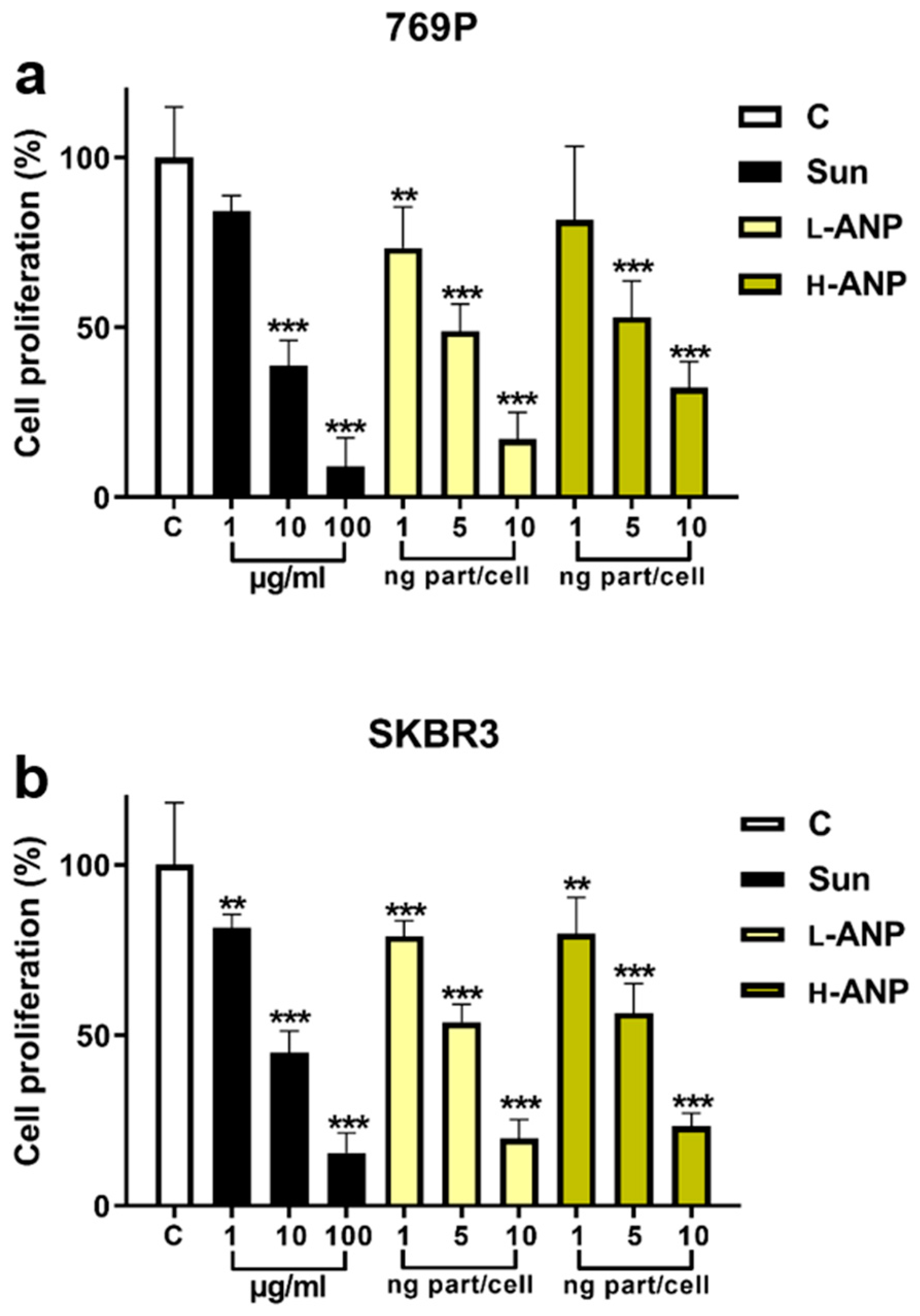
2.4. Determination of Intracellular Drug Release
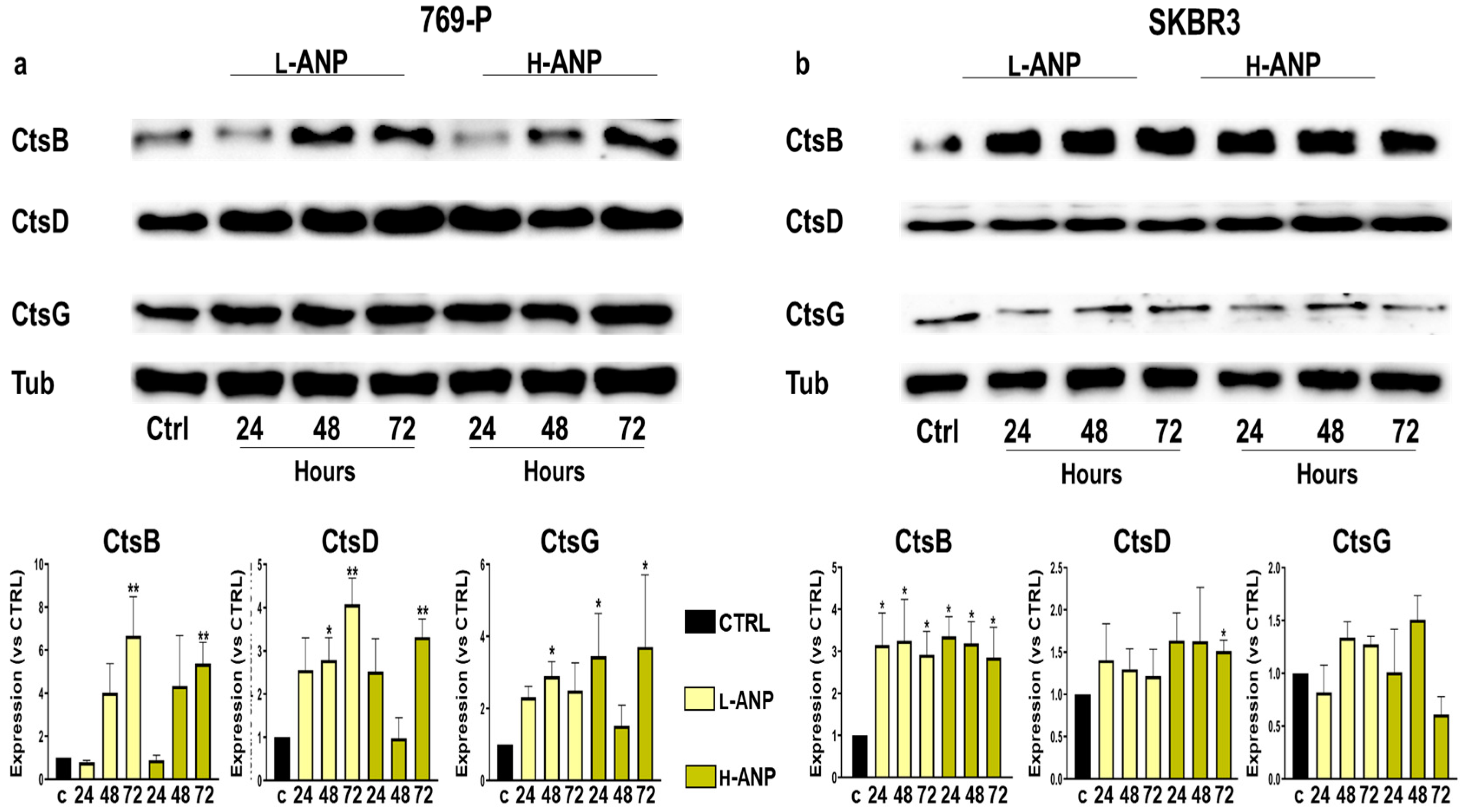
2.5. Determination of Lysosomal Cathepsin B, D, and G Expression
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Nanoparticle Synthesis and Determination of Glutaraldehyde Concentrations to Generate Lightly Cross-Linked ANPs (L-ANPs) and Heavily Cross-Linked ANPs (H-ANPs)
4.3. Nanoparticle Size and Surface Charge Characterization
4.4. Electron Microscopy and UV-Vis Spectral Analysis
4.5. Particle Loading and Release
4.6. Determination of Particle Degradation and Release in Proteolytic Conditions
4.7. MTT Assay
4.8. Flow Cytometry Analysis
4.9. Western Blot Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bond, J.S. Proteases: History, discovery, and roles in health and disease. J. Biol. Chem. 2019, 294, 1643–1651. [Google Scholar] [CrossRef] [PubMed]
- Rudzińska, M.; Parodi, A.; Soond, S.M.; Vinarov, A.Z.; Korolev, D.O.; Morozov, A.O.; Daglioglu, C.; Tutar, Y.; Zamyatnin, A.A., Jr. The role of cysteine cathepsins in cancer progression and drug resistance. Int. J. Mol. Sci. 2019, 20, 3602. [Google Scholar] [CrossRef] [PubMed]
- Ballabio, A.; Bonifacino, J.S. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Biol. 2020, 21, 101–118. [Google Scholar] [CrossRef] [PubMed]
- Yadati, T.; Houben, T.; Bitorina, A.; Shiri-Sverdlov, R. The ins and outs of cathepsins: Physiological function and role in disease management. Cells 2020, 9, 1679. [Google Scholar] [CrossRef] [PubMed]
- Rudzińska, M.; Parodi, A.; Maslova, V.D.; Efremov, Y.M.; Gorokhovets, N.V.; Makarov, V.A.; Popkov, V.A.; Golovin, A.V.; Zernii, E.Y.; Zamyatnin, A.A., Jr. Cysteine cathepsins inhibition affects their expression and human renal cancer cell phenotype. Cancers 2020, 12, 1310. [Google Scholar] [CrossRef]
- Breznik, B.; Mitrović, A.; Lah, T.T.; Kos, J. Cystatins in cancer progression: More than just cathepsin inhibitors. Biochimie 2019, 166, 233–250. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, M.; Song, S. Cathepsin D enhances breast cancer invasion and metastasis through promoting hepsin ubiquitin-proteasome degradation. Cancer Lett. 2018, 438, 105–115. [Google Scholar] [CrossRef]
- Lawrence, R.E.; Zoncu, R. The lysosome as a cellular centre for signalling, metabolism and quality control. Nat. Cell Biol. 2019, 21, 133–142. [Google Scholar] [CrossRef]
- Soond, S.M.; Savvateeva, L.V.; Makarov, V.A.; Gorokhovets, N.V.; Townsend, P.A.; Zamyatnin, A.A., Jr. Cathepsin S cleaves BAX as a novel and therapeutically important regulatory mechanism for apoptosis. Pharmaceutics 2021, 13, 339. [Google Scholar] [CrossRef]
- Seo, S.U.; Woo, S.M.; Im, S.-S.; Jang, Y.; Han, E.; Kim, S.H.; Lee, H.; Lee, H.-S.; Nam, J.-O.; Gabrielson, E. Cathepsin D as a potential therapeutic target to enhance anticancer drug-induced apoptosis via RNF183-mediated destabilization of Bcl-xL in cancer cells. Cell Death Dis. 2022, 13, 1–13. [Google Scholar] [CrossRef]
- Parodi, A.; Voronina, M.V.; Zamyatnin, A.A. The importance of nanocarriers' intra- and extracellular degradation: What we know and should know about it? Curr. Med. Chem. 2023. [Google Scholar] [CrossRef]
- Manzanares, D.; Ceña, V. Endocytosis: The nanoparticle and submicron nanocompounds gateway into the cell. Pharmaceutics 2020, 12, 371. [Google Scholar] [CrossRef]
- Parodi, A.; Buzaeva, P.; Nigovora, D.; Baldin, A.; Kostyushev, D.; Chulanov, V.; Savvateeva, L.V.; Zamyatnin, A.A. Nanomedicine for increasing the oral bioavailability of cancer treatments. J. Nanobiotechnol. 2021, 19, 354. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.L.; Ho, H.K. Navigating albumin-based nanoparticles through various drug delivery routes. Drug Discov. Today 2018, 23, 1108–1114. [Google Scholar] [CrossRef] [PubMed]
- Parodi, A.; Miao, J.; Soond, S.M.; Rudzińska, M.; Zamyatnin, A.A., Jr. Albumin nanovectors in cancer therapy and imaging. Biomolecules 2019, 9, 218. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, J.; Cho, J.; Malik, A.B. Prevention of vascular inflammation by nanoparticle targeting of adherent neutrophils. Nat. Nanotechnol. 2014, 9, 204–210. [Google Scholar] [CrossRef]
- Wang, T.; Xue, J.; Hu, Q.; Zhou, M.; Chang, C.; Luo, Y. Synthetic surfactant-and cross-linker-free preparation of highly stable lipid-polymer hybrid nanoparticles as potential oral delivery vehicles. Sci. Rep. 2017, 7, 2750. [Google Scholar] [CrossRef]
- Merodio, M.; Arnedo, A.; Renedo, M.J.; Irache, J.M. Ganciclovir-loaded albumin nanoparticles: Characterization and in vitro release properties. Eur. J. Pharm. Sci. 2001, 12, 251–259. [Google Scholar] [CrossRef]
- Amighi, F.; Emam-Djomeh, Z.; Labbafi-Mazraeh-Shahi, M. Effect of different cross-linking agents on the preparation of bovine serum albumin nanoparticles. J. Iran. Chem. Soc. 2020, 17, 1223–1235. [Google Scholar] [CrossRef]
- Jahanban-Esfahlan, A.; Dastmalchi, S.; Davaran, S. A simple improved desolvation method for the rapid preparation of albumin nanoparticles. Int. J. Biol. Macromol. 2016, 91, 703–709. [Google Scholar] [CrossRef]
- Fei, J.; Zhang, H.; Wang, A.; Qin, C.; Xue, H.; Li, J. Biofluid-Triggered Burst Release from an Adaptive Covalently Assembled Dipeptide Nanocontainer for Emergency Treatment. Adv. Healthc. Mater. 2017, 6, 1601198. [Google Scholar] [CrossRef] [PubMed]
- Islam, N.; Dmour, I.; Taha, M.O. Degradability of chitosan micro/nanoparticles for pulmonary drug delivery. Heliyon 2019, 5, e01684. [Google Scholar] [CrossRef]
- Raja, M.A.G.; Amjad, M.W. Development and Characterization of Mitoxantrone-Loaded Glutaraldehyde Crosslinked Sodium Alginate Nanoparticles for the Delivery of Anticancer Drugs. J. Pharm. Res. Int. 2021, 33, 18–24. [Google Scholar]
- Langer, K.; Anhorn, M.; Steinhauser, I.; Dreis, S.; Celebi, D.; Schrickel, N.; Faust, S.; Vogel, V. Human serum albumin (HSA) nanoparticles: Reproducibility of preparation process and kinetics of enzymatic degradation. Int. J. Pharm. 2008, 347, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Fratto, M.; Imperatori, M.; Vincenzi, B.; Tomao, F.; Santini, D.; Tonini, G. New perspectives: Role of Sunitinib in breast cancer. La Clin. Ter. 2011, 162, 251–257. [Google Scholar]
- Golovastova, M.O.; Korolev, D.O.; Tsoy, L.V.; Varshavsky, V.A.; Xu, W.-H.; Vinarov, A.Z.; Zernii, E.Y.; Philippov, P.P.; Zamyatnin, A.A. Biomarkers of renal tumors: The current state and clinical perspectives. Curr. Urol. Rep. 2017, 18, 3. [Google Scholar] [CrossRef]
- Johnson, L.V.; Walsh, M.L.; Chen, L.B. Localization of mitochondria in living cells with rhodamine 123. Proc. Natl. Acad. Sci. USA 1980, 77, 990–994. [Google Scholar] [CrossRef]
- Parodi, A.; Kostyushev, D.; Brezgin, S.; Kostyusheva, A.; Borodina, T.; Akasov, R.; Frolova, A.; Chulanov, V.; Zamyatnin, A.A., Jr. Biomimetic approaches for targeting tumor inflammation. Semin. Cancer Biol. 2022, 86, 555–567. [Google Scholar] [CrossRef]
- Polo, E.; Collado, M.; Pelaz, B.; Del Pino, P. Advances toward more efficient targeted delivery of nanoparticles in vivo: Understanding interactions between nanoparticles and cells. ACS Nano 2017, 11, 2397–2402. [Google Scholar] [CrossRef]
- Busatto, C.; Pesoa, J.; Helbling, I.; Luna, J.; Estenoz, D. Effect of particle size, polydispersity and polymer degradation on progesterone release from PLGA microparticles: Experimental and mathematical modeling. Int. J. Pharm. 2018, 536, 360–369. [Google Scholar] [CrossRef]
- Möller, K.; Bein, T. Degradable drug carriers: Vanishing mesoporous silica nanoparticles. Chem. Mater. 2019, 31, 4364–4378. [Google Scholar] [CrossRef]
- Su, S.; Kang, P.M. Systemic review of biodegradable nanomaterials in nanomedicine. Nanomaterials 2020, 10, 656. [Google Scholar] [CrossRef] [PubMed]
- Qiu, M.; Singh, A.; Wang, D.; Qu, J.; Swihart, M.; Zhang, H.; Prasad, P.N. Biocompatible and biodegradable inorganic nanostructures for nanomedicine: Silicon and black phosphorus. Nano Today 2019, 25, 135–155. [Google Scholar] [CrossRef]
- Blomhoff, R.; Eskild, W.; Berg, T. Endocytosis of formaldehyde-treated serum albumin via scavenger pathway in liver endothelial cells. Biochem. J. 1984, 218, 81–86. [Google Scholar] [CrossRef]
- Wan, X.; Zheng, X.; Pang, X.; Zhang, Z.; Zhang, Q. Incorporation of lapatinib into human serum albumin nanoparticles with enhanced anti-tumor effects in HER2-positive breast cancer. Colloids Surf. B Biointerfaces 2015, 136, 817–827. [Google Scholar] [CrossRef]
- Zhang, Y.; Yue, X.; Kim, B.; Yao, S.; Bondar, M.V.; Belfield, K.D. Bovine serum albumin nanoparticles with fluorogenic near-IR-emitting squaraine dyes. ACS Appl. Mater. Interfaces 2013, 5, 8710–8717. [Google Scholar] [CrossRef]
- Look, J.; Wilhelm, N.; von Briesen, H.; Noske, N.; Günther, C.; Langer, K.; Gorjup, E. Ligand-modified human serum albumin nanoparticles for enhanced gene delivery. Mol. Pharm. 2015, 12, 3202–3213. [Google Scholar] [CrossRef]
- Bootz, A.; Vogel, V.; Schubert, D.; Kreuter, J. Comparison of scanning electron microscopy, dynamic light scattering and analytical ultracentrifugation for the sizing of poly (butyl cyanoacrylate) nanoparticles. Eur. J. Pharm. Biopharm. 2004, 57, 369–375. [Google Scholar] [CrossRef]
- Souza, T.G.; Ciminelli, V.S.; Mohallem, N.D.S. A comparison of TEM and DLS methods to characterize size distribution of ceramic nanoparticles. J. Phys. Conf. Ser. 2016, 733, 012039. [Google Scholar] [CrossRef]
- Zeeshan, F.; Madheswaran, T.; Panneerselvam, J.; Taliyan, R.; Kesharwani, P. Human serum albumin as multifunctional nanocarrier for cancer therapy. J. Pharm. Sci. 2021, 110, 3111–3117. [Google Scholar] [CrossRef]
- Takigawa, T.; Endo, Y. Effects of glutaraldehyde exposure on human health. J. Occup. Health 2006, 48, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Rudzinska-Radecka, M.; Frolova, A.S.; Balakireva, A.V.; Gorokhovets, N.V.; Pokrovsky, V.S.; Sokolova, D.V.; Korolev, D.O.; Potoldykova, N.V.; Vinarov, A.Z.; Parodi, A. In Silico, In Vitro, and Clinical Investigations of Cathepsin B and Stefin A mRNA Expression and a Correlation Analysis in Kidney Cancer. Cells 2022, 11, 1455. [Google Scholar] [CrossRef] [PubMed]
- Parodi, A.; Evangelopoulos, M.; Arrighetti, N.; Cevenini, A.; Livingston, M.; Khaled, S.Z.; Brown, B.S.; Yazdi, I.K.; Paradiso, F.; Campa-Carranza, J.N. Endosomal Escape of Polymer-Coated Silica Nanoparticles in Endothelial Cells. Small 2020, 16, 1907693. [Google Scholar] [CrossRef] [PubMed]
- Mijanović, O.; Branković, A.; Panin, A.N.; Savchuk, S.; Timashev, P.; Ulasov, I.; Lesniak, M.S. Cathepsin B: A sellsword of cancer progression. Cancer Lett. 2019, 449, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.I.; Yang, S.; Shim, M.K.; Kim, K. Cathepsin B-responsive prodrugs for cancer-targeted therapy: Recent advances and progress for clinical translation. Nano Res. 2022, 15, 7247–7266. [Google Scholar] [CrossRef]
- Ohshita, T.; Hiroi, Y. Degradation of serum albumin by rat liver and kidney lysosomes. J. Nutr. Sci. Vitaminol. 1998, 44, 641–653. [Google Scholar] [CrossRef]
- Di Spiezio, A.; Marques, A.R.; Schmidt, L.; Thießen, N.; Gallwitz, L.; Fogh, J.; Bartsch, U.; Saftig, P. Analysis of cathepsin B and cathepsin L treatment to clear toxic lysosomal protein aggregates in neuronal ceroid lipofuscinosis. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2021, 1867, 166205. [Google Scholar] [CrossRef]
- Präbst, K.; Engelhardt, H.; Ringgeler, S.; Hübner, H. Basic colorimetric proliferation assays: MTT, WST, and resazurin. In Cell Viability Assays; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1–17. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kolesova, E.P.; Egorova, V.S.; Syrocheva, A.O.; Frolova, A.S.; Kostyushev, D.; Kostyusheva, A.; Brezgin, S.; Trushina, D.B.; Fatkhutdinova, L.; Zyuzin, M.; et al. Proteolytic Resistance Determines Albumin Nanoparticle Drug Delivery Properties and Increases Cathepsin B, D, and G Expression. Int. J. Mol. Sci. 2023, 24, 10245. https://doi.org/10.3390/ijms241210245
Kolesova EP, Egorova VS, Syrocheva AO, Frolova AS, Kostyushev D, Kostyusheva A, Brezgin S, Trushina DB, Fatkhutdinova L, Zyuzin M, et al. Proteolytic Resistance Determines Albumin Nanoparticle Drug Delivery Properties and Increases Cathepsin B, D, and G Expression. International Journal of Molecular Sciences. 2023; 24(12):10245. https://doi.org/10.3390/ijms241210245
Chicago/Turabian StyleKolesova, Ekaterina P., Vera S. Egorova, Anastasiia O. Syrocheva, Anastasiia S. Frolova, Dmitry Kostyushev, Anastasiia Kostyusheva, Sergey Brezgin, Daria B. Trushina, Landysh Fatkhutdinova, Mikhail Zyuzin, and et al. 2023. "Proteolytic Resistance Determines Albumin Nanoparticle Drug Delivery Properties and Increases Cathepsin B, D, and G Expression" International Journal of Molecular Sciences 24, no. 12: 10245. https://doi.org/10.3390/ijms241210245
APA StyleKolesova, E. P., Egorova, V. S., Syrocheva, A. O., Frolova, A. S., Kostyushev, D., Kostyusheva, A., Brezgin, S., Trushina, D. B., Fatkhutdinova, L., Zyuzin, M., Demina, P. A., Khaydukov, E. V., Zamyatnin, A. A., Jr., & Parodi, A. (2023). Proteolytic Resistance Determines Albumin Nanoparticle Drug Delivery Properties and Increases Cathepsin B, D, and G Expression. International Journal of Molecular Sciences, 24(12), 10245. https://doi.org/10.3390/ijms241210245










