A Novel Introgression Line Library Derived from a Wild Melon Gives Insights into the Genetics of Melon Domestication, Uncovering New Genetic Variability Useful for Breeding
Abstract
1. Introduction
2. Results
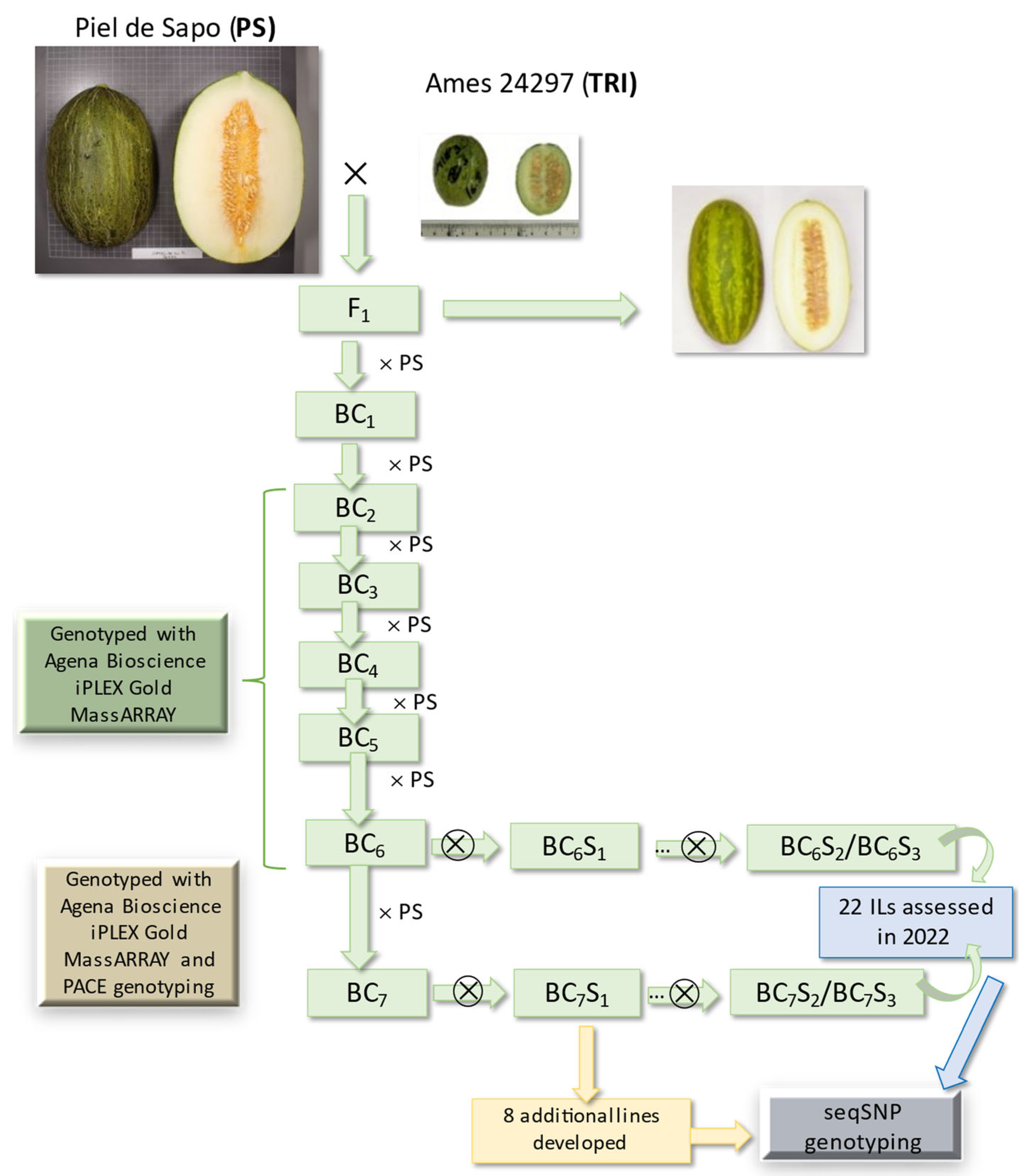
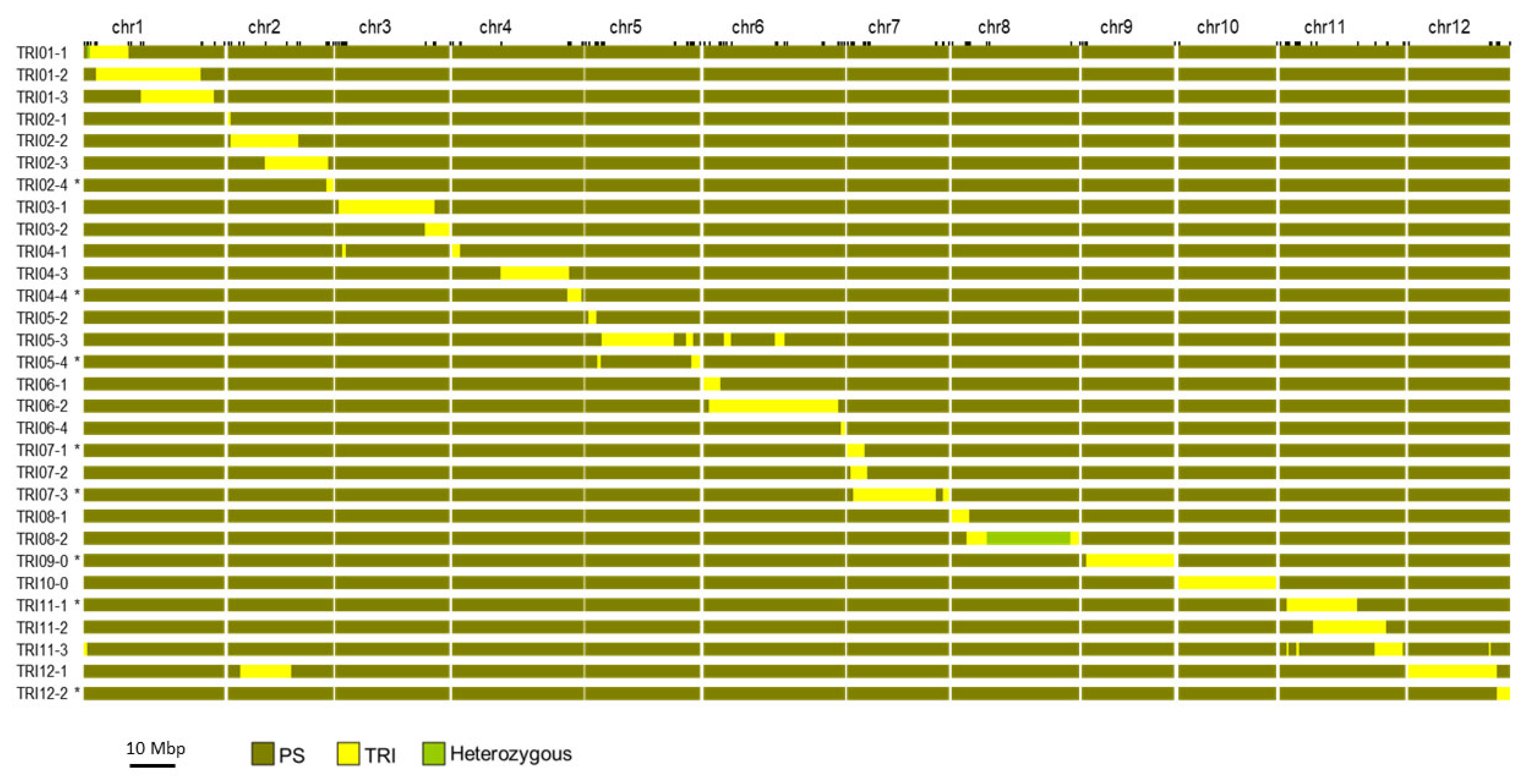
2.1. Development of the Introgression Line (IL) Collection
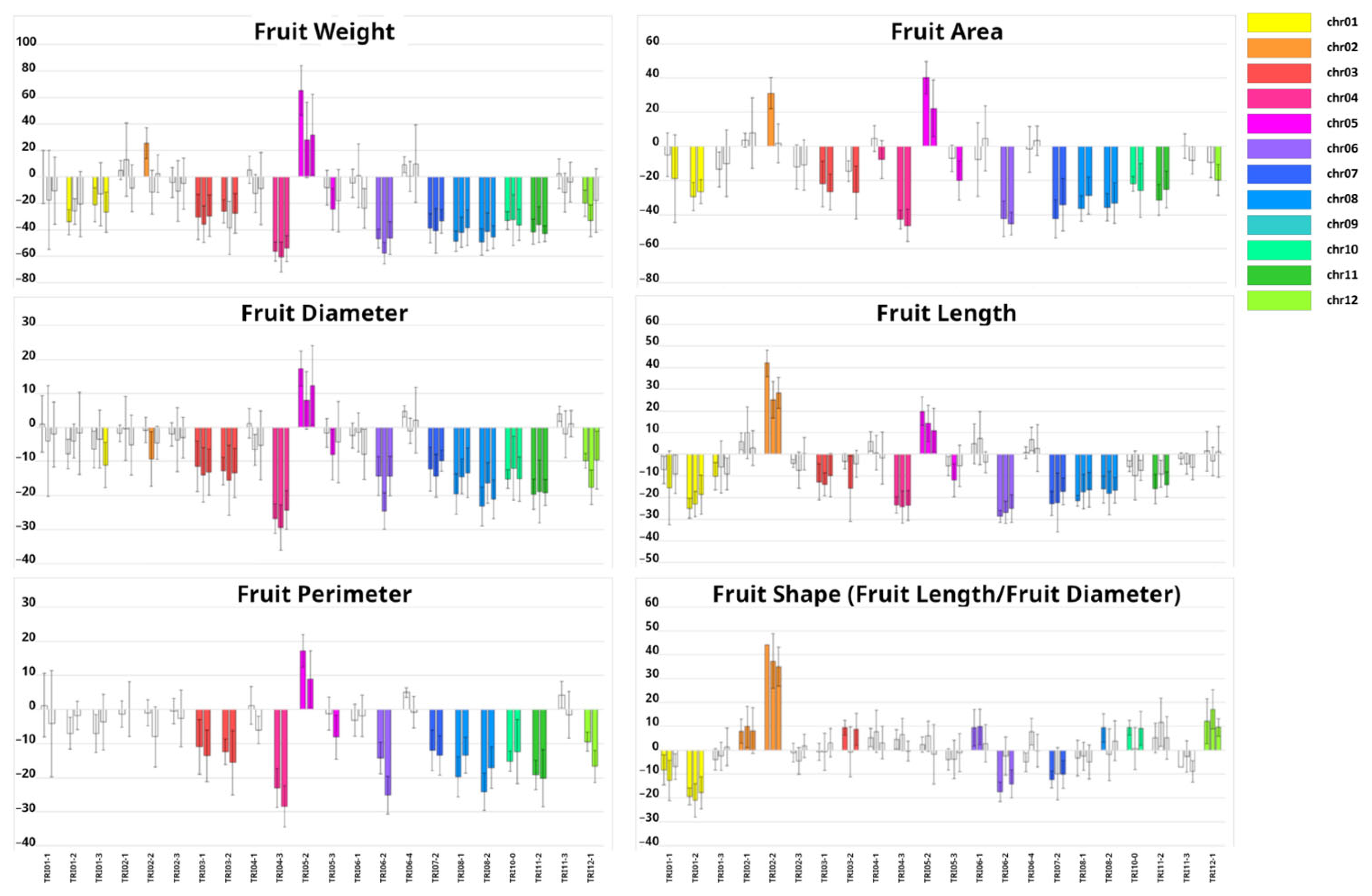
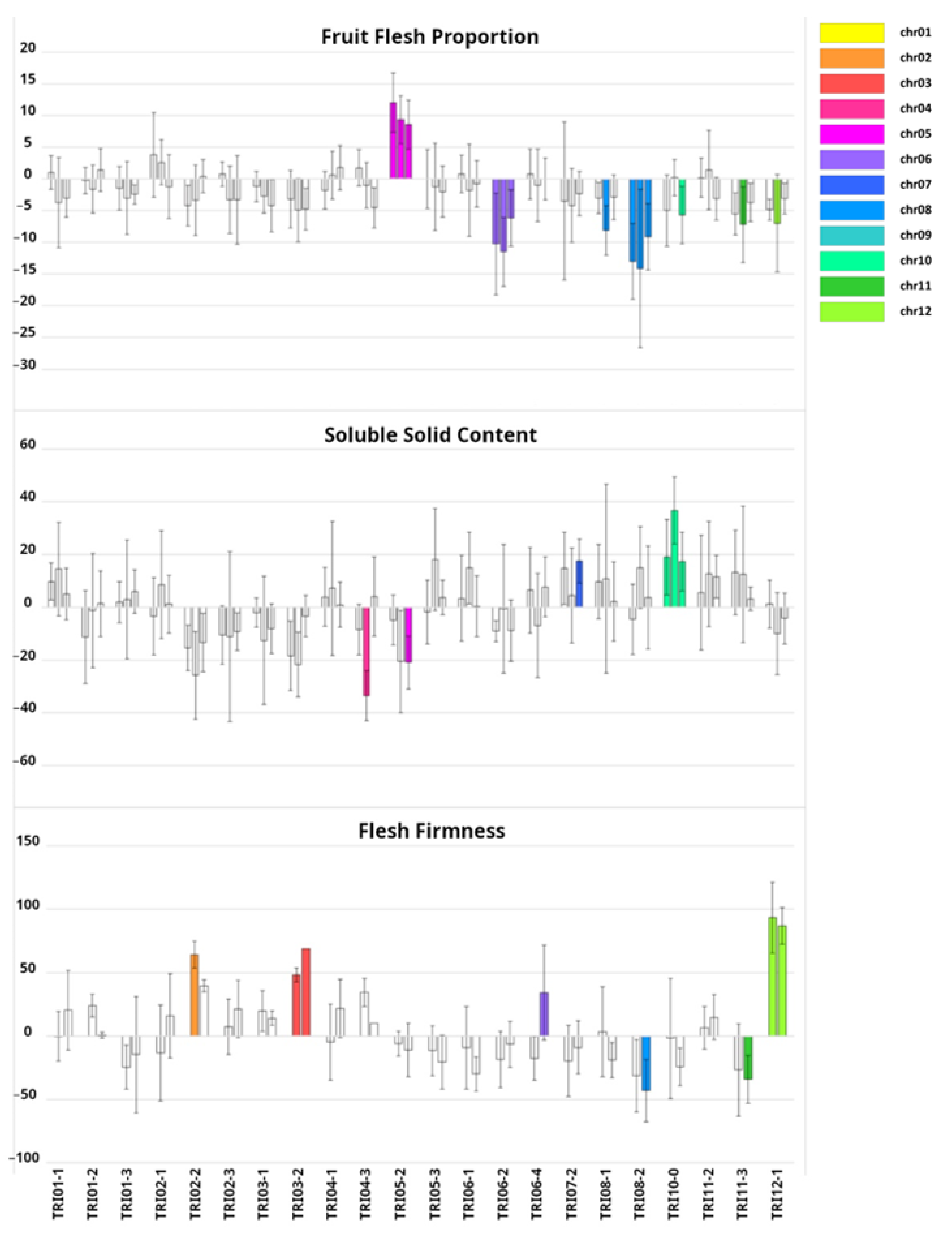
2.2. Phenotypic Characterization of Introgression Lines (ILs)
3. Discussion
3.1. Development of the Introgression Line (IL) Collection
3.2. Genetic Dissection of Agronomic Traits with the Introgression Line (IL) Collection
4. Materials and Methods
4.1. Plant Material and Introgression Line (IL) Development
4.2. Genotyping Methods
4.3. Phenotypic Characterization of the Introgression Line (IL) Collection
4.4. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pitrat, M. Melon Genetic Resources: Phenotypic Diversity and Horticultural Taxonomy. In Genetics and Genomics of Cucurbitaceae; Plant Genetics and Genomics: Crops and Models; Grumet, R., Katzir, N., Garcia-Mas, J., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 25–60. ISBN 978-3-319-49332-9. [Google Scholar]
- Pitrat, M.; Hanelt, P.; Hammer, K. Some comments on infraspecific classification of cultivars of melon. Acta Hortic. 2000, 510, 29–36. [Google Scholar] [CrossRef]
- Esteras, C.; Formisano, G.; Roig, C.; Díaz, A.; Blanca, J.; Garcia-Mas, J.; Gómez-Guillamón, M.L.; López-Sesé, A.I.; Lázaro, A.; Monforte, A.J.; et al. SNP genotyping in melons: Genetic variation, population structure, and linkage disequilibrium. Theor. Appl. Genet. 2013, 126, 1285–1303. [Google Scholar] [CrossRef]
- Gonzalo, M.J.; Díaz, A.; Dhillon, N.P.S.; Reddy, U.K.; Picó, B.; Monforte, A.J. Re-evaluation of the role of Indian germplasm as center of melon diversification based on genotyping-by-sequencing analysis. BMC Genom. 2019, 20, 448. [Google Scholar] [CrossRef]
- Endl, J.; Achigan-Dako, E.G.; Pandey, A.K.; Monforte, A.J.; Pico, B.; Schaefer, H. Repeated domestication of melon (Cucumis melo L.) in Africa and Asia and a new close relative from India. Am. J. Bot. 2018, 105, 1662–1671. [Google Scholar] [CrossRef]
- Zhao, G.; Lian, Q.; Zhang, Z.; Fu, Q.; He, Y.; Ma, S.; Ruggieri, V.; Monforte, A.J.; Wang, P.; Julca, I.; et al. A comprehensive genome variation map of melon identifies multiple domestication events and loci influencing agronomic traits. Nat. Genet. 2019, 51, 1607–1615. [Google Scholar] [CrossRef]
- Purugganan, M.D. Evolutionary Insights into the Nature of Plant Domestication. Curr. Biol. 2019, 29, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Turner-Hissong, S.D.; Mabry, M.E.; Beissinger, T.M.; Ross-Ibarra, J.; Pires, J.C. Evolutionary insights into plant breeding. Curr. Opin. Plant Biol. 2020, 54, 93–100. [Google Scholar] [CrossRef]
- Díaz, A.; Martín-Hernández, A.M.; Dolcet-Sanjuan, R.; Garcés-Claver, A.; Álvarez, J.M.; Garcia-Mas, J.; Picó, B.; Monforte, A.J. Quantitative trait loci analysis of melon (Cucumis melo L.) domestication-related traits. Theor. Appl. Genet. 2017, 130, 1837–1856. [Google Scholar] [CrossRef] [PubMed]
- Riahi, C.; Reig-Valiente, J.L.; Picó, B.; Díaz, A.; Gonzalo, M.J.; Monforte, A.J. Evidence of the role of QTL epistatic interactions in the increase of melon fruit flesh content during domestication. Agronomy 2020, 10, 1064. [Google Scholar] [CrossRef]
- Lian, Q.; Fu, Q.; Xu, Y.; Hu, Z.; Zheng, J.; Zhang, A.; He, Y.; Wang, C.; Xu, C.; Chen, B.; et al. QTLs and candidate genes analyses for fruit size under domestication and differentiation in melon (Cucumis melo L.) based on high resolution maps. BMC Plant Biol. 2021, 21, 126. [Google Scholar] [CrossRef]
- Dempewolf, H.; Baute, G.; Anderson, J.; Kilian, B.; Smith, C.; Guarino, L. Past and future use of wild relatives in crop breeding. Crop Sci. 2017, 57, 1070–1082. [Google Scholar] [CrossRef]
- Xu, L.; He, Y.; Tang, L.; Xu, Y.; Zhao, G. Genetics, Genomics, and Breeding in Melon. Agronomy 2022, 12, 2891. [Google Scholar] [CrossRef]
- Tanksley, S.D.; Nelson, J.C. Advanced backcross QTL analysis: A method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor. Appl. Genet. 1996, 92, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Monforte, A.J.; Diaz, A.; Caño-Delgado, A.; Van der Knaap, E. The genetic basis of fruit morphology in horticultural crops: Lessons from tomato and melon. J. Exp. Bot. 2014, 65, 4625–4637. [Google Scholar] [CrossRef]
- Zamir, D. Improving plant breeding with exotic genetic libraries. Nat. Rev. Genet. 2001, 2, 983–989. [Google Scholar] [CrossRef]
- Eshed, Y.; Zamir, D. An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 1995, 141, 1147–1162. [Google Scholar] [CrossRef]
- Monforte, A.J.; Tanksley, S.D. Development of a set of near isogenic and backcross recombinant inbred lines containing most of the Lycopersicon hirsutum genome in a L. esculentum genetic background: A tool for gene mapping and gene discovery. Genome 2000, 43, 803–813. [Google Scholar] [CrossRef] [PubMed]
- Canady, M.A.; Meglic, V.; Chetelat, R.T. A library of Solanum lycopersicoides introgression lines in cultivated tomato. Genome 2005, 48, 685–697. [Google Scholar] [CrossRef] [PubMed]
- Barrantes, W.; Fernández-del-Carmen, A.; López-Casado, G.; González-Sánchez, M.Á.; Fernández-Muñoz, R.; Granell, A.; Monforte, A.J. Highly efficient genomics-assisted development of a library of introgression lines of Solanum pimpinellifolium. Mol. Breed. 2014, 34, 1817–1831. [Google Scholar] [CrossRef]
- Zhang, B.; Ma, L.; Wu, B.; Xing, Y.; Qiu, X. Introgression Lines: Valuable Resources for Functional Genomics Research and Breeding in Rice (Oryza sativa L.). Front. Plant Sci. 2022, 13, 863789. [Google Scholar] [CrossRef] [PubMed]
- Eduardo, I.; Arús, P.; Monforte, A.J. Development of a genomic library of near isogenic lines (NILs) in melon (Cucumis melo L.) from the exotic accession PI161375. Theor. Appl. Genet. 2005, 112, 139–148. [Google Scholar] [CrossRef]
- Santo Domingo, M.; Mayobre, C.; Pereira, L.; Argyris, J.; Valverde, L.; Martín-Hernández, A.M.; Garcia-Mas, J.; Pujol, M. Fruit morphology and ripening-related QTLs in a newly developed introgression line collection of the elite varieties “Védrantais” and “Piel de Sapo”. Plants 2022, 11, 3120. [Google Scholar] [CrossRef] [PubMed]
- Castro, G.; Perpiñá, G.; Monforte, A.J.; Picó, B.; Esteras, C. New melon introgression lines in a Piel de Sapo genetic background with desirable agronomical traits from dudaim melons. Euphytica 2019, 215, 169. [Google Scholar] [CrossRef]
- Díaz, A.; Zarouri, B.; Fergany, M.; Eduardo, I.; Alvarez, J.M.; Picó, B.; Monforte, A.J. Mapping and introgression of QTL involved in fruit shape transgressive segregation into ‘piel de sapo’ melon (Cucumis melo L.). PLoS ONE 2014, 9, e104188. [Google Scholar] [CrossRef]
- Perpiñá, G.; Esteras, C.; Gibon, Y.; Monforte, A.J.; Picó, B. A new genomic library of melon introgression lines in a cantaloupe genetic background for dissecting desirable agronomical traits. BMC Plant Biol. 2016, 16, 154. [Google Scholar] [CrossRef]
- Pereira, L.; Santo Domingo, M.; Argyris, J.; Mayobre, C.; Valverde, L.; Martín-Hernández, A.M.; Pujol, M.; Garcia-Mas, J. A novel introgression line collection to unravel the genetics of climacteric ripening and fruit quality in melon. Sci. Rep. 2021, 11, 11364. [Google Scholar] [CrossRef]
- Díaz, A.; Fergany, M.; Formisano, G.; Ziarsolo, P.; Blanca, J.; Fei, Z.; Staub, J.E.; Zalapa, J.E.; Cuevas, H.E.; Dace, G.; et al. A consensus linkage map for molecular markers and quantitative trait loci associated with economically important traits in melon (Cucumis melo L.). BMC Plant Biol. 2011, 11, 111. [Google Scholar] [CrossRef] [PubMed]
- Giner, A.; Pascual, L.; Bourgeois, M.; Gyetvai, G.; Rios, P.; Picó, B.; Troadec, C.; Bendahmane, A.; Garcia-Mas, J.; Martín-Hernández, A.M. A mutation in the melon Vacuolar Protein Sorting 41 prevents systemic infection of Cucumber mosaic virus. Sci. Rep. 2017, 7, 10471. [Google Scholar] [CrossRef]
- Ríos, P.; Argyris, J.; Vegas, J.; Leida, C.; Kenigswald, M.; Tzuri, G.; Troadec, C.; Bendahmane, A.; Katzir, N.; Picó, B.; et al. ETHQV6.3 is involved in melon climacteric fruit ripening and is encoded by a NAC domain transcription factor. Plant J. Cell Mol. Biol. 2017, 91, 671–683. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Martínez, C.; Gonzalo, M.J.; Sipowicz, P.; Campos, M.; Martínez-Fernández, I.; Leida, C.; Zouine, M.; Alexiou, K.G.; Garcia-Mas, J.; Gómez, M.D.; et al. A cryptic variation in a member of the Ovate Family Proteins is underlying the melon fruit shape QTL fsqs8.1. Theor. Appl. Genet. 2022, 135, 785–801. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhao, Q.; Du, P.; Xu, C.; Wang, B.; Feng, Q.; Liu, Q.; Tang, S.; Gu, M.; Han, B.; et al. Developing high throughput genotyped chromosome segment substitution lines based on population whole-genome re-sequencing in rice (Oryza sativa L.). BMC Genom. 2010, 11, 656. [Google Scholar] [CrossRef] [PubMed]
- Finkers, R.; van Heusden, A.W.; Meijer-Dekens, F.; van Kan, J.A.L.; Maris, P.; Lindhout, P. The construction of a Solanum habrochaites LYC4 introgression line population and the identification of QTLs for resistance to Botrytis cinerea. Theor. Appl. Genet. 2007, 114, 1071–1080. [Google Scholar] [CrossRef] [PubMed]
- Castanera, R.; Ruggieri, V.; Pujol, M.; Garcia-Mas, J.; Casacuberta, J.M. An improved melon reference genome with single-molecule sequencing uncovers a recent burst of transposable elements with potential impact on genes. Front. Plant Sci. 2020, 10, 1815. [Google Scholar] [CrossRef] [PubMed]
- Pereira, L.; Ruggieri, V.; Pérez, S.; Alexiou, K.G.; Fernández, M.; Jahrmann, T.; Pujol, M.; Garcia-Mas, J. QTL mapping of melon fruit quality traits using a high-density GBS-based genetic map. BMC Plant Biol. 2018, 18, 324. [Google Scholar] [CrossRef] [PubMed]
- Eduardo, I.; Arús, P.; Monforte, A.J.; Obando, J.; Fernández-Trujillo, J.P.; Martínez, J.A.; Alarcón, A.L.; Álvarez, J.M.; van der Knaap, E. Estimating the genetic architecture of fruit quality traits in melon using a genomic library of near isogenic lines. J. Am. Soc. Hortic. Sci. 2007, 132, 80–89. [Google Scholar] [CrossRef]
- Barrantes, W.; López-Casado, G.; García-Martínez, S.; Alonso, A.; Rubio, F.; Ruiz, J.J.; Fernández-Muñoz, R.; Granell, A.; Monforte, A.J. Exploring new alleles involved in tomato fruit quality in an introgression line library of Solanum pimpinellifolium. Front. Plant Sci. 2016, 7, 1172. [Google Scholar] [CrossRef]
- Pan, Y.; Wang, Y.; McGregor, C.; Liu, S.; Luan, F.; Gao, M.; Weng, Y. Genetic architecture of fruit size and shape variation in cucurbits: A comparative perspective. Theor. Appl. Genet. 2020, 133, 1–21. [Google Scholar] [CrossRef]
- Boualem, A.; Fergany, M.; Fernandez, R.; Troadec, C.; Martin, A.; Morin, H.; Sari, M.-A.; Collin, F.; Flowers, J.M.; Pitrat, M.; et al. A conserved mutation in an ethylene biosynthesis enzyme leads to andromonoecy in melons. Science 2008, 321, 836–838. [Google Scholar] [CrossRef]
- Argyris, J.M.; Díaz, A.; Ruggieri, V.; Fernández, M.; Jahrmann, T.; Gibon, Y.; Picó, B.; Martín-Hernández, A.M.; Monforte, A.J.; Garcia-Mas, J. QTL analyses in multiple populations employed for the fine mapping and identification of candidate genes at a locus affecting sugar accumulation in melon (Cucumis melo) L. Front. Plant Sci. 2017, 8, 1679. [Google Scholar] [CrossRef]
- Roy, A.; Bal, S.S.; Fergany, M.; Kaur, S.; Singh, H.; Malik, A.A.; Singh, J.; Monforte, A.J.; Dhillon, N.P.S. Wild melon diversity in India (Punjab State). Genet. Resour. Crop Evol. 2012, 59, 755–767. [Google Scholar] [CrossRef]
- Malik, A.A.; Vashisht, V.K.; Singh, K.; Sharma, A.; Singh, D.K.; Singh, H.; Monforte, A.J.; McCreight, J.D.; Dhillon, N.P.S. Diversity among melon (Cucumis melo L.) landraces from the Indo-Gangetic plains of India and their genetic relationship with USA melon cultivars. Genet. Resour. Crop Evol. 2014, 61, 1189–1208. [Google Scholar] [CrossRef]
- Oren, E.; Tzuri, G.; Dafna, A.; Meir, A.; Kumar, R.; Katzir, N.; Elkind, Y.; Freilich, S.; Schaffer, A.A.; Tadmor, Y.; et al. High-density NGS-based map construction and genetic dissection of fruit shape and rind netting in Cucumis melo. Theor. Appl. Genet. 2020, 133, 1927–1945. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhang, B.; Keyhaninejad, N.; Rodríguez, G.R.; Kim, H.J.; Chakrabarti, M.; Illa-Berenguer, E.; Taitano, N.K.; Gonzalo, M.J.; Díaz, A.; et al. A common genetic mechanism underlies morphological diversity in fruits and other plant organs. Nat. Commun. 2018, 9, 4734. [Google Scholar] [CrossRef] [PubMed]
- Pereira, L.; Santo Domingo, M.; Ruggieri, V.; Argyris, J.; Phillips, M.A.; Zhao, G.; Lian, Q.; Xu, Y.; He, Y.; Huang, S.; et al. Genetic dissection of climacteric fruit ripening in a melon population segregating for ripening behavior. Hortic. Res. 2020, 7, 187. [Google Scholar] [CrossRef] [PubMed]
- Monforte, A.J.; Oliver, M.; Gonzalo, M.J.; Alvarez, J.M.; Dolcet-Sanjuan, R.; Arús, P. Identification of quantitative trait loci involved in fruit quality traits in melon (Cucumis melo L.). Theor. Appl. Genet. 2004, 108, 750–758. [Google Scholar] [CrossRef]
- Feder, A.; Burger, J.; Gao, S.; Lewinsohn, E.; Katzir, N.; Schaffer, A.A.; Meir, A.; Davidovich-Rikanati, R.; Portnoy, V.; Gal-On, A.; et al. A kelch domain-containing F-Box coding gene negatively regulates flavonoid accumulation in Muskmelon. Plant Physiol. 2015, 169, 1714–1726. [Google Scholar] [CrossRef]
- Doyle, J.; Doyle, J. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Garcia-Mas, J.; Benjak, A.; Sanseverino, W.; Bourgeois, M.; Mir, G.; González, V.M.; Hénaff, E.; Câmara, F.; Cozzuto, L.; Lowy, E.; et al. The genome of melon (Cucumis melo L.). Proc. Natl. Acad. Sci. USA 2012, 109, 11872–11877. [Google Scholar] [CrossRef]
- Sanseverino, W.; Hénaff, E.; Vives, C.; Pinosio, S.; Burgos-Paz, W.; Morgante, M.; Ramos-Onsins, S.E.; Garcia-Mas, J.; Casacuberta, J.M. Transposon insertions, structural variations, and SNPs contribute to the evolution of the melon genome. Mol. Biol. Evol. 2015, 32, 2760–2774. [Google Scholar] [CrossRef]
- Van Berloo, R. GGT 2.0: Versatile software for visualization and analysis of genetic data. J. Hered. 2008, 99, 232–236. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Berg, S.; Kutra, D.; Kroeger, T.; Straehle, C.N.; Kausler, B.X.; Haubold, C.; Schiegg, M.; Ales, J.; Beier, T.; Rudy, M.; et al. ilastik: Interactive machine learning for (bio)image analysis. Nat. Methods 2019, 16, 1226–1232. [Google Scholar] [CrossRef] [PubMed]

| Trait 1 | Alcàsser | Meliana | Algarrobo |
|---|---|---|---|
| FW (g) | 2514.48 ± 324.73 a | 2637.36 ± 508.29 a | 1752.68 ± 459.23 b |
| FP (mm) | 483.56 ± 23.18 a | 473.68 ± 31.47 a | n.d. 2 |
| FS | 1.39 ± 0.06 a | 1.52 ± 0.13 b | 1.41 ± 0.12 a |
| FL (mm) | 215.08 ± 11.32 a | 231.08 ± 19.98 b | 185.16 ± 14.97 c |
| FD (mm) | 154.72 ± 7.19 a | 152.36 ± 9.99 a | 132.74 ± 12.72 b |
| FA (cm2) | 263.16 ± 22.80 a | 286.22 ± 38.39 b | n.d. |
| FFP (%) | 70.32 ± 3.00 a | 70.42 ± 3.21 a | 70.88 ± 3.53 a |
| SSC (ºBrix) | 12.24 ± 1.30 a | 10.40 ± 2.72 b | 13.92 ± 1.60 c |
| FF (kg/cm2) | 2.12 ± 0.43 a | 2.55 ± 0.85 b | n.d. |
| Location | IL | Location x IL | ||||
|---|---|---|---|---|---|---|
| 3 L 1 | 2 L 2 | 3 L | 2 L | 3 L | 2 L | |
| Trait 3 | Snedecor’s F | |||||
| FW (g) | 163.89 ** | 2.23 | 37.24 ** | 25.55 ** | 2.71 ** | 1.76 |
| FP (mm) | n.d. 4 | 27.45 ** | n.d. | 23.67 ** | n.d. | 1.75 |
| FS | 85.69 ** | 104.06 ** | 42.38 ** | 27.59 ** | 2.27 ** | 2.10 * |
| FL (mm) | 260.46 ** | 19.7 ** | 50.69 ** | 32.99 ** | 2.16 ** | 1.30 |
| FD (mm) | 173.73 ** | 20.09 ** | 31.42 ** | 21.95 ** | 1.57 | 1.60 |
| FA (cm2) | n.d. | 4.68 | n.d. | 25.62 ** | n.d. | 1.64 |
| FFP (%) | 3.62 | 2.97 | 14.39 ** | 9.29 ** | 1.22 | 0.52 |
| SSC (ºBrix) | 186.17 ** | 51.84 ** | 9.27 ** | 5.74 ** | 1.79 * | 1.20 |
| FF (kg/cm2) | n.d. | 31.06 ** | n.d. | 14.42 ** | n.d. | 1.83 |
| Trait | Alcàsser | Meliana | Algarrobo |
|---|---|---|---|
| Yellow background in rind | TRI03-2 | TRI03-2 | TRI03-2 |
| Pale background in rind | TRI04-1 | TRI04-1 | |
| TRI08-2 | TRI08-2 | TRI08-2 | |
| Abscission layer | TRI08-1 | TRI08-1 | n.d 1 |
| TRI08-2 | TRI08-2 | n.d. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campos, M.; Gonzalo, M.J.; Díaz, A.; Picó, B.; Gómez-Guillamón, M.L.; Monforte, A.J.; Esteras, C. A Novel Introgression Line Library Derived from a Wild Melon Gives Insights into the Genetics of Melon Domestication, Uncovering New Genetic Variability Useful for Breeding. Int. J. Mol. Sci. 2023, 24, 10099. https://doi.org/10.3390/ijms241210099
Campos M, Gonzalo MJ, Díaz A, Picó B, Gómez-Guillamón ML, Monforte AJ, Esteras C. A Novel Introgression Line Library Derived from a Wild Melon Gives Insights into the Genetics of Melon Domestication, Uncovering New Genetic Variability Useful for Breeding. International Journal of Molecular Sciences. 2023; 24(12):10099. https://doi.org/10.3390/ijms241210099
Chicago/Turabian StyleCampos, Manuel, Maria José Gonzalo, Aurora Díaz, Belén Picó, Maria Luisa Gómez-Guillamón, Antonio José Monforte, and Cristina Esteras. 2023. "A Novel Introgression Line Library Derived from a Wild Melon Gives Insights into the Genetics of Melon Domestication, Uncovering New Genetic Variability Useful for Breeding" International Journal of Molecular Sciences 24, no. 12: 10099. https://doi.org/10.3390/ijms241210099
APA StyleCampos, M., Gonzalo, M. J., Díaz, A., Picó, B., Gómez-Guillamón, M. L., Monforte, A. J., & Esteras, C. (2023). A Novel Introgression Line Library Derived from a Wild Melon Gives Insights into the Genetics of Melon Domestication, Uncovering New Genetic Variability Useful for Breeding. International Journal of Molecular Sciences, 24(12), 10099. https://doi.org/10.3390/ijms241210099








