p53 Dysregulation in Breast Cancer: Insights on Mutations in the TP53 Network and p53 Isoform Expression
Abstract
:1. Introduction
2. Results
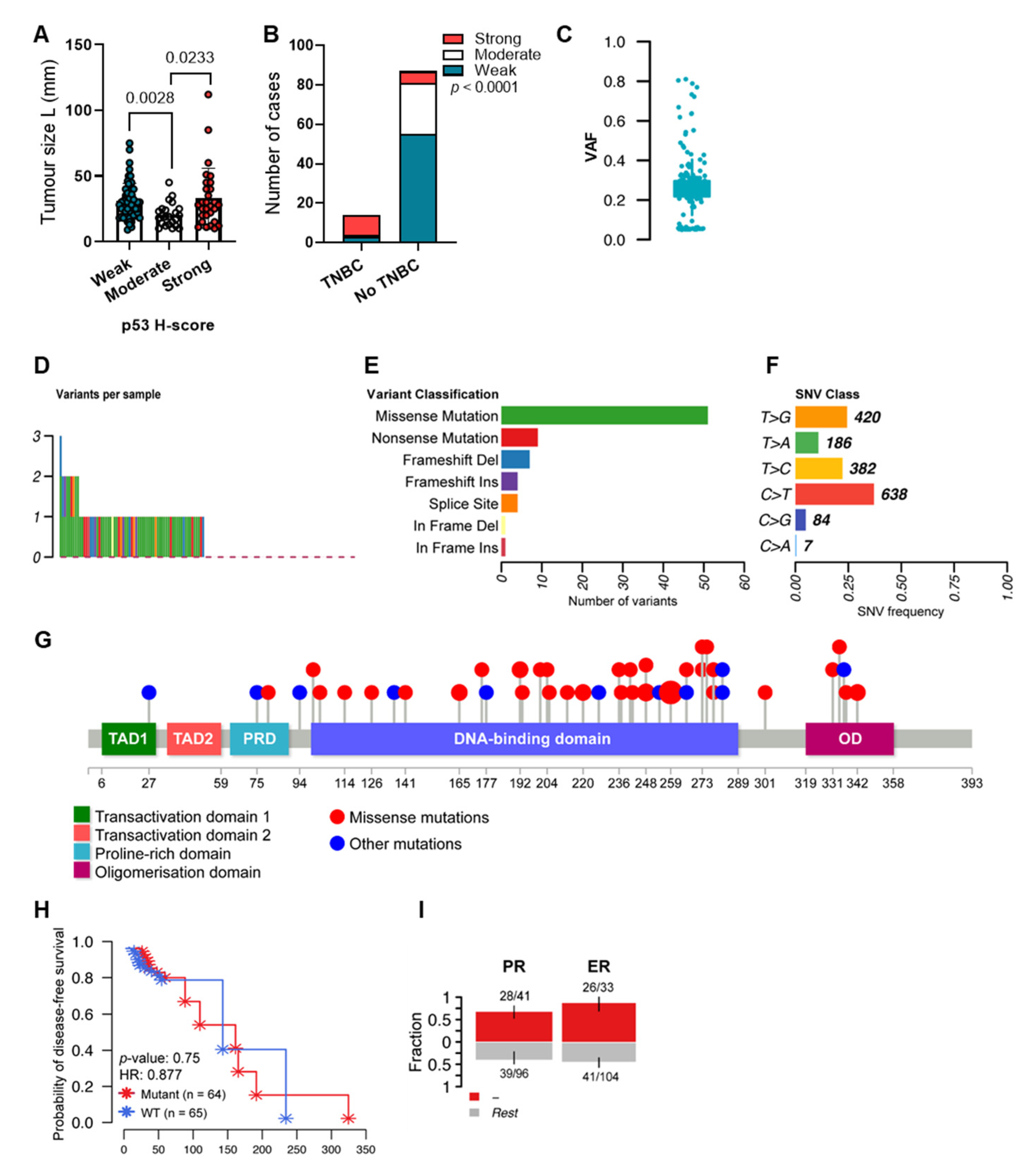
2.1. The Mutation Landscape of TP53 in Breast Cancers with Variable Levels of p53
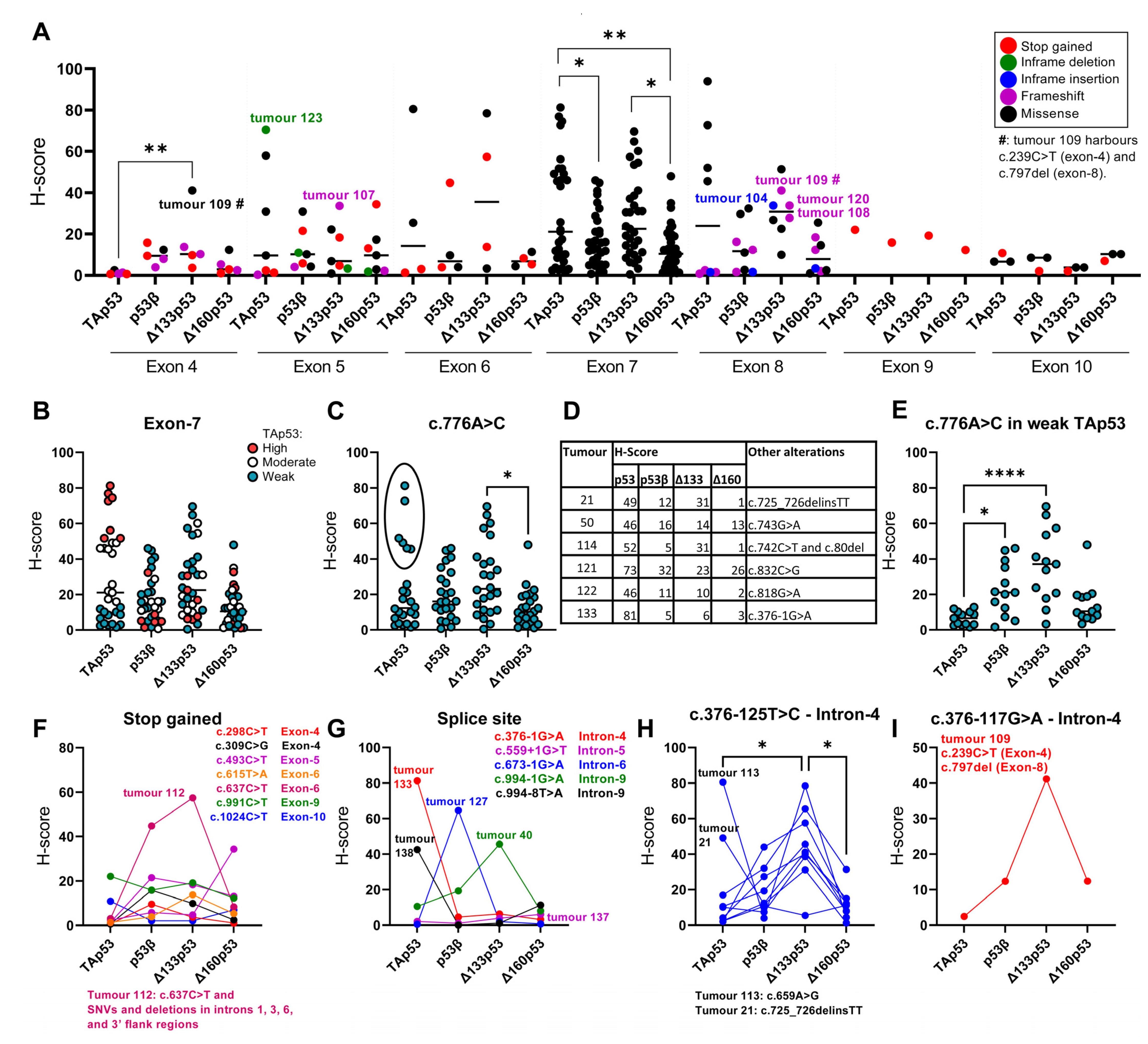
2.2. TP53 Mutations and p53 Isoform Expression
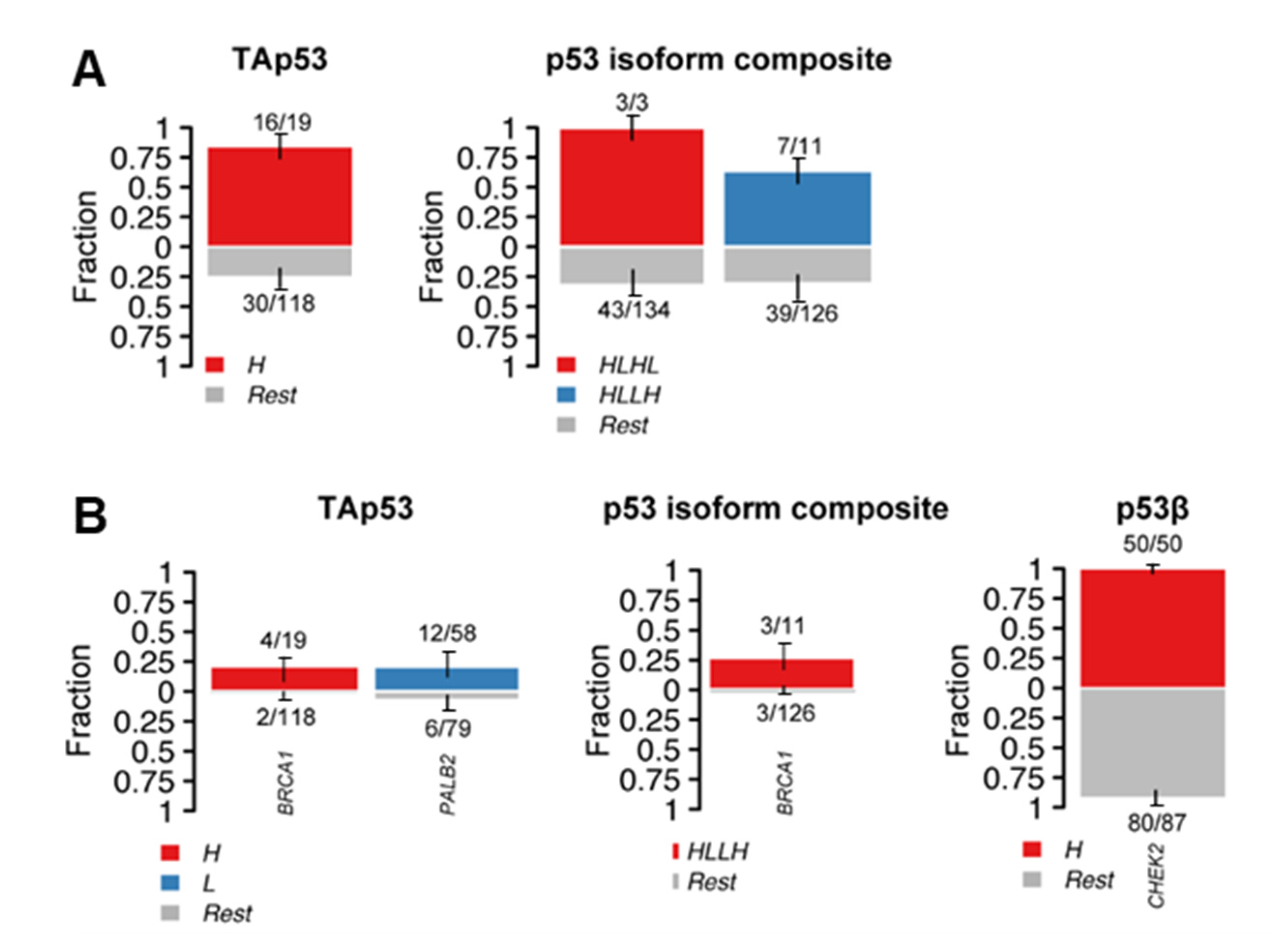
2.3. Alterations in p53 Interactors and p53 Isoform Expression
3. Discussion
4. Materials and Methods
4.1. Study Cohort
4.2. Next-Generation Sequencing
4.3. Bioinformatics
4.4. Immunohistochemistry of p53 Isoforms
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Olivier, M.; Hollstein, M.; Hainaut, P. TP53 mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2010, 2, a001008. [Google Scholar] [CrossRef] [Green Version]
- Dumay, A.; Feugeas, J.P.; Wittmer, E.; Lehmann-Che, J.; Bertheau, P.; Espie, M.; Plassa, L.F.; Cottu, P.; Marty, M.; Andre, F.; et al. Distinct tumor protein p53 mutants in breast cancer subgroups. Int. J. Cancer 2013, 132, 1227–1231. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef] [Green Version]
- Shahbandi, A.; Nguyen, H.D.; Jackson, J.G. TP53 Mutations and Outcomes in Breast Cancer: Reading beyond the Headlines. Trends Cancer 2020, 6, 98–110. [Google Scholar] [CrossRef]
- Olivier, M.; Langerød, A.; Carrieri, P.; Bergh, J.; Klaar, S.; Eyfjord, J.; Theillet, C.; Rodriguez, C.; Lidereau, R.; Bièche, I.; et al. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin. Cancer Res. 2006, 12, 1157–1167. [Google Scholar] [CrossRef] [Green Version]
- Ghatak, D.; Das Ghosh, D.; Roychoudhury, S. Cancer Stemness: p53 at the Wheel. Front. Oncol. 2020, 10, 604124. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, C.; Feng, Z. Tumor suppressor p53 and its gain-of-function mutants in cancer. Acta Biochim. Biophys. Sin. 2014, 46, 170–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, L.D.; Smeds, J.; George, J.; Vega, V.B.; Vergara, L.; Ploner, A.; Pawitan, Y.; Hall, P.; Klaar, S.; Liu, E.T.; et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc. Natl. Acad. Sci. USA 2005, 102, 13550–13555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coutant, C.; Rouzier, R.; Qi, Y.; Lehmann-Che, J.; Bianchini, G.; Iwamoto, T.; Hortobagyi, G.N.; Symmans, W.F.; Uzan, S.; Andre, F.; et al. Distinct p53 gene signatures are needed to predict prognosis and response to chemotherapy in ER-positive and ER-negative breast cancers. Clin. Cancer Res. 2011, 17, 2591–2601. [Google Scholar] [CrossRef] [Green Version]
- Dookeran, K.A.; Dignam, J.J.; Ferrer, K.; Sekosan, M.; McCaskill-Stevens, W.; Gehlert, S. p53 as a marker of prognosis in African-American women with breast cancer. Ann. Surg. Oncol. 2010, 17, 1398–1405. [Google Scholar] [CrossRef] [PubMed]
- Biganzoli, E.; Coradini, D.; Ambrogi, F.; Garibaldi, J.M.; Lisboa, P.; Soria, D.; Green, A.R.; Pedriali, M.; Piantelli, M.; Querzoli, P.; et al. p53 status identifies two subgroups of triple-negative breast cancers with distinct biological features. Jpn. J. Clin. Oncol. 2011, 41, 172–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bae, S.Y.; Nam, S.J.; Jung, Y.; Lee, S.B.; Park, B.W.; Lim, W.; Jung, S.H.; Yang, H.W.; Jung, S.P. Differences in prognosis and efficacy of chemotherapy by p53 expression in triple-negative breast cancer. Breast Cancer Res. Treat. 2018, 172, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Chae, B.J.; Bae, J.S.; Lee, A.; Park, W.C.; Seo, Y.J.; Song, B.J.; Kim, J.S.; Jung, S.S. p53 as a specific prognostic factor in triple-negative breast cancer. Jpn. J. Clin. Oncol. 2009, 39, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Petitjean, A.; Mathe, E.; Kato, S.; Ishioka, C.; Tavtigian, S.V.; Hainaut, P.; Olivier, M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: Lessons from recent developments in the IARC TP53 database. Hum. Mutat. 2007, 28, 622–629. [Google Scholar] [CrossRef]
- Knappskog, S.; Chrisanthar, R.; Løkkevik, E.; Anker, G.; Østenstad, B.; Lundgren, S.; Risberg, T.; Mjaaland, I.; Leirvaag, B.; Miletic, H.; et al. Low expression levels of ATM may substitute for CHEK2/TP53 mutations predicting resistance towards anthracycline and mitomycin chemotherapy in breast cancer. Breast Cancer Res. 2012, 14, R47. [Google Scholar] [CrossRef] [Green Version]
- Haupt, S.; Buckley, D.; Pang, J.M.; Panimaya, J.; Paul, P.J.; Gamell, C.; Takano, E.A.; Ying Lee, Y.; Hiddingh, S.; Rogers, T.M.; et al. Targeting Mdmx to treat breast cancers with wild-type p53. Cell Death Dis. 2015, 6, e1821. [Google Scholar] [CrossRef] [Green Version]
- Yu, Q.; Li, Y.; Mu, K.; Li, Z.; Meng, Q.; Wu, X.; Wang, Y.; Li, L. Amplification of Mdmx and overexpression of MDM2 contribute to mammary carcinogenesis by substituting for p53 mutations. Diagn. Pathol. 2014, 9, 71. [Google Scholar] [CrossRef] [Green Version]
- Mayo, L.D.; Donner, D.B. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc. Natl. Acad. Sci. USA 2001, 98, 11598–11603. [Google Scholar] [CrossRef] [Green Version]
- Iacobucci, I.; Ferrari, A.; Lonetti, A.; Papayannidis, C.; Paoloni, F.; Trino, S.; Storlazzi, C.T.; Ottaviani, E.; Cattina, F.; Impera, L.; et al. CDKN2A/B alterations impair prognosis in adult BCR-ABL1-positive acute lymphoblastic leukemia patients. Clin. Cancer Res. 2011, 17, 7413–7423. [Google Scholar] [CrossRef] [Green Version]
- Kamijo, T.; Weber, J.D.; Zambetti, G.; Zindy, F.; Roussel, M.F.; Sherr, C.J. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc. Natl. Acad. Sci. USA 1998, 95, 8292–8297. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.L.; Sang, B.; Liu, G.Z.; Li, J.M.; Zhang, X.D.; Liu, L.X.; Thorne, R.F.; Wu, M. SENEBLOC, a long non-coding RNA suppresses senescence via p53-dependent and independent mechanisms. Nucleic Acids Res. 2020, 48, 3089–3102. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.C.; Liu, X.Y.; Teng, L.; Ji, Q.; Wu, Y.; Li, J.M.; Gao, W.; Zhang, Y.Y.; La, T.; Tabatabaee, H.; et al. c-Myc inactivation of p53 through the pan-cancer lncRNA MILIP drives cancer pathogenesis. Nat. Commun. 2020, 11, 4980. [Google Scholar] [CrossRef]
- Zang, Y.; Shi, Y.; Liu, K.; Qiao, L.; Guo, X.; Chen, D. Δ40p53 is involved in the inactivation of autophagy and contributes to inhibition of cell death in HCT116-Δ40p53 cells. Oncotarget 2017, 8, 12754–12763. [Google Scholar] [CrossRef] [Green Version]
- Horikawa, I.; Park, K.Y.; Isogaya, K.; Hiyoshi, Y.; Li, H.; Anami, K.; Robles, A.I.; Mondal, A.M.; Fujita, K.; Serrano, M.; et al. Δ133p53 represses p53-inducible senescence genes and enhances the generation of human induced pluripotent stem cells. Cell Death Differ. 2017, 24, 1017–1028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, H.; Zhang, Y.; Jiang, K.; Ye, S.; Chen, S.; Zhang, Q.; Peng, J.; Chen, J. p73 coordinates with Δ133p53 to promote DNA double-strand break repair. Cell Death Differ. 2018, 25, 1063–1079. [Google Scholar] [CrossRef]
- Campbell, H.; Fleming, N.; Roth, I.; Mehta, S.; Wiles, A.; Williams, G.; Vennin, C.; Arsic, N.; Parkin, A.; Pajic, M.; et al. ∆133p53 isoform promotes tumour invasion and metastasis via interleukin-6 activation of JAK-STAT and RhoA-ROCK signalling. Nat. Commun. 2018, 9, 254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.-X.; Pan, W.-Y.; Chen, J. p53 and its isoforms in DNA double-stranded break repair. J. Zhejiang Univ. Sci. B 2019, 20, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Pan, X.; Abali, G.K.; Little, J.B.; Yuan, Z.M. Functional interplay between p53 and Δ133p53 in adaptive stress response. Cell Death Differ. 2020, 27, 1618–1632. [Google Scholar] [CrossRef]
- Levandowski, C.B.; Jones, T.; Gruca, M.; Ramamoorthy, S.; Dowell, R.D.; Taatjes, D.J. The Δ40p53 isoform inhibits p53-dependent eRNA transcription and enables regulation by signal-specific transcription factors during p53 activation. PLoS Biol. 2021, 19, e3001364. [Google Scholar] [CrossRef]
- Zhang, X.; Groen, K.; Morten, B.C.; Steffens Reinhardt, L.; Campbell, H.G.; Braithwaite, A.W.; Bourdon, J.C.; Avery-Kiejda, K.A. The effect of p53 and its N-terminally truncated isoform, Δ40p53, on breast cancer migration and invasion. Mol. Oncol. 2021, 16, 447–465. [Google Scholar] [CrossRef]
- Guo, Y.; Rall-Scharpf, M.; Bourdon, J.-C.; Wiesmüller, L.; Biber, S. p53 isoforms differentially impact on the POLι dependent DNA damage tolerance pathway. Cell Death Dis. 2021, 12, 941. [Google Scholar] [CrossRef]
- Tadijan, A.; Precazzini, F.; Hanžić, N.; Radić, M.; Gavioli, N.; Vlašić, I.; Ozretić, P.; Pinto, L.; Škreblin, L.; Barban, G.; et al. Altered Expression of Shorter p53 Family Isoforms Can Impact Melanoma Aggressiveness. Cancers 2021, 13, 5231. [Google Scholar] [CrossRef]
- Arsic, N.; Slatter, T.; Gadea, G.; Villain, E.; Fournet, A.; Kazantseva, M.; Allemand, F.; Sibille, N.; Seveno, M.; de Rossi, S.; et al. Δ133p53β isoform pro-invasive activity is regulated through an aggregation-dependent mechanism in cancer cells. Nat. Commun. 2021, 12, 5463. [Google Scholar] [CrossRef]
- Bourdon, J.C.; Fernandes, K.; Murray-Zmijewski, F.; Liu, G.; Diot, A.; Xirodimas, D.P.; Saville, M.K.; Lane, D.P. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005, 19, 2122–2137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourdon, J.C. p53 and its isoforms in cancer. Br. J. Cancer 2007, 97, 277–282. [Google Scholar] [CrossRef]
- Anensen, N.; Oyan, A.M.; Bourdon, J.-C.; Kalland, K.H.; Bruserud, O.; Gjertsen, B.T. A Distinct p53 Protein Isoform Signature Reflects the Onset of Induction Chemotherapy for Acute Myeloid Leukemia. Clin. Cancer Res. 2006, 12, 3985–3992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boldrup, L.; Bourdon, J.-C.; Coates, P.J.; Sjöström, B.; Nylander, K. Expression of p53 isoforms in squamous cell carcinoma of the head and neck. Eur. J. Cancer 2007, 43, 617–623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourdon, J.-C.; Khoury, M.; Diot, A.; Baker, L.; Fernandes, K.; Aoubala, M.; Quinlan, P.; Purdie, C.; Jordan, L.; Prats, A.-C.; et al. p53 mutant breast cancer patients expressing p53gamma have as good a prognosis as wild-type p53 breast cancer patients. Breast Cancer Res. 2011, 13, R7. [Google Scholar] [CrossRef] [PubMed]
- Ånensen, N.; Hjelle, S.M.; Van Belle, W.; Haaland, I.; Silden, E.; Bourdon, J.C.; Hovland, R.; Taskén, K.; Knappskog, S.; Lønning, P.E.; et al. Correlation analysis of p53 protein isoforms with NPM1/FLT3 mutations and therapy response in acute myeloid leukemia. Oncogene 2012, 31, 1533–1545. [Google Scholar] [CrossRef] [Green Version]
- Surget, S.; Khoury, M.P.; Bourdon, J.C. Uncovering the role of p53 splice variants in human malignancy: A clinical perspective. OncoTargets Ther. 2014, 7, 57–68. [Google Scholar]
- Hayman, L.; Chaudhry, W.R.; Revin, V.V.; Zhelev, N.; Bourdon, J.C. What is the potential of p53 isoforms as a predictive biomarker in the treatment of cancer? Expert Rev. Mol. Diagn. 2019, 19, 149–159. [Google Scholar] [CrossRef] [Green Version]
- Avery-Kiejda, K.A.; Morten, B.; Wong-Brown, M.W.; Mathe, A.; Scott, R.J. The relative mRNA expression of p53 isoforms in breast cancer is associated with clinical features and outcome. Carcinogenesis 2014, 35, 586–596. [Google Scholar] [CrossRef]
- Bischof, K.; Knappskog, S.; Stefansson, I.; McCormack, E.M.; Trovik, J.; Werner, H.M.J.; Woie, K.; Gjertsen, B.T.; Bjorge, L. High expression of the p53 isoform gamma is associated with reduced progression-free survival in uterine serous carcinoma. BMC Cancer 2018, 18, 684. [Google Scholar] [CrossRef] [Green Version]
- Kazantseva, M.; Eiholzer, R.A.; Mehta, S.; Taha, A.; Bowie, S.; Roth, I.; Zhou, J.; Joruiz, S.M.; Royds, J.A.; Hung, N.A.; et al. Elevation of the TP53 isoform Δ133p53β in glioblastomas: An alternative to mutant p53 in promoting tumor development. J. Pathol. 2018, 246, 77–88. [Google Scholar] [CrossRef] [Green Version]
- Knezovic Florijan, M.; Ozretic, P.; Bujak, M.; Pezze, L.; Ciribilli, Y.; Kastelan, Z.; Slade, N.; Hudolin, T. The role of p53 isoforms’ expression and p53 mutation status in renal cell cancer prognosis. Urol. Oncol. 2019, 37, 578.e1–578.e10. [Google Scholar] [CrossRef]
- Melo Dos Santos, N.; de Oliveira, G.A.P.; Ramos Rocha, M.; Pedrote, M.M.; Diniz da Silva Ferretti, G.; Pereira Rangel, L.; Morgado-Diaz, J.A.; Silva, J.L.; Rodrigues Pereira Gimba, E. Loss of the p53 transactivation domain results in high amyloid aggregation of the Delta40p53 isoform in endometrial carcinoma cells. J. Biol. Chem. 2019, 294, 9430–9439. [Google Scholar] [CrossRef] [Green Version]
- Kazantseva, M.; Mehta, S.; Eiholzer, R.A.; Gimenez, G.; Bowie, S.; Campbell, H.; Reily-Bell, A.L.; Roth, I.; Ray, S.; Drummond, C.J.; et al. The Δ133p53β isoform promotes an immunosuppressive environment leading to aggressive prostate cancer. Cell Death Dis. 2019, 10, 631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, L.; Hainaut, P.; Blanchet, S.; Ariffin, H. Expression of p53 N-terminal isoforms in B-cell precursor acute lymphoblastic leukemia and its correlation with clinicopathological profiles. BMC Cancer 2020, 20, 110. [Google Scholar] [CrossRef] [Green Version]
- Steffens Reinhardt, L.; Zhang, X.; Wawruszak, A.; Groen, K.; De Iuliis, G.N.; Avery-Kiejda, K.A. Good Cop, Bad Cop: Defining the Roles of Delta40p53 in Cancer and Aging. Cancers 2020, 12, 1659. [Google Scholar] [CrossRef] [PubMed]
- Eiholzer, R.A.; Mehta, S.; Kazantseva, M.; Drummond, C.J.; McKinney, C.; Young, K.; Slater, D.; Morten, B.C.; Avery-Kiejda, K.A.; Lasham, A.; et al. Intronic TP53 Polymorphisms Are Associated with Increased Δ133TP53 Transcript, Immune Infiltration and Cancer Risk. Cancers 2020, 12, 2472. [Google Scholar] [CrossRef] [PubMed]
- Hofstetter, G.; Berger, A.; Fiegl, H.; Slade, N.; Zoric, A.; Holzer, B.; Schuster, E.; Mobus, V.J.; Reimer, D.; Daxenbichler, G.; et al. Alternative splicing of p53 and p73: The novel p53 splice variant p53delta is an independent prognostic marker in ovarian cancer. Oncogene 2010, 29, 1997–2004. [Google Scholar] [CrossRef] [PubMed]
- Hofstetter, G.; Berger, A.; Schuster, E.; Wolf, A.; Hager, G.; Vergote, I.; Cadron, I.; Sehouli, J.; Braicu, E.I.; Mahner, S.; et al. [Delta]133p53 is an independent prognostic marker in p53 mutant advanced serous ovarian cancer. Br. J. Cancer 2011, 105, 1593–1599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofstetter, G.; Berger, A.; Berger, R.; Zoric, A.; Braicu, E.I.; Reimer, D.; Fiegl, H.; Marth, C.; Zeimet, A.G.; Ulmer, H.; et al. The N-terminally truncated p53 isoform Delta40p53 influences prognosis in mucinous ovarian cancer. Int. J. Gynecol. Cancer 2012, 22, 372–379. [Google Scholar] [CrossRef]
- Morten, B.C.; Wong-Brown, M.W.; Scott, R.J.; Avery-Kiejda, K.A. The presence of the intron 3 16 bp duplication polymorphism of p53 (rs17878362) in breast cancer is associated with a low Delta40p53:p53 ratio and better outcome. Carcinogenesis 2016, 37, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Marcel, V.; Perrier, S.; Aoubala, M.; Ageorges, S.; Groves, M.J.; Diot, A.; Fernandes, K.; Tauro, S.; Bourdon, J.C. Delta160p53 is a novel N-terminal p53 isoform encoded by Delta133p53 transcript. FEBS Lett. 2010, 584, 4463–4468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, A.; Stewart, D.; Matlashewski, G. Regulation of human p53 activity and cell localization by alternative splicing. Mol. Cell Biol. 2004, 24, 7987–7997. [Google Scholar] [CrossRef] [Green Version]
- Courtois, S.; Verhaegh, G.; North, S.; Luciani, M.G.; Lassus, P.; Hibner, U.; Oren, M.; Hainaut, P. DeltaN-p53, a natural isoform of p53 lacking the first transactivation domain, counteracts growth suppression by wild-type p53. Oncogene 2002, 21, 6722–6728. [Google Scholar] [CrossRef] [Green Version]
- Ray, P.S.; Grover, R.; Das, S. Two internal ribosome entry sites mediate the translation of p53 isoforms. EMBO Rep. 2006, 7, 404–410. [Google Scholar] [CrossRef] [Green Version]
- Candeias, M.M.; Powell, D.J.; Roubalova, E.; Apcher, S.; Bourougaa, K.; Vojtesek, B.; Bruzzoni-Giovanelli, H.; Fåhraeus, R. Expression of p53 and p53/47 are controlled by alternative mechanisms of messenger RNA translation initiation. Oncogene 2006, 25, 6936–6947. [Google Scholar] [CrossRef] [Green Version]
- Solomon, H.; Bräuning, B.; Fainer, I.; Ben-Nissan, G.; Rabani, S.; Goldfinger, N.; Moscovitz, O.; Shakked, Z.; Rotter, V.; Sharon, M. Post-translational regulation of p53 function through 20S proteasome-mediated cleavage. Cell Death Differ. 2017, 24, 2187–2198. [Google Scholar] [CrossRef]
- Marcel, V.; Petit, I.; Murray-Zmijewski, F.; de Rugy, T.G.; Fernandes, K.; Meuray, V.; Diot, A.; Lane, D.P.; Aberdam, D.; Bourdon, J.C. Diverse p63 and p73 isoforms regulate Delta 133p53 expression through modulation of the internal TP53 promoter activity. Cell Death Differ. 2012, 19, 816–826. [Google Scholar] [CrossRef]
- Bellini, I.; Pitto, L.; Marini, M.G.; Porcu, L.; Moi, P.; Garritano, S.; Boldrini, L.; Rainaldi, G.; Fontanini, G.; Chiarugi, M.; et al. DeltaN133p53 expression levels in relation to haplotypes of the TP53 internal promoter region. Hum. Mutat. 2010, 31, 456–465. [Google Scholar] [CrossRef]
- Leroy, B.; Ballinger, M.L.; Baran-Marszak, F.; Bond, G.L.; Braithwaite, A.; Concin, N.; Donehower, L.A.; El-Deiry, W.S.; Fenaux, P.; Gaidano, G.; et al. Recommended Guidelines for Validation, Quality Control, and Reporting of TP53 Variants in Clinical Practice. Cancer Res. 2017, 77, 1250–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steffens Reinhardt, L.; Groen, K.; Morten, B.C.; Bourdon, J.-C.; Avery-Kiejda, K.A. Cytoplasmic p53β Isoforms Are Associated with Worse Disease-Free Survival in Breast Cancer. Int. J. Mol. Sci. 2022, 23, 6670. [Google Scholar] [PubMed]
- Bankhead, P.; Fernández, J.A.; McArt, D.G.; Boyle, D.P.; Li, G.; Loughrey, M.B.; Irwin, G.W.; Harkin, D.P.; James, J.A.; McQuaid, S.; et al. Integrated tumor identification and automated scoring minimizes pathologist involvement and provides new insights to key biomarkers in breast cancer. Lab. Investig. 2018, 98, 15–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, P.; Du, C.W.; Kwan, M.; Liang, S.X.; Zhang, G.J. The impact of p53 in predicting clinical outcome of breast cancer patients with visceral metastasis. Sci. Rep. 2013, 3, 2246. [Google Scholar] [CrossRef] [Green Version]
- Mayakonda, A.; Lin, D.C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef] [Green Version]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Albarca Aguilera, M.; Meyer, R.; Massouras, A. VarSome: The human genomic variant search engine. Bioinformatics 2018, 35, 1978–1980. [Google Scholar] [CrossRef] [Green Version]
- Baugh, E.H.; Ke, H.; Levine, A.J.; Bonneau, R.A.; Chan, C.S. Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differ. 2018, 25, 154–160. [Google Scholar] [CrossRef]
- Kennedy, M.C.; Lowe, S.W. Mutant p53: It’s not all one and the same. Cell Death Differ. 2022, 29, 983–987. [Google Scholar] [CrossRef]
- Wu, S.; Zhou, J.; Zhang, K.; Chen, H.; Luo, M.; Lu, Y.; Sun, Y.; Chen, Y. Molecular Mechanisms of PALB2 Function and Its Role in Breast Cancer Management. Front. Oncol. 2020, 10, 301. [Google Scholar] [CrossRef] [Green Version]
- Mehrgou, A.; Akouchekian, M. The importance of BRCA1 and BRCA2 genes mutations in breast cancer development. Med. J. Islam. Repub. Iran 2016, 30, 369. [Google Scholar] [PubMed]
- Nik-Zainal, S.; Davies, H.; Staaf, J.; Ramakrishna, M.; Glodzik, D.; Zou, X.; Martincorena, I.; Alexandrov, L.B.; Martin, S.; Wedge, D.C.; et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016, 534, 47–54. [Google Scholar] [CrossRef] [Green Version]
- Holstege, H.; Joosse, S.A.; van Oostrom, C.T.; Nederlof, P.M.; de Vries, A.; Jonkers, J. High incidence of protein-truncating TP53 mutations in BRCA1-related breast cancer. Cancer Res. 2009, 69, 3625–3633. [Google Scholar] [CrossRef] [Green Version]
- Mahdi, A.H.; Huo, Y.; Chen, Y.; Selenica, P.; Sharma, A.; Merritt, E.; Barnard, N.; Chan, C.; Ganesan, S.; Reis-Filho, J.S.; et al. Loss of the BRCA1-PALB2 interaction accelerates p53-associated tumor development in mice. Genes Dis. 2022, 9, 807–813. [Google Scholar] [CrossRef]
- Vieler, M.; Sanyal, S. p53 Isoforms and Their Implications in Cancer. Cancers 2018, 10, 288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehta, S.; Campbell, H.; Drummond, C.J.; Li, K.; Murray, K.; Slatter, T.; Bourdon, J.-C.; Braithwaite, A.W. Adaptive homeostasis and the p53 isoform network. EMBO Rep. 2021, 22, e53085. [Google Scholar] [CrossRef] [PubMed]
- Steffens Reinhardt, L.; Groen, K.; Newton, C.; Avery-Kiejda, K.A. The role of truncated p53 isoforms in the DNA damage response. Biochim. Et Biophys. Acta (BBA)-Rev. Cancer 2023, 1878, 188882. [Google Scholar] [CrossRef]
- Candeias, M.M.; Hagiwara, M.; Matsuda, M. Cancer-specific mutations in p53 induce the translation of Δ160p53 promoting tumorigenesis. EMBO Rep. 2016, 17, 1542–1551. [Google Scholar] [CrossRef]
- Gadea, G.; Arsic, N.; Fernandes, K.; Diot, A.; Joruiz, S.M.; Abdallah, S.; Meuray, V.; Vinot, S.; Anguille, C.; Remenyi, J.; et al. TP53 drives invasion through expression of its Δ133p53β variant. eLife 2016, 5, e14734. [Google Scholar] [CrossRef]
- Rainville, I.; Hatcher, S.; Rosenthal, E.; Larson, K.; Bernhisel, R.; Meek, S.; Gorringe, H.; Mundt, E.; Manley, S. High risk of breast cancer in women with biallelic pathogenic variants in CHEK2. Breast Cancer Res. Treat. 2020, 180, 503–509. [Google Scholar] [CrossRef] [Green Version]
- Mathe, A.; Wong-Brown, M.; Morten, B.; Forbes, J.F.; Braye, S.G.; Avery-Kiejda, K.A.; Scott, R.J. Novel genes associated with lymph node metastasis in triple negative breast cancer. Sci. Rep. 2015, 5, 15832. [Google Scholar] [CrossRef] [Green Version]
- Kandoth, J.G.C.; Mattioni, M.; Struck, A.; Boursin, Y.; Penson, A.; Chavan, S. skcc/vcf2maf: vcf2maf v1.6.16 (v1.6.16). Available online: https://zenodo.org/record/1185418#.YxF6F3ZByUk (accessed on 5 September 2022).
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng, P.C.; Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef] [Green Version]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2017, 46, D1062–D1067. [Google Scholar] [CrossRef] [Green Version]
- IARC. TP53 Database. Available online: https://tp53.isb-cgc.org/ (accessed on 23 September 2022).
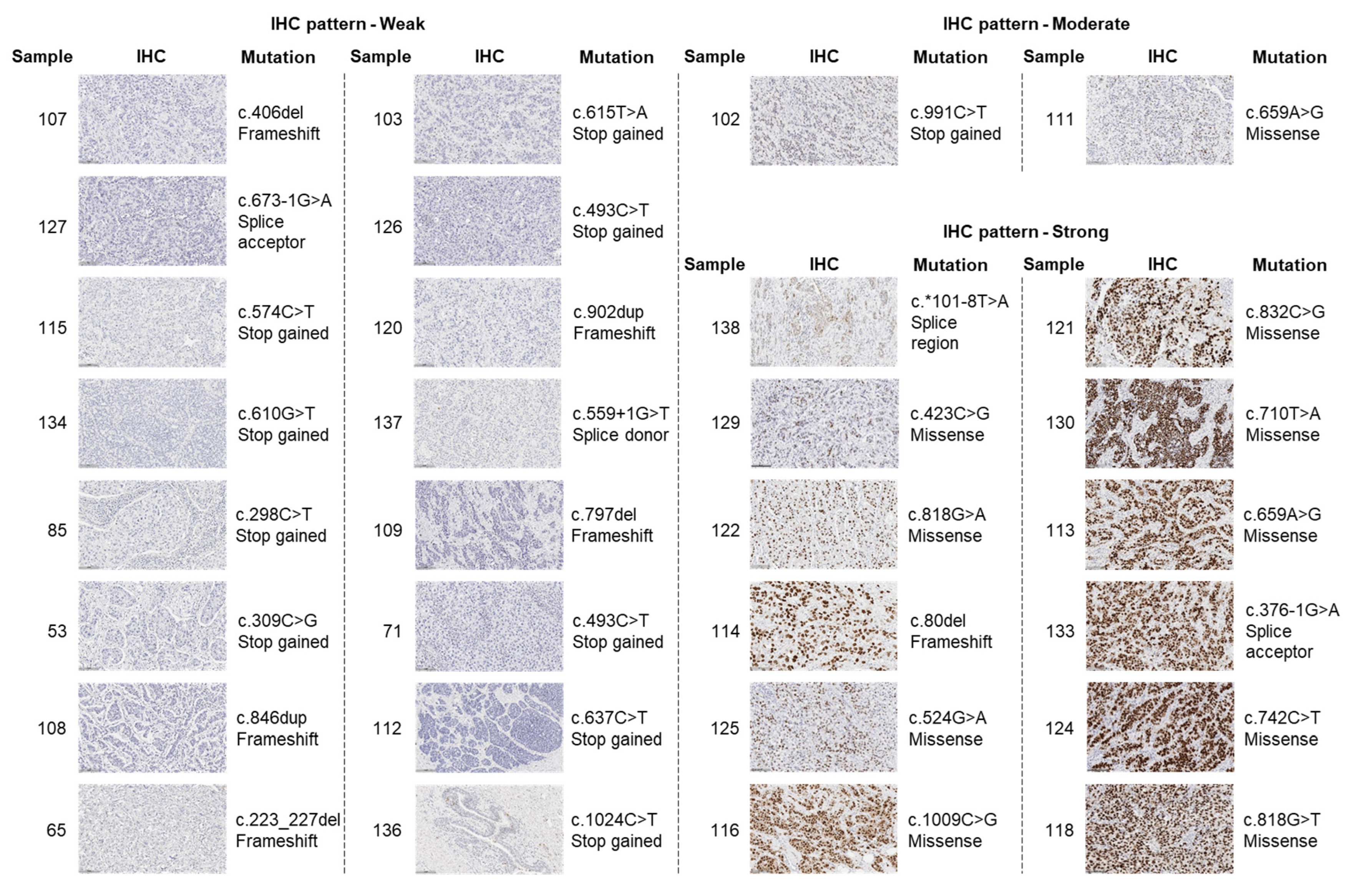
| Alteration | Tumours | IHC Pattern | ||
|---|---|---|---|---|
| Variant | Outcomes | |||
| c.406del | deletion | - | 107 | Weak |
| c.673-1G>A | SNV | RNA splicing disruption and likely results in an absent or disrupted protein product. | 127 | |
| c.574C>T | SNV | Expected to result in loss of function by premature protein truncation or nonsense-mediated mRNA decay. | 115 77 (IHC NA) | |
| c.610G>T | SNV | - | 134 | |
| c.298C>T | SNV | Predicted to cause nonsense-mediated decay and result in an absent protein. | 85 | |
| c.309C>G | SNV | Expected to result in loss of function by premature protein truncation or nonsense-mediated mRNA decay. This alteration is deficient at growth suppression. | 53 | |
| c.846dup | insertion | - | 108 | |
| c.223_227del | deletion | Novel variant. | 65 | |
| c.615T>A | SNV | Interpreted as a disease-causing mutation. | 103 | |
| c.493C>T | Expected to result in loss of function by premature protein truncation or nonsense-mediated mRNA decay. | 126 71 | ||
| c.902dup | insertion | Expected to result in an absent or disrupted protein product. | 120 | |
| c.559+1G>T | splice donor | Expected to result in an absent or non-functional TP53 protein. | 137 | |
| c.797del | deletion | - | 109 | |
| c.637C>T | SNV | Predicted to cause a truncated or absent TP53 protein due to nonsense mediated decay. | 112 | |
| c.1024C>T | SNV | Predicted to cause a truncation of the encoded protein or absence of the protein due to nonsense mediated decay. | 136 131 (IHC NA) | |
| c.991C>T | SNV | Prediction tools suggest that this variant may not impact RNA splicing. | 102 | Moderate |
| c.659A>G | SNV | Computational prediction suggests that this variant may have deleterious impact on protein structure and function. | 111 | |
| c.*101-8T>A | SNV | - | 138 | Strong |
| c.423C>G | SNV | Reported to have loss of transactivation capacity in yeast-based assays and is predicted to affect several p53 isoforms. | 129 | |
| c.818G>A | SNV | Computational prediction suggests that this variant may have deleterious impact on protein structure and function. | 122 | |
| c.80del | deletion | Expected to result in an absent or disrupted protein product. | 114 | |
| c.524G>A | SNV | A well-characterised mutation “hotspot” in the functionally critical DNA-binding domain. | 125 | |
| c.1009C>G | SNV | - | 116 | |
| c.832C>G | SNV | Reported to have loss of transactivation capacity in functional studies in yeast and human cell lines. | 121 | |
| c.710T>A | SNV | - | 130 | |
| c.659A>G | SNV | Computational prediction suggests that this variant may have deleterious impact on protein structure and function. | 113 | |
| c.376-1G>A | SNV | Expected to disrupt RNA splicing and lead to a loss of protein function. | 133 | |
| c.742C>T | SNV | Computational prediction suggests that this variant may have deleterious impact on protein structure and function. | 124 | |
| c.818G>T | SNV | A well-characterised mutation “hotspot” located within the functionally critical DNA-binding domain. | 118 | |
| c.1007del | deletion | - | 91 | NA |
| c.282del | deletion | - | 135 | |
| c.341dup | insertion | - | 18 | |
| c.602T>A | SNV | - | 105 | |
| c.681dup | insertion | - | 128 | |
| c.760_761del | deletion | - | 106 | |
| c.994-1G>A | SNV | This variant induces altered splicing and may result in an absent or disrupted protein product. | 40 | |
| Gene | Variant | dbSNP | Impact | N * | p53 Isoform Expression ** |
|---|---|---|---|---|---|
| BRCA1 | p.Ile562Thr | rs1555591375 | Moderate | 1 | High TAp53 and Δ160p53 |
| p.Ser768Arg | novel | Moderate | 1 | ||
| p.Ala194Val | novel | Moderate | 1 | ||
| p.Asp295His | novel | Moderate | 1 | ||
| p.Glu881Ter | rs397508988 | High | 1 | NA | |
| p.Glu1570IlefsTer2 | novel | High | 1 | ||
| p.Asp219His | rs273902779 | Moderate | 1 | ||
| CHEK2 | p.Thr421Pro | novel | Moderate | 130 | NA (n = 35) High p53β (n = 50) Low p53β (n = 45) |
| p.Val146Phe | rs907433465 | Moderate | 1 | High p53β | |
| p.Val25Ile | rs142243299 | Moderate | 1 | NA | |
| PALB2 | p.Tyr551Asn | novel | Moderate | 13 | NA (n = 2) High TAp53 (n = 3) Low TAp53 (n = 5) |
| p.Thr721Ile | novel | Moderate | 1 | High TAp53 | |
| p.Thr193AsnfsTer2 | rs875989790 | High | 1 | ||
| p.Ser779Ter | rs764509489 | High | 1 | Low TAp53 | |
| p.Leu100Phe | rs61756147 | Moderate | 1 | ||
| p.Leu1143ThrfsTer14 | rs587776425 | High | 1 | NA | |
| p.Glu384Ter | - | High | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Steffens Reinhardt, L.; Groen, K.; Xavier, A.; Avery-Kiejda, K.A. p53 Dysregulation in Breast Cancer: Insights on Mutations in the TP53 Network and p53 Isoform Expression. Int. J. Mol. Sci. 2023, 24, 10078. https://doi.org/10.3390/ijms241210078
Steffens Reinhardt L, Groen K, Xavier A, Avery-Kiejda KA. p53 Dysregulation in Breast Cancer: Insights on Mutations in the TP53 Network and p53 Isoform Expression. International Journal of Molecular Sciences. 2023; 24(12):10078. https://doi.org/10.3390/ijms241210078
Chicago/Turabian StyleSteffens Reinhardt, Luiza, Kira Groen, Alexandre Xavier, and Kelly A. Avery-Kiejda. 2023. "p53 Dysregulation in Breast Cancer: Insights on Mutations in the TP53 Network and p53 Isoform Expression" International Journal of Molecular Sciences 24, no. 12: 10078. https://doi.org/10.3390/ijms241210078





