Delta/Notch-like Epidermal Growth Factor-Related Receptor (DNER), a Potential Prognostic Marker of Gastric Cancer Regulates Cell Survival and Cell Cycle Progression
Abstract
1. Introduction
2. Results
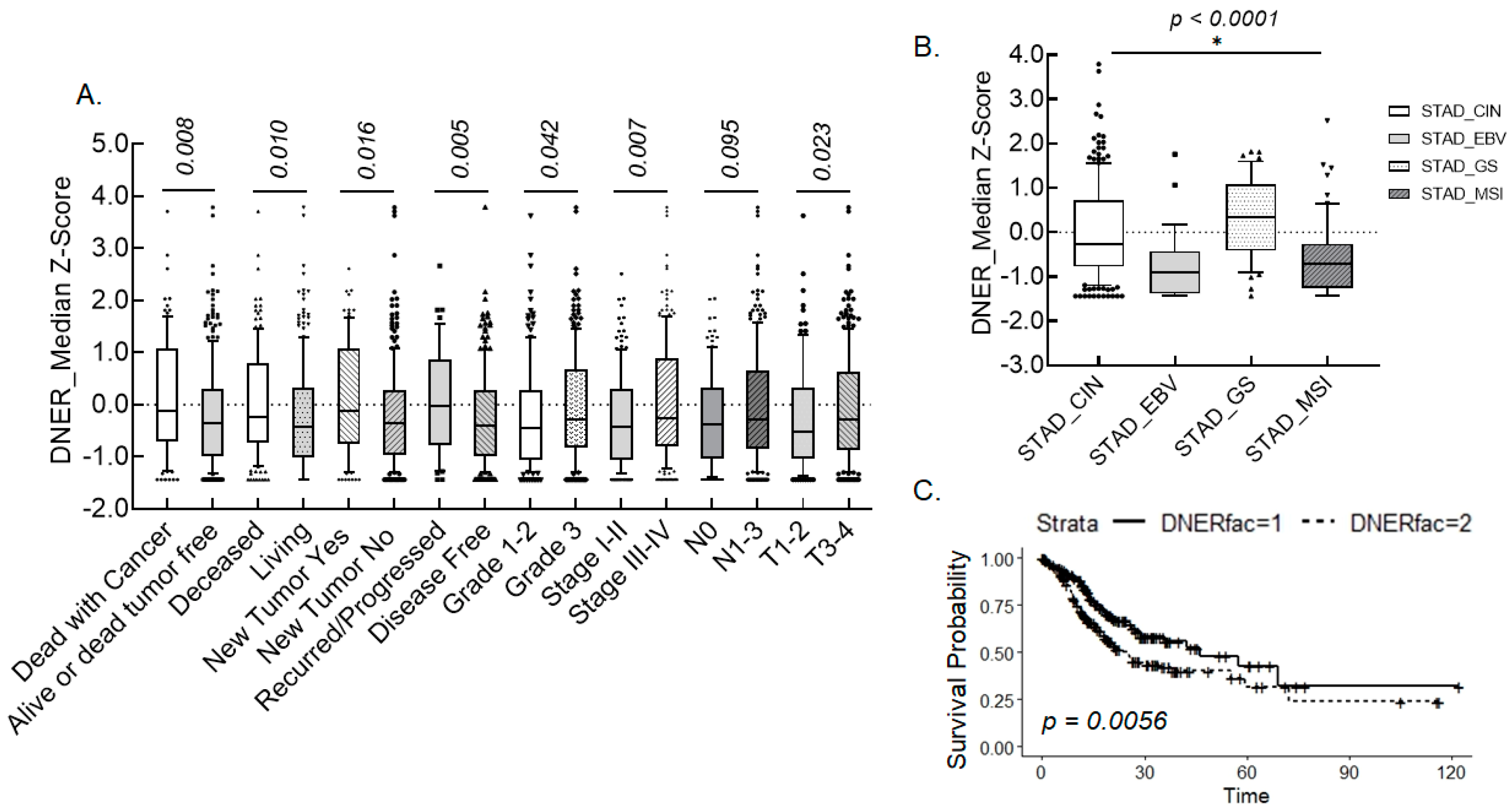
2.1. Pathological Association of DNER Expression in Gastric Cancer
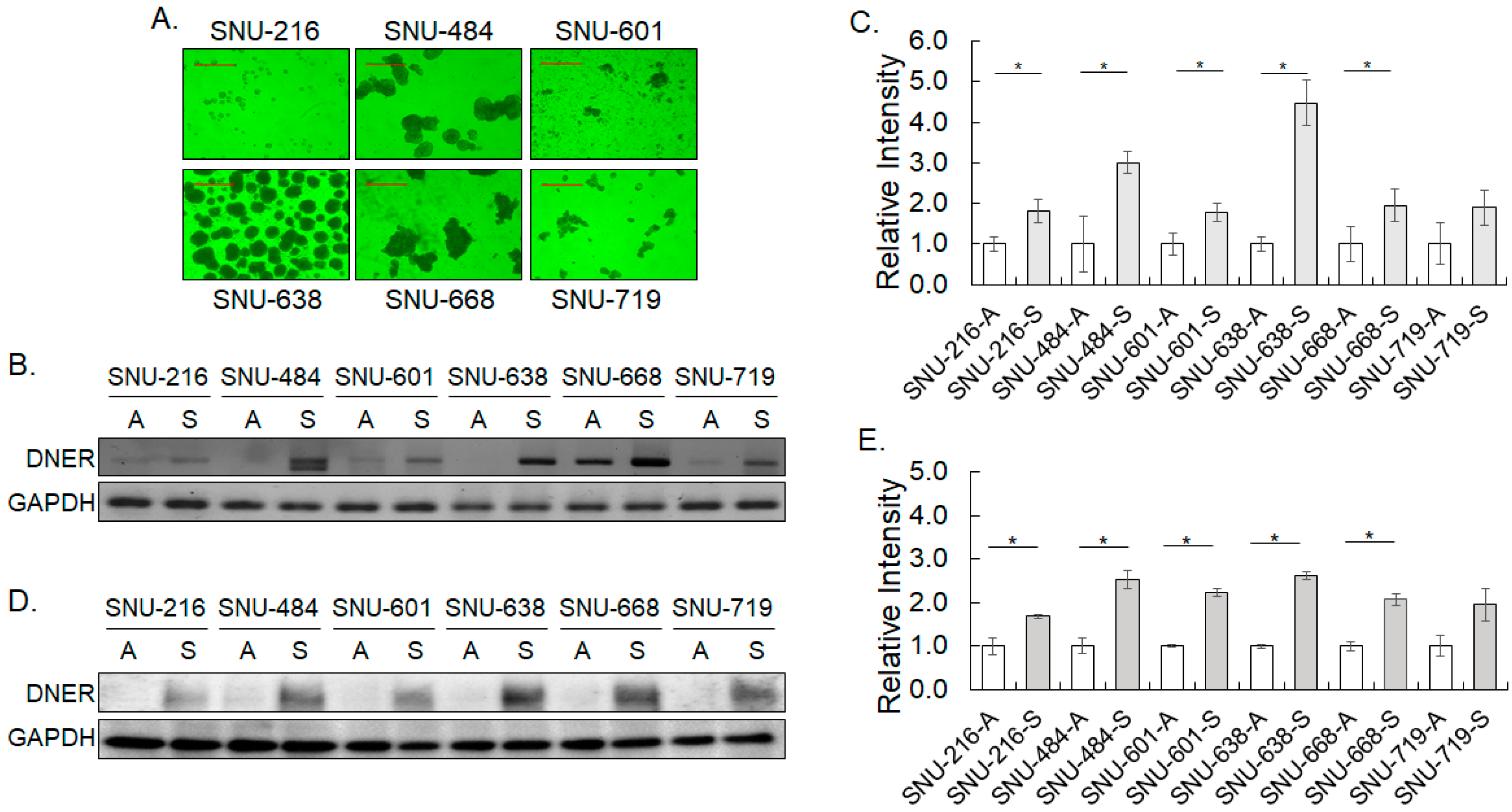
2.2. DNER Expression Increased in the Tumor Spheroid Culture of Gastric Cancer Cells
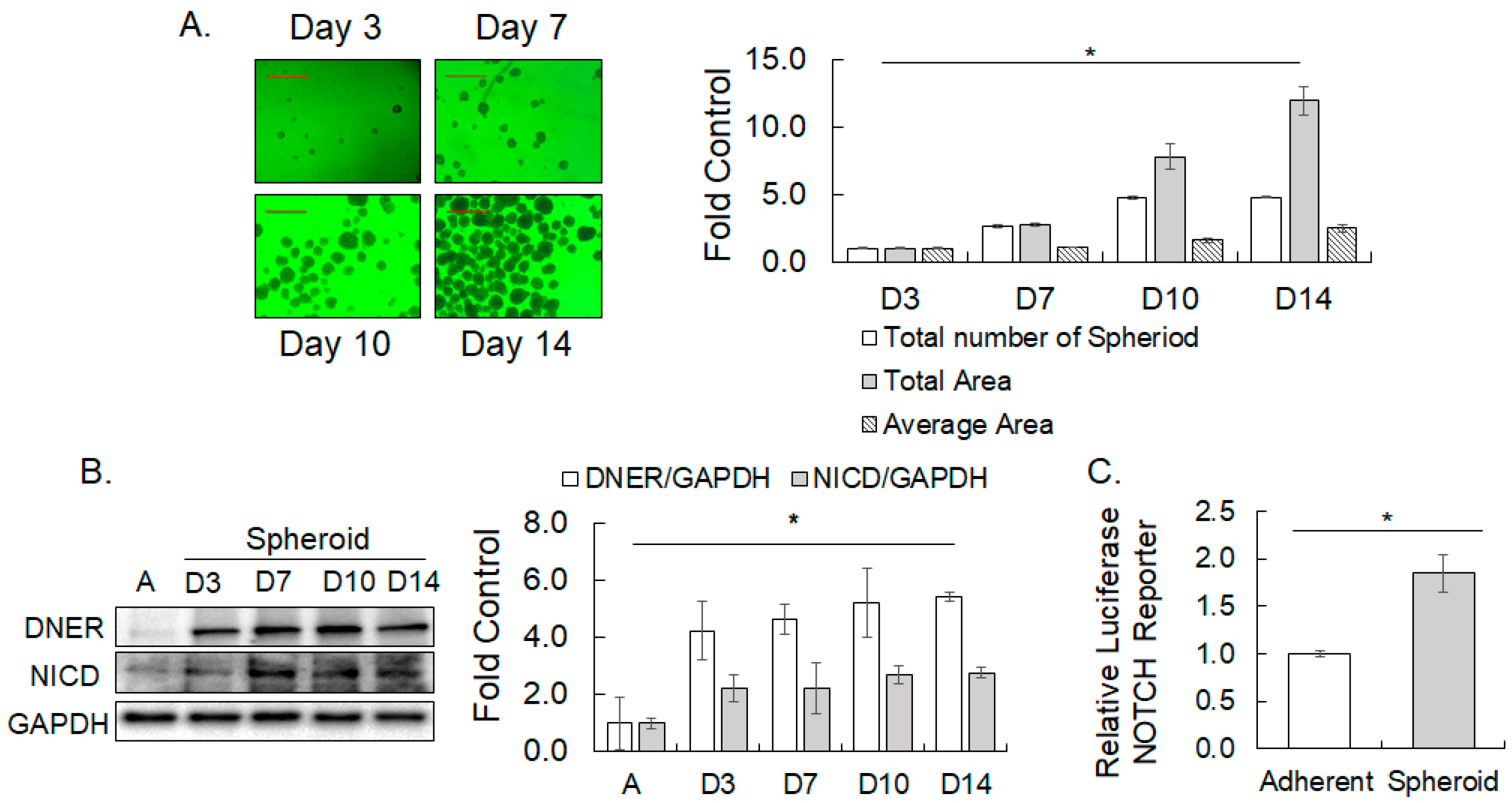
2.3. DNER and NICD Expression Increased in Spheroid Formation
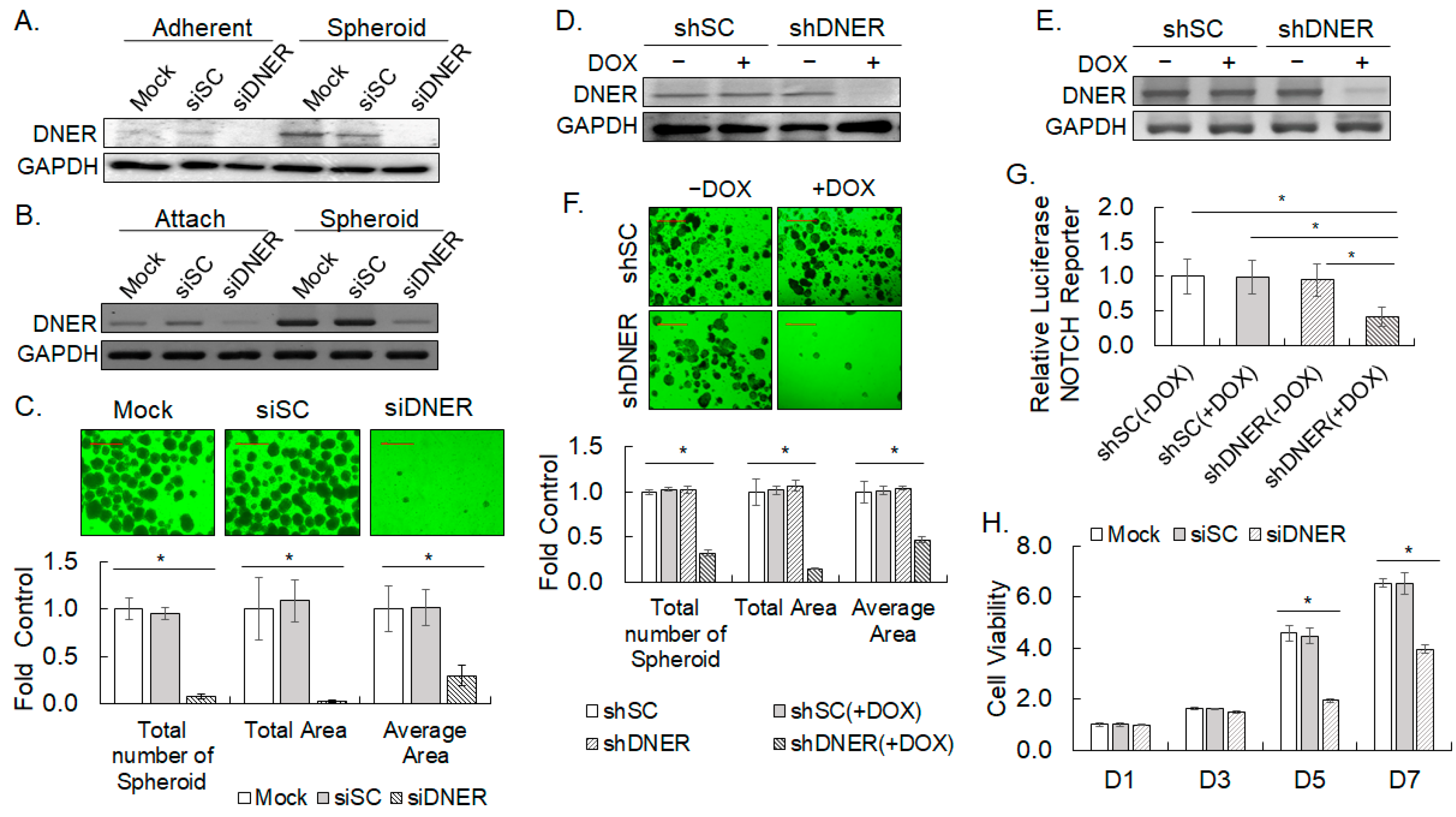
2.4. Silencing DNER Expression Reduced Spheroid Formation and Cell Viability
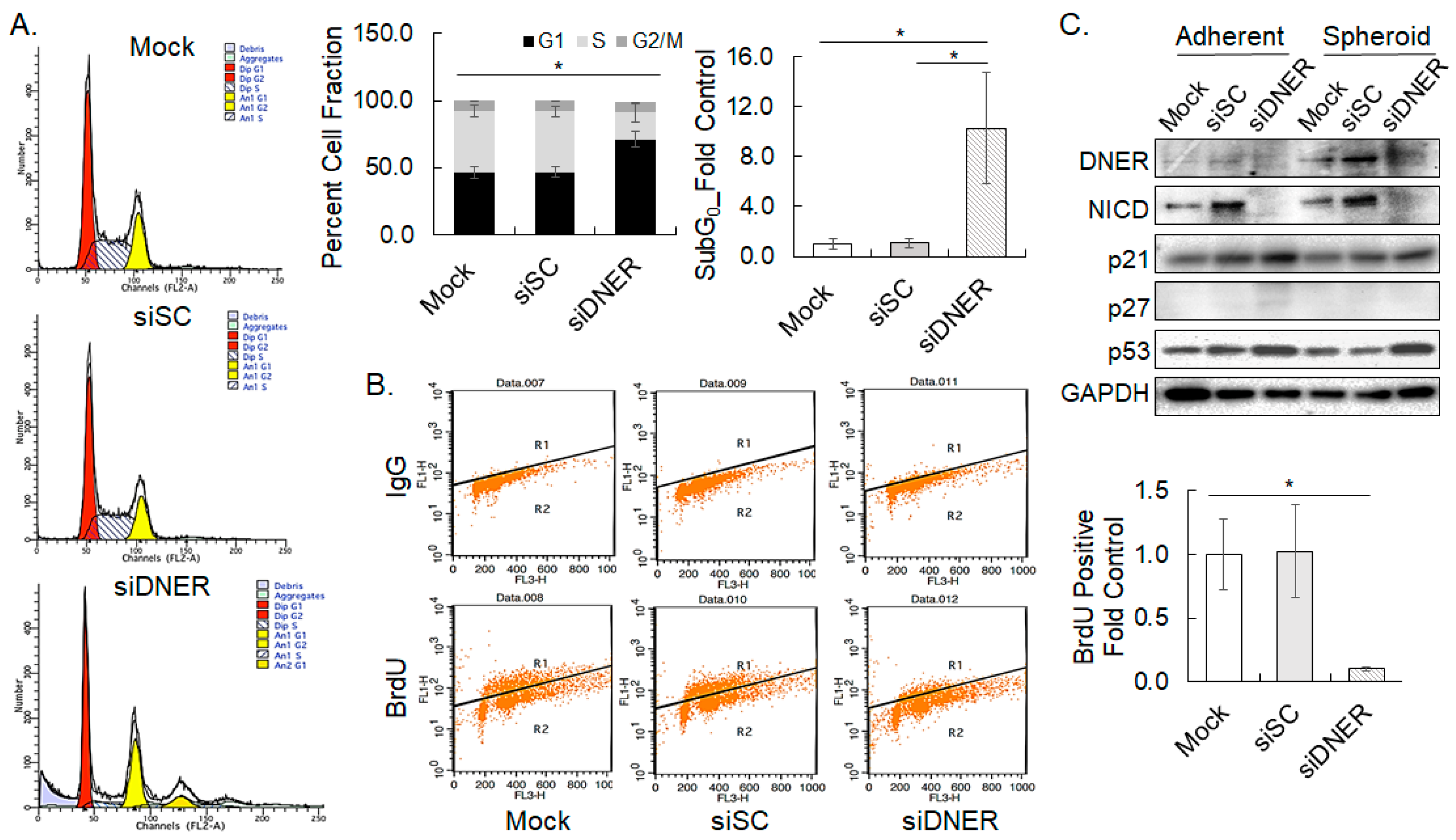
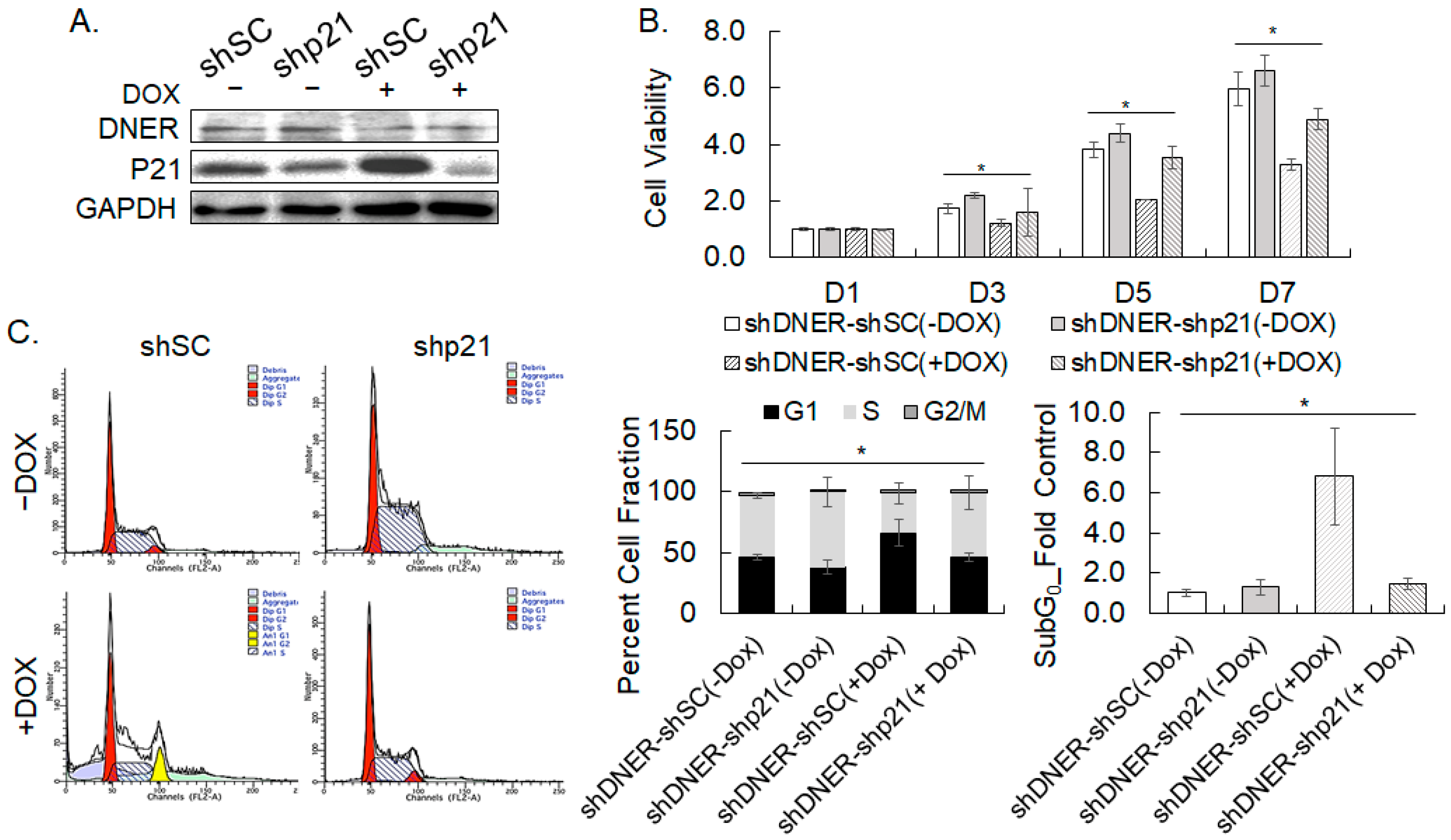
2.5. Silencing DNER Expression Inhibited Cell Cycle Progression at the G1/S Phase
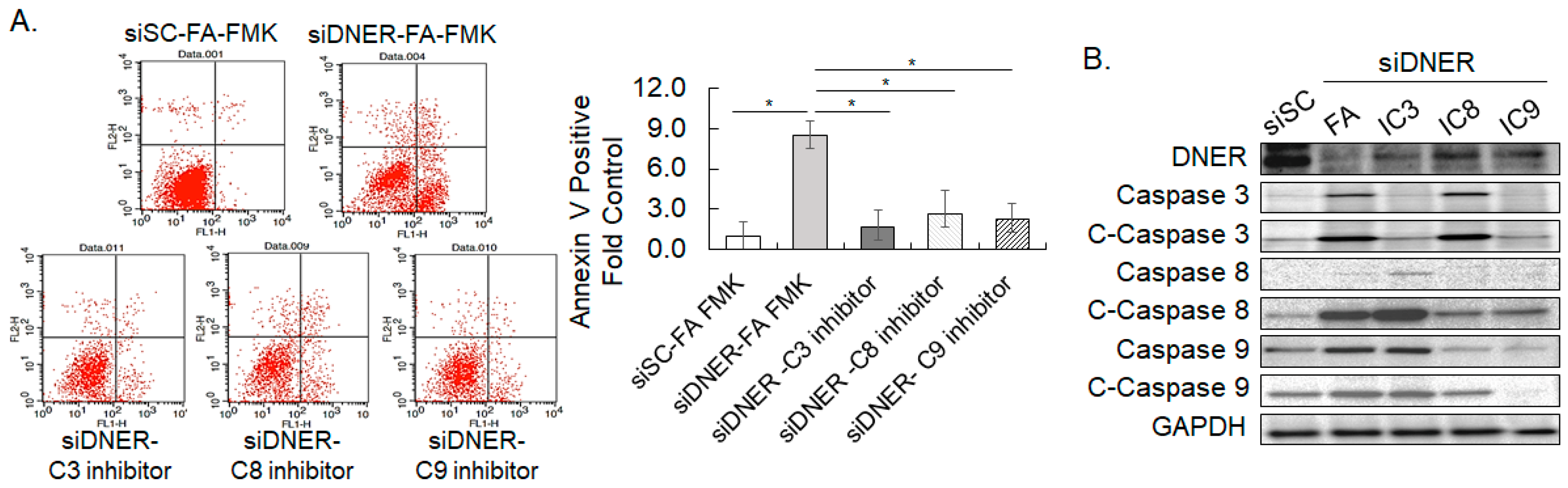
2.6. Silencing DNER Expression Increased Apoptotic Cell Death
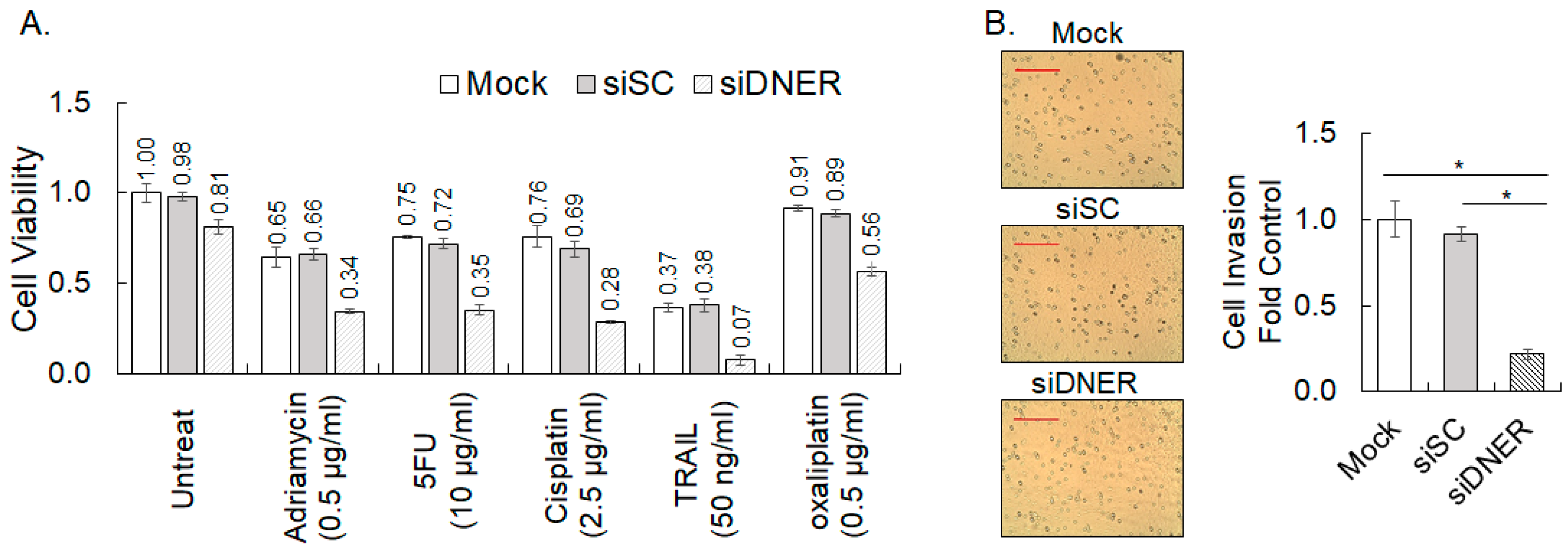
2.7. Silencing DNER Expression Enhanced Chemosensitivity and Reduced Invasion of Gastric Cancer Cells
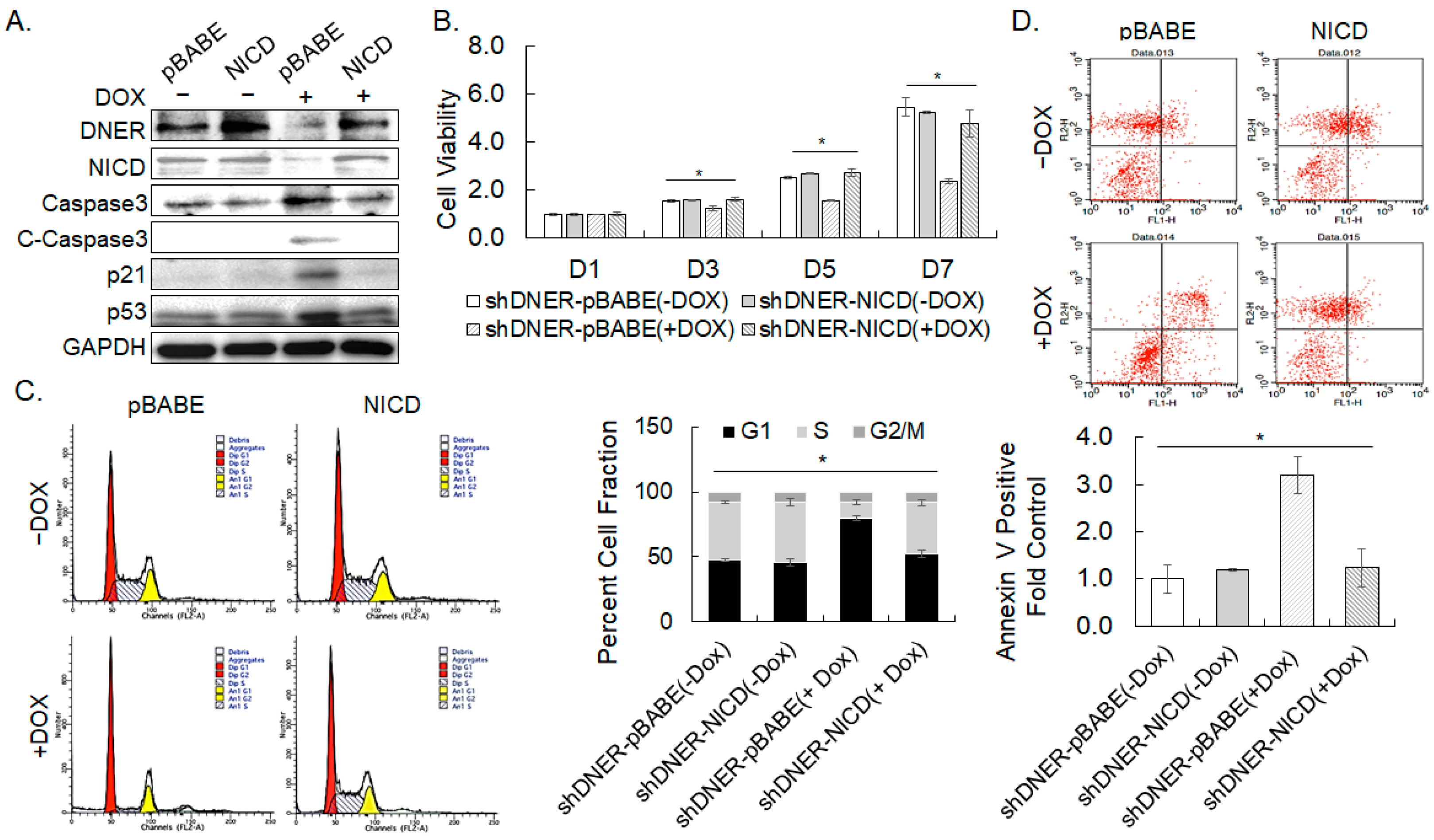
2.8. Overexpression of NICD Rescued DNER-Silenced Cells from Growth Arrest and Apoptosis
2.9. Depletion of p21cip/waf Rescued DNER-Silenced Cells from Cell Cycle Arrest
2.10. Silencing p53 Decreased the Apoptosis of DNER-Silenced Cells
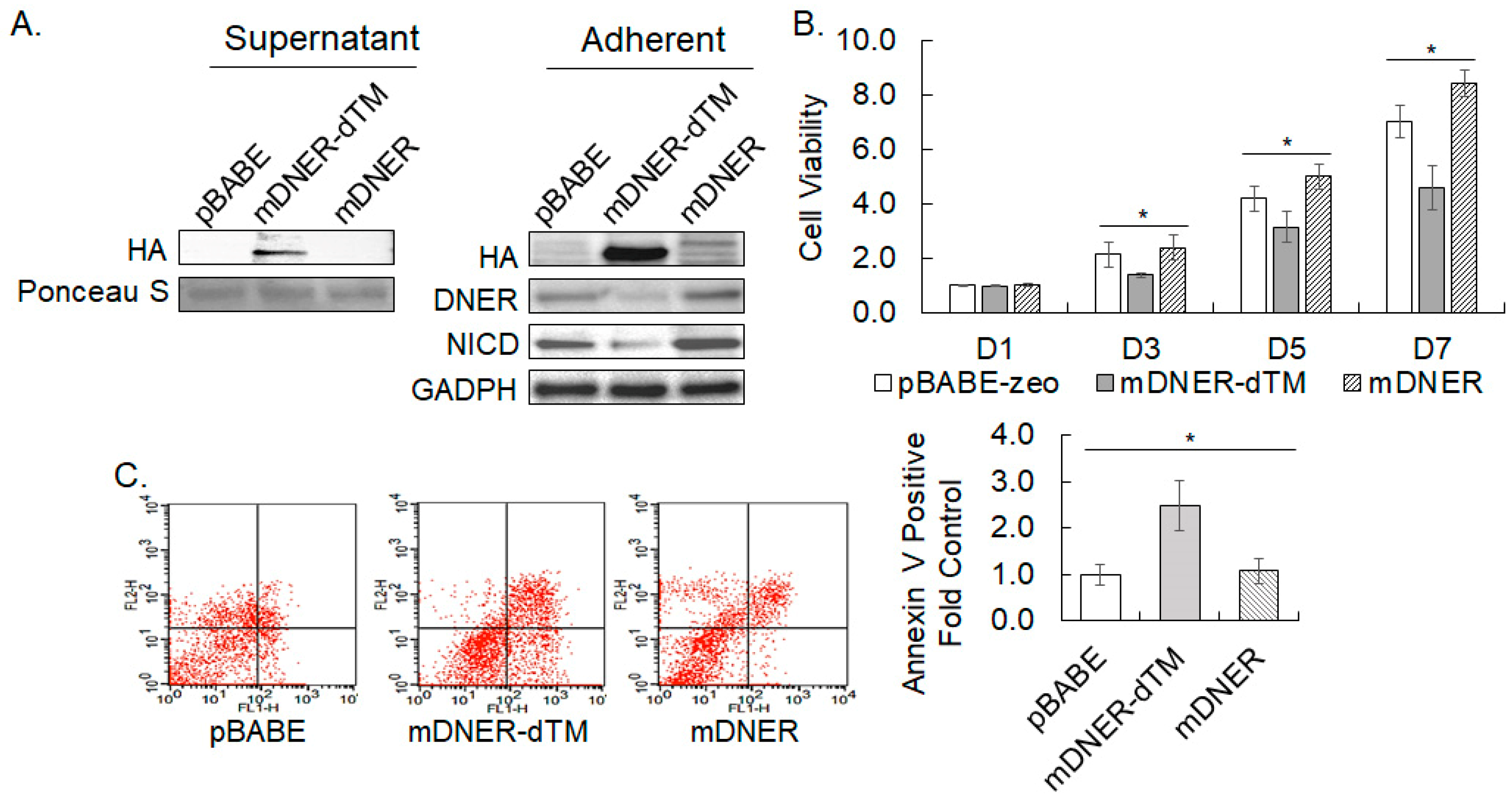
2.11. Secreted Mouse DNER Interfered with Notch Signaling and Causes Reduction in Cell Viability
2.12. TGF-β Signaling Pathway Modulated Spheroid Formation of Gastric Cancer by Targeting DNER
3. Discussion
3.1. Association of DNER Expression with Clinicopathology of Gastric Cancer
3.2. A Role of DNER in Regulation of Tumor Spheroid Formation, Cell Viability, Cell Proliferation and Apoptosis
3.3. Regulation of DNER Expression by the TGF-β Signaling
3.4. DNER in the Notch Signaling
4. Materials and Methods
4.1. Gastric Cancer Cell Lines and Cell Culture
4.2. Spheroid Formation Assay
4.3. Luciferase Assay
4.4. Invasion Assay
4.5. Cell Viability Assay
4.6. Western Blot Analysis
4.7. Flow Cytometry
4.8. Knockdown of Gene Expression by Lentiviral Transduction or RNA Interference
4.9. Overexpression of mDNER, mDNER-dTM, and NICD by Retroviral Transduction
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Artavanis-Tsakonas, S.; Rand, M.D.; Lake, R.J. Notch Signaling: Cell Fate Control and Signal Integration in Development. Science 1999, 284, 770–776. [Google Scholar] [CrossRef] [PubMed]
- Blaumueller, C.M.; Qi, H.; Zagouras, P.; Artavanis-Tsakonas, S. Intracellular Cleavage of Notch Leads to a Heterodimeric Receptor on the Plasma Membrane. Cell 1997, 90, 281–291. [Google Scholar] [CrossRef]
- Logeat, F.; Bessia, C.; Brou, C.; LeBail, O.; Jarriault, S.; Seidah, N.G.; Israël, A. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl. Acad. Sci. USA 1998, 95, 8108–8112. [Google Scholar] [CrossRef]
- Bray, S. Notch signalling in context. Nat. Rev. Mol. Cell Biol. 2016, 17, 722–735. [Google Scholar] [CrossRef] [PubMed]
- Majumder, S.; Crabtree, J.S.; Golde, T.E.; Minter, L.M.; Osborne, B.A.; Miele, L. Targeting Notch in oncology: The path forward. Nat. Rev. Drug Discov. 2021, 20, 125–144. [Google Scholar] [CrossRef]
- Kumar, S.; Nandi, A.; Singh, S.; Regulapati, R.; Li, N.; Tobias, J.W.; Siebel, C.W.; Blanco, M.A.; Klein-Szanto, A.J.; Lengner, C.; et al. Dll1(+) quiescent tumor stem cells drive chemoresistance in breast cancer through NF-kappaB survival pathway. Nat. Commun. 2021, 12, 432. [Google Scholar] [CrossRef]
- Nowell, C.S.; Radtke, F. Notch as a tumour suppressor. Nat. Rev. Cancer 2017, 17, 145–159. [Google Scholar] [CrossRef]
- Roper, N.; Velez, M.J.; Chiappori, A.; Kim, Y.S.; Wei, J.S.; Sindiri, S.; Takahashi, N.; Mulford, D.; Kumar, S.; Ylaya, K.; et al. Notch signaling and efficacy of PD-1/PD-L1 blockade in relapsed small cell lung cancer. Nat. Commun. 2021, 12, 3880. [Google Scholar] [CrossRef]
- Cheng, P.; Zlobin, A.; Volgina, V.; Gottipati, S.; Osborne, B.; Simel, E.J.; Miele, L.; Gabrilovich, D.I. Notch-1 regulates NF-kappaB activity in hemopoietic progenitor cells. J. Immunol. 2001, 167, 4458–4467. [Google Scholar] [CrossRef]
- Ronchini, C.; Capobianco, A.J. Induction of cyclin D1 transcription and CDK2 activity by Notch(ic): Implication for cell cycle disruption in transformation by Notch(ic). Mol. Cell Biol. 2001, 21, 5925–5934. [Google Scholar] [CrossRef] [PubMed]
- Weng, A.P.; Millholland, J.M.; Yashiro-Ohtani, Y.; Arcangeli, M.L.; Lau, A.; Wai, C.; del Bianco, C.; Rodriguez, C.G.; Sai, H.; Tobias, J.; et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006, 20, 2096–2109. [Google Scholar] [CrossRef]
- D’Angelo, R.C.; Ouzounova, M.; Davis, A.; Choi, D.; Tchuenkam, S.M.; Kim, G.; Luther, T.; Quraishi, A.A.; Senbabaoglu, Y.; Conley, S.J.; et al. Notch Reporter Activity in Breast Cancer Cell Lines Identifies a Subset of Cells with Stem Cell Activity. Mol. Cancer Ther. 2015, 14, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Ahmad, A.; Li, Y.; Azmi, A.S.; Miele, L.; Sarkar, F.H. Targeting notch to eradicate pancreatic cancer stem cells for cancer therapy. Anticancer Res. 2011, 31, 1105–1113. [Google Scholar] [PubMed]
- Hibdon, E.S.; Razumilava, N.; Keeley, T.M.; Wong, G.; Solanki, S.; Shah, Y.M.; Samuelson, L.C. Notch and mTOR Signaling Pathways Promote Human Gastric Cancer Cell Proliferation. Neoplasia 2019, 21, 702–712. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xie, F.; Zhang, H.; Wu, X.; Ji, G.; Li, J.; Hong, L. Effects of mRNA expression of five Notch ligands on prognosis of gastric carcinoma. Sci. Rep. 2022, 12, 15141. [Google Scholar] [CrossRef]
- Yao, Y.; Ni, Y.; Zhang, J.; Wang, H.; Shao, S. The role of Notch signaling in gastric carcinoma: Molecular pathogenesis and novel therapeutic targets. Oncotarget 2017, 8, 53839–53853. [Google Scholar] [CrossRef]
- Ma, H.; Li, N.; Mo, Z. Elevated Notch-1 expression promotes the lymph node metastasis of gastric cancer and the Notch-1-PTEN-ERK1/2 signalling axis promotes the progression of gastric cancer. Cytokine 2022, 159, 156013. [Google Scholar] [CrossRef]
- Yeh, T.S.; Wu, C.W.; Hsu, K.W.; Liao, W.J.; Yang, M.C.; Li, A.F.Y.; Wang, A.M.; Kuo, M.L.; Chi, C.W. The activated Notch1 signal pathway is associated with gastric cancer progression through cyclooxygen-ase-2. Cancer Res. 2009, 69, 5039–5048. [Google Scholar] [CrossRef]
- Eiraku, M.; Tohgo, A.; Ono, K.; Kaneko, M.; Fujishima, K.; Hirano, T.; Kengaku, M. DNER acts as a neuron-specific Notch ligand during Bergmann glial development. Nat. Neurosci. 2005, 8, 873–880. [Google Scholar] [CrossRef]
- Greene, M.; Lai, Y.; Pajcini, K.; Bailis, W.; Pear, W.S.; Lancaster, E. Delta/Notch-Like EGF-Related Receptor (DNER) Is Not a Notch Ligand. PLoS ONE 2016, 11, e0161157. [Google Scholar] [CrossRef] [PubMed]
- Eiraku, M.; Hirata, Y.; Takeshima, H.; Hirano, T.; Kengaku, M. Delta/notch-like epidermal growth factor (EGF)-related receptor, a novel EGF-like repeat-containing pro-tein targeted to dendrites of developing and adult central nervous system neurons. J. Biol. Chem. 2002, 277, 25400–25407. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Wang, X.; Zhang, X.; Zhang, X.; Jiang, H. DNER modulates the length, polarity and synaptogenesis of spiral ganglion neurons via the Notch signaling pathway. Mol. Med. Rep. 2018, 17, 2357–2365. [Google Scholar] [CrossRef] [PubMed]
- Campana, I.G.; Silva, G.D. Anti-Tr/DNER Antibody–Associated Cerebellar Ataxia: A Systematic Review. Cerebellum 2022, 21, 1085–1091. [Google Scholar] [CrossRef]
- Wang, L.; Wu, Q.; Li, Z.; Sun, S.; Yuan, J.; Li, J.; Zhang, Y.; Yu, D.; Wang, C.; Sun, S. Delta/notch-like epidermal growth factor-related receptor promotes stemness to facilitate breast cancer progression. Cell Signal. 2019, 63, 109389. [Google Scholar] [CrossRef]
- Wang, L.; Wu, Q.; Zhu, S.; Li, Z.; Yuan, J.; Yu, D.; Xu, Z.; Li, J.; Sun, S.; Wang, C. Delta/notch-like epidermal growth factor-related receptor (DNER) orchestrates stemness and cancer progression in prostate cancer. Am. J. Transl. Res. 2017, 9, 5031–5039. [Google Scholar]
- Liang, Y.; Luo, H.; Zhang, H.; Dong, Y.; Bao, Y. Oncogene Delta/Notch-Like EGF-Related Receptor Promotes Cell Proliferation, Invasion, and Migration in Hepatocellular Carcinoma and Predicts a Poor Prognosis. Cancer Biother. Radiopharm. 2018, 33, 380–386. [Google Scholar] [CrossRef]
- Tao, H.; Wang, C.; Zhu, Y.; Lu, C.; Zhou, X. Role of Delta/Notch-like epidermal growth factor-related receptor in gastric cancer patients and cells and its clinical significance. Anti-Cancer Drugs 2022, 33, 1175–1181. [Google Scholar] [CrossRef]
- Wang, H.; Shen, L.; Li, Y.; Lv, J. Integrated characterisation of cancer genes identifies key molecular biomarkers in stomach adenocarcinoma. J. Clin. Pathol. 2020, 73, 579–586. [Google Scholar] [CrossRef]
- Engeland, K. Cell cycle regulation: p53-p21-RB signaling. Cell Death Differ. 2022, 29, 946–960. [Google Scholar] [CrossRef]
- Hernandez Borrero, L.J.; El-Deiry, W.S. Tumor suppressor p53: Biology, signaling pathways, and therapeutic targeting. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188556. [Google Scholar] [CrossRef] [PubMed]
- Mungamuri, S.K.; Yang, X.; Thor, A.D.; Somasundaram, K. Survival signaling by Notch1: Mammalian target of rapamycin (mTOR)-dependent inhibition of p53. Cancer Res. 2006, 66, 4715–4724. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-W.; Yang, S.-T.; Chien, M.-H.; Hua, K.-T.; Wu, C.-J.; Hsiao, S.M.; Lin, H.; Hsiao, M.; Su, J.-L.; Wei, L.-H. The STAT3-miRNA-92-Wnt Signaling Pathway Regulates Spheroid Formation and Malignant Progression in Ovarian Cancer. Cancer Res. 2017, 77, 1955–1967. [Google Scholar] [CrossRef] [PubMed]
- Rafehi, S.; Valdes, Y.R.; Bertrand, M.; McGee, J.; Prefontaine, M.; Sugimoto, A.; DiMattia, G.E.; Shepherd, T.G. TGFbeta signaling regulates epithelial-mesenchymal plasticity in ovarian cancer ascites-derived spheroids. Endocr. Relat. Cancer 2016, 23, 147–159. [Google Scholar] [CrossRef]
- Small, D.; Kovalenko, D.; Soldi, R.; Mandinova, A.; Kolev, V.; Trifonova, R.; Bagala, C.; Kacer, D.; Battelli, C.; Liaw, L.; et al. Notch Activation Suppresses Fibroblast Growth Factor-dependent Cellular Transformation. J. Biol. Chem. 2003, 278, 16405–16413. [Google Scholar] [CrossRef]
- Vorwald, C.E.; Joshee, S.; Leach, J.K. Spatial localization of endothelial cells in heterotypic spheroids influences Notch signaling. J. Mol. Med. 2020, 98, 425–435. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Z.; Wu, Q.; Li, C.; Li, J.; Zhang, Y.; Wang, C.; Sun, S.; Sun, S. DNER promotes epithelial-mesenchymal transition and prevents chemosensitivity through the Wnt/beta-catenin pathway in breast cancer. Cell Death Dis. 2020, 11, 642. [Google Scholar] [CrossRef]
- To, H.T.N.; Le, Q.A.; Bui, H.T.T.; Park, J.-H.; Kang, D. Modulation of Spheroid Forming Capacity and TRAIL Sensitivity by KLF4 and Nanog in Gastric Cancer Cells. Curr. Issues Mol. Biol. 2022, 45, 233–248. [Google Scholar] [CrossRef]
- Lee, J.W.; Sung, J.S.; Park, Y.S.; Chung, S.; Kim, Y.H. Identification of different gene expressions between diffuse- and intestinal-type spheroid-forming gastric cancer cells. Gastric Cancer 2019, 22, 967–979. [Google Scholar] [CrossRef]
- Sun, P.; Xia, S.; Lal, B.; Eberhart, C.G.; Quinones-Hinojosa, A.; Maciaczyk, J.; Matsui, W.; DiMeco, F.; Piccirillo, S.M.; Vescovi, A.L.; et al. DNER, an Epigenetically Modulated Gene, Regulates Glioblastoma-Derived Neurosphere Cell Differentiation and Tumor Propagation. Stem Cells 2009, 27, 1473–1486. [Google Scholar] [CrossRef]
- Philipp-Staheli, J.; Kim, K.-H.; Liggitt, D.; Gurley, K.E.; Longton, G.; Kemp, C.J. Distinct roles for p53, p27Kip1, and p21Cip1 during tumor development. Oncogene 2004, 23, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Heavey, S.; O’byrne, K.J.; Gately, K. Strategies for co-targeting the PI3K/AKT/mTOR pathway in NSCLC. Cancer Treat. Rev. 2014, 40, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, T.; Steiner, R.; Lengerke, C. SOX2 and p53 Expression Control Converges in PI3K/AKT Signaling with Versa-tile Implications for Stemness and Cancer. Int. J. Mol. Sci. 2020, 21, 4902. [Google Scholar] [CrossRef] [PubMed]
- Kokaji, E.; Shimomura, A.; Minamisaka, T.; Nakajima, T.; Miwa, S.; Hatta, H.; Nishida, T.; Kiya, C.; Imura, J. Endoglin (CD105) and SMAD4 regulate spheroid formation and the suppression of the invasive ability of human pancreatic cancer cells. Int. J. Oncol. 2018, 52, 892–900. [Google Scholar] [CrossRef] [PubMed]
- Naber, H.P.; Wiercinska, E.; Ten Dijke, P.; van Laar, T. Spheroid assay to measure TGF-beta-induced invasion. J. Vis. Exp. 2011, 57, e3337. [Google Scholar]
- Son, B.K.; Kim, D.H.; Min, K.W.; Kim, E.K.; Kwon, M.J. Smad4/Fascin index is highly prognostic in patients with diffuse type EBV-associated gastric cancer. Pathol. Res. Pract. 2018, 214, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Dotto, G.P. Crosstalk of Notch with p53 and p63 in cancer growth control. Nat. Rev. Cancer 2009, 9, 587–595. [Google Scholar] [CrossRef]
- Liu, L.; Charville, G.W.; Cheung, T.H.; Yoo, B.; Santos, P.J.; Schroeder, M.; Rando, T.A. Impaired Notch Signaling Leads to a Decrease in p53 Activity and Mitotic Catastrophe in Aged Muscle Stem Cells. Cell Stem Cell 2018, 23, 544–556.e4. [Google Scholar] [CrossRef]
- Roemer, K. Notch and the p53 Clan of Transcription Factors. Notch Signal. Embryol. Cancer 2012, 727, 223–240. [Google Scholar]
- Hu, S.; Chen, Q.; Lin, T.; Hong, W.; Wu, W.; Wu, M.; Du, X.; Jin, R. The function of Notch1 intracellular domain in the differentiation of gastric cancer. Oncol. Lett. 2018, 15, 6171–6178. [Google Scholar] [CrossRef]
- Schmidl, B.; Siegl, M.; Boxberg, M.; Stögbauer, F.; Jira, D.; Winter, C.; Stark, L.; Pickhard, A.; Wollenberg, B.; Wirth, M. NOTCH1 Intracellular Domain and the Tumor Microenvironment as Prognostic Markers in HNSCC. Cancers 2022, 14, 1080. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Xing, R.; Liu, J.; Xing, F. Role of CSL-dependent and independent Notch signaling pathways in cell apoptosis. Apoptosis 2016, 21, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Khan, S.K.; Gvozdenovic-Jeremic, J.; Kim, Y.; Dahlman, J.; Kim, H.; Park, O.; Ishitani, T.; Jho, E.H.; Gao, B.; et al. Hippo signaling interactions with Wnt/beta-catenin and Notch signaling repress liver tumorigenesis. J. Clin. Investig. 2017, 127, 137–152. [Google Scholar] [CrossRef]
- Bui, H.T.T.; Le, N.H.; Le, Q.A.; Kim, S.E.; Lee, S.; Kang, D. Synergistic apoptosis of human gastric cancer cells by bortezomib and TRAIL. Int. J. Med. Sci. 2019, 16, 1412–1423. [Google Scholar] [CrossRef]
- Choi, E.K.; Kim, H.D.; Park, E.J.; Song, S.Y.; Phan, T.T.; Nam, M.; Kim, M.; Kim, D.-U.; Hoe, K.-L. 8-Methoxypsoralen Induces Apoptosis by Upregulating p53 and Inhibits Metastasis by Downregulating MMP-2 and MMP-9 in Human Gastric Cancer Cells. Biomol. Ther. 2023, 31, 219–226. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
To, H.T.N.; Park, J.-H.; Kim, J.W.; Kang, D. Delta/Notch-like Epidermal Growth Factor-Related Receptor (DNER), a Potential Prognostic Marker of Gastric Cancer Regulates Cell Survival and Cell Cycle Progression. Int. J. Mol. Sci. 2023, 24, 10077. https://doi.org/10.3390/ijms241210077
To HTN, Park J-H, Kim JW, Kang D. Delta/Notch-like Epidermal Growth Factor-Related Receptor (DNER), a Potential Prognostic Marker of Gastric Cancer Regulates Cell Survival and Cell Cycle Progression. International Journal of Molecular Sciences. 2023; 24(12):10077. https://doi.org/10.3390/ijms241210077
Chicago/Turabian StyleTo, Han Thi Ngoc, Ji-Hong Park, Jeong Won Kim, and Dongchul Kang. 2023. "Delta/Notch-like Epidermal Growth Factor-Related Receptor (DNER), a Potential Prognostic Marker of Gastric Cancer Regulates Cell Survival and Cell Cycle Progression" International Journal of Molecular Sciences 24, no. 12: 10077. https://doi.org/10.3390/ijms241210077
APA StyleTo, H. T. N., Park, J.-H., Kim, J. W., & Kang, D. (2023). Delta/Notch-like Epidermal Growth Factor-Related Receptor (DNER), a Potential Prognostic Marker of Gastric Cancer Regulates Cell Survival and Cell Cycle Progression. International Journal of Molecular Sciences, 24(12), 10077. https://doi.org/10.3390/ijms241210077





