PCTAIRE Protein Kinase 1 (PCTK1) Suppresses Proliferation, Stemness, and Chemoresistance in Colorectal Cancer through the BMPR1B-Smad1/5/8 Signaling Pathway
Abstract
1. Introduction
2. Results
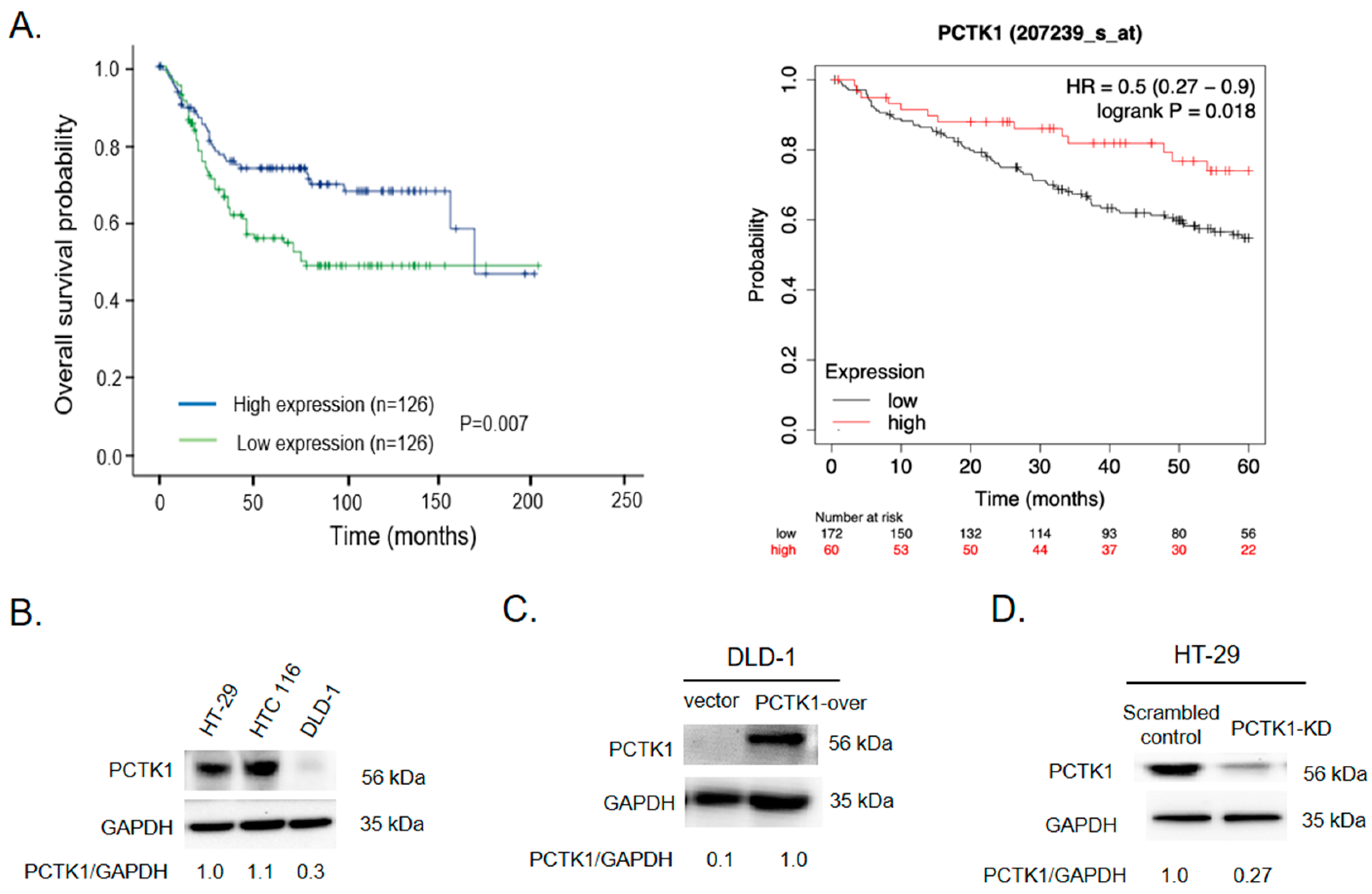
2.1. High PCTK1 Expression Was Associated with a More Favorable CRC Prognosis
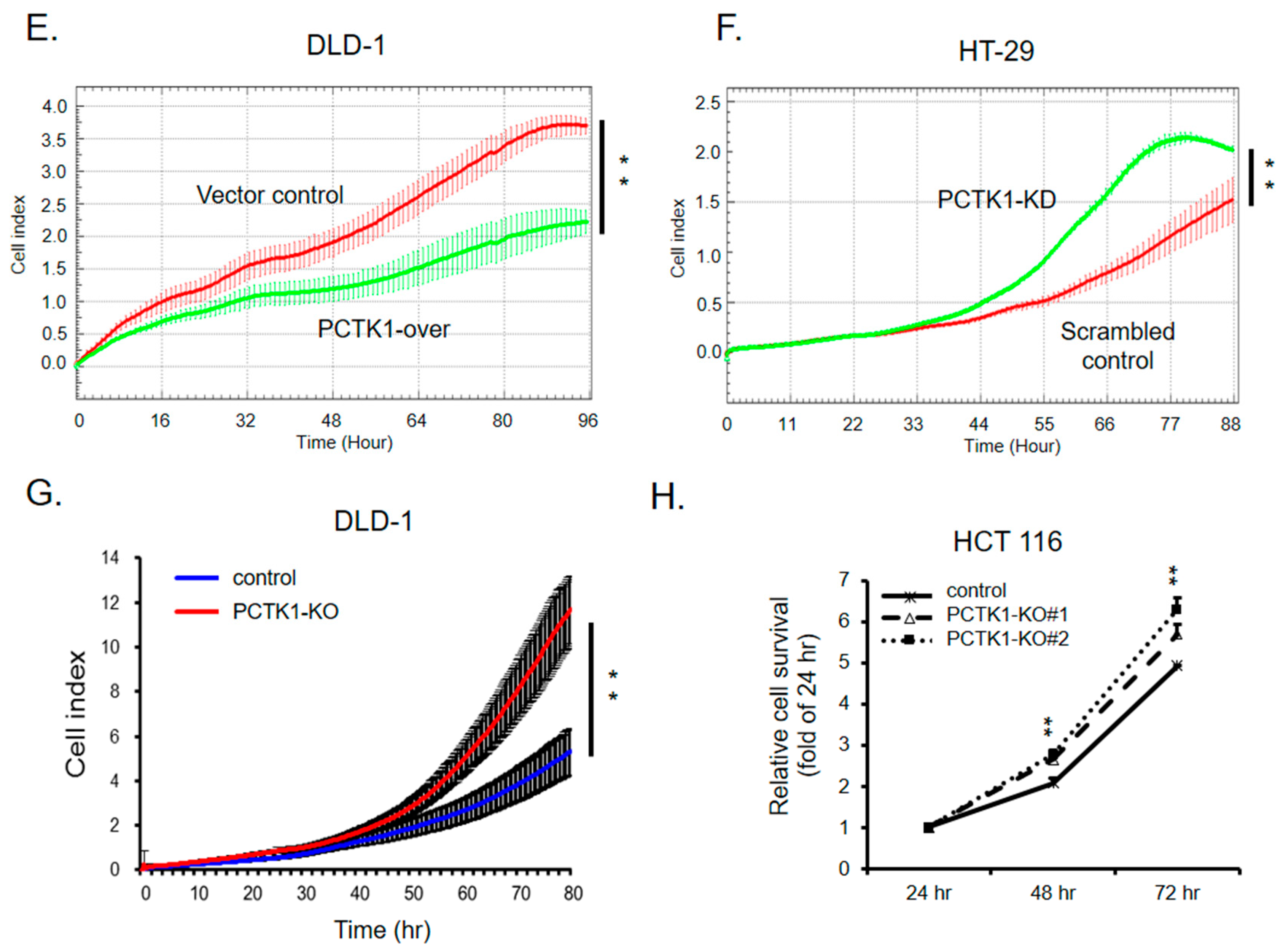
2.2. PCTK1 Expression Suppressed CRC Cell Proliferation
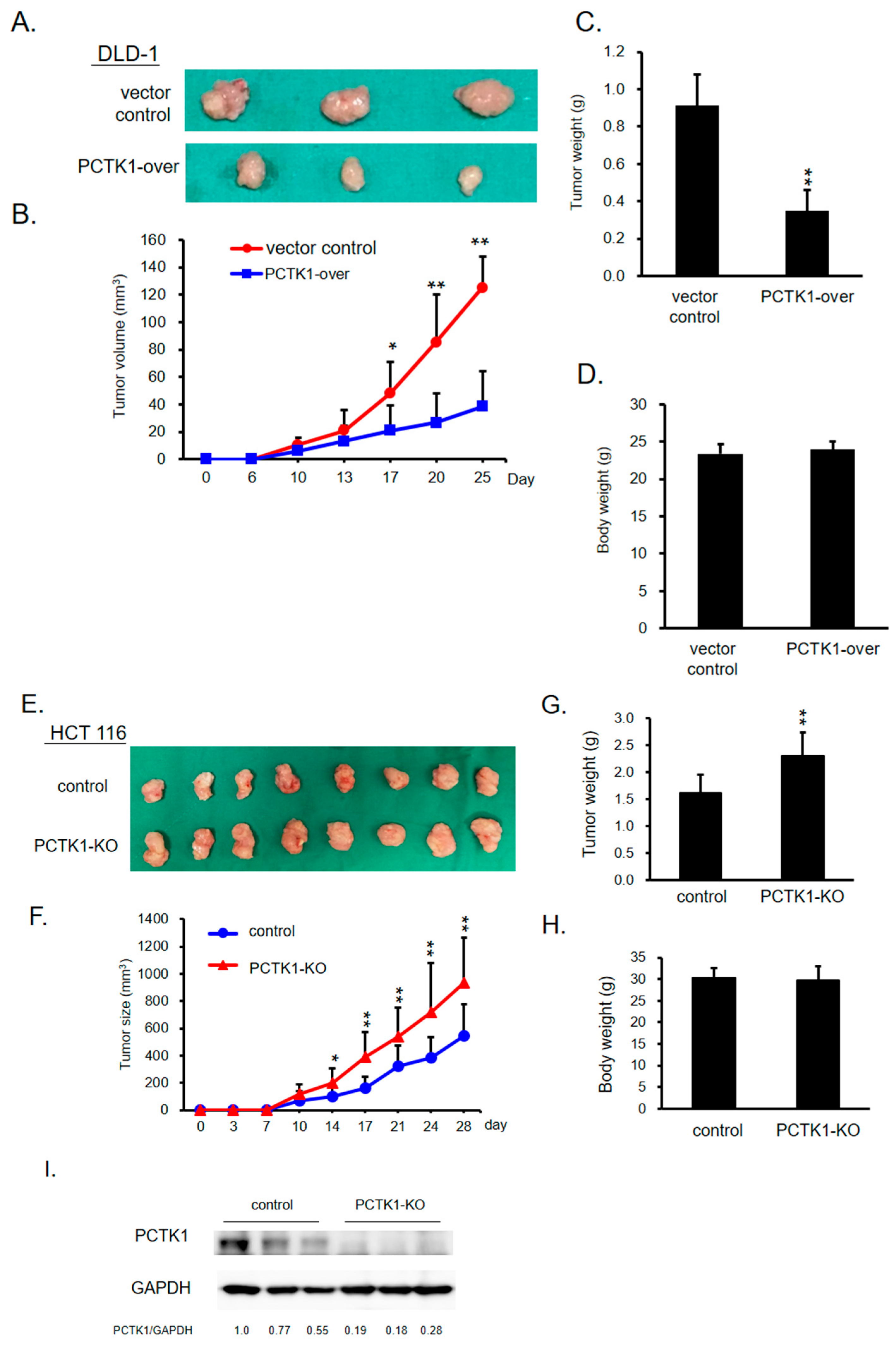
2.3. PCTK1 Inhibited Tumorigenesis and Tumor Growth In Vivo
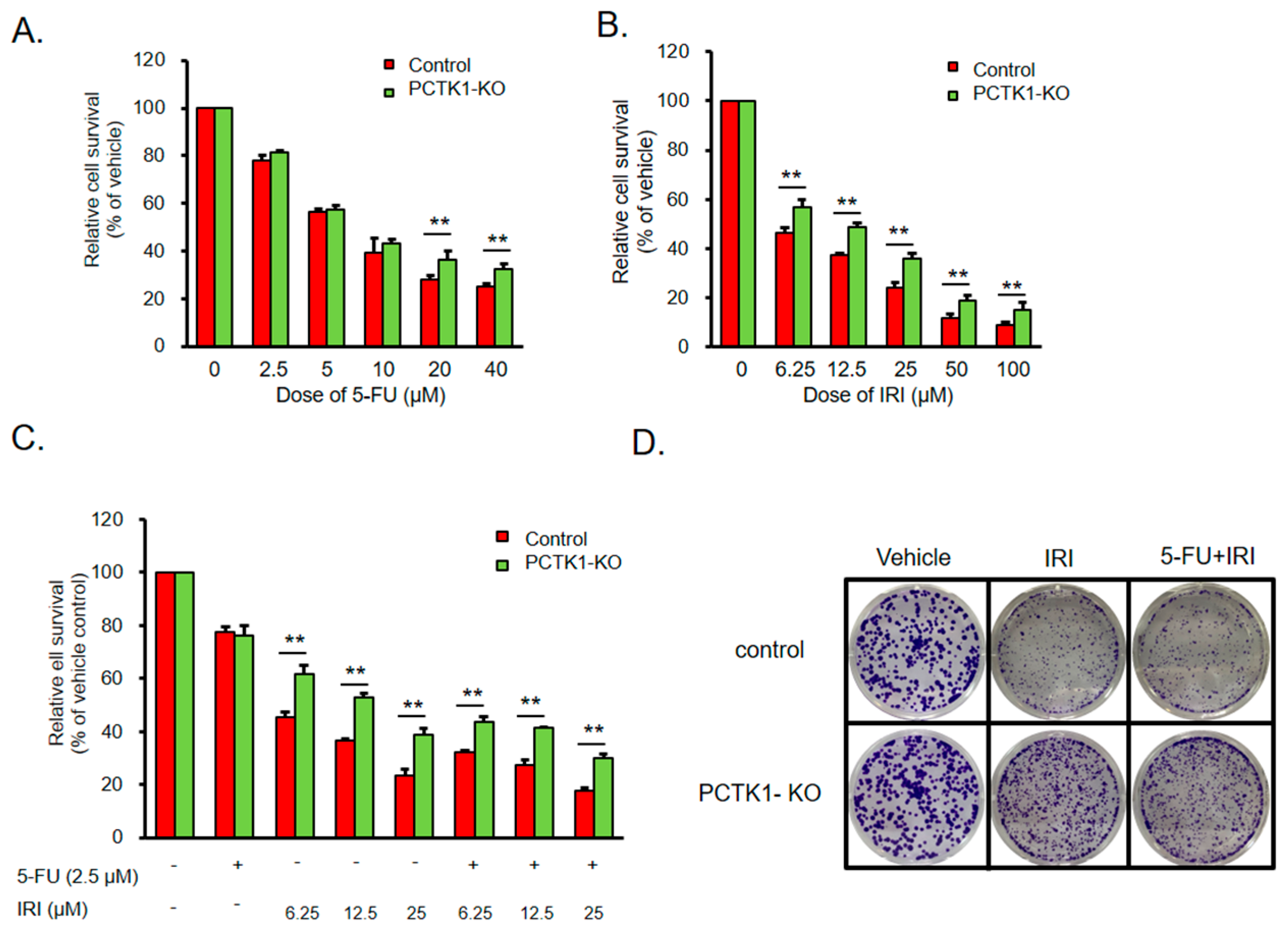
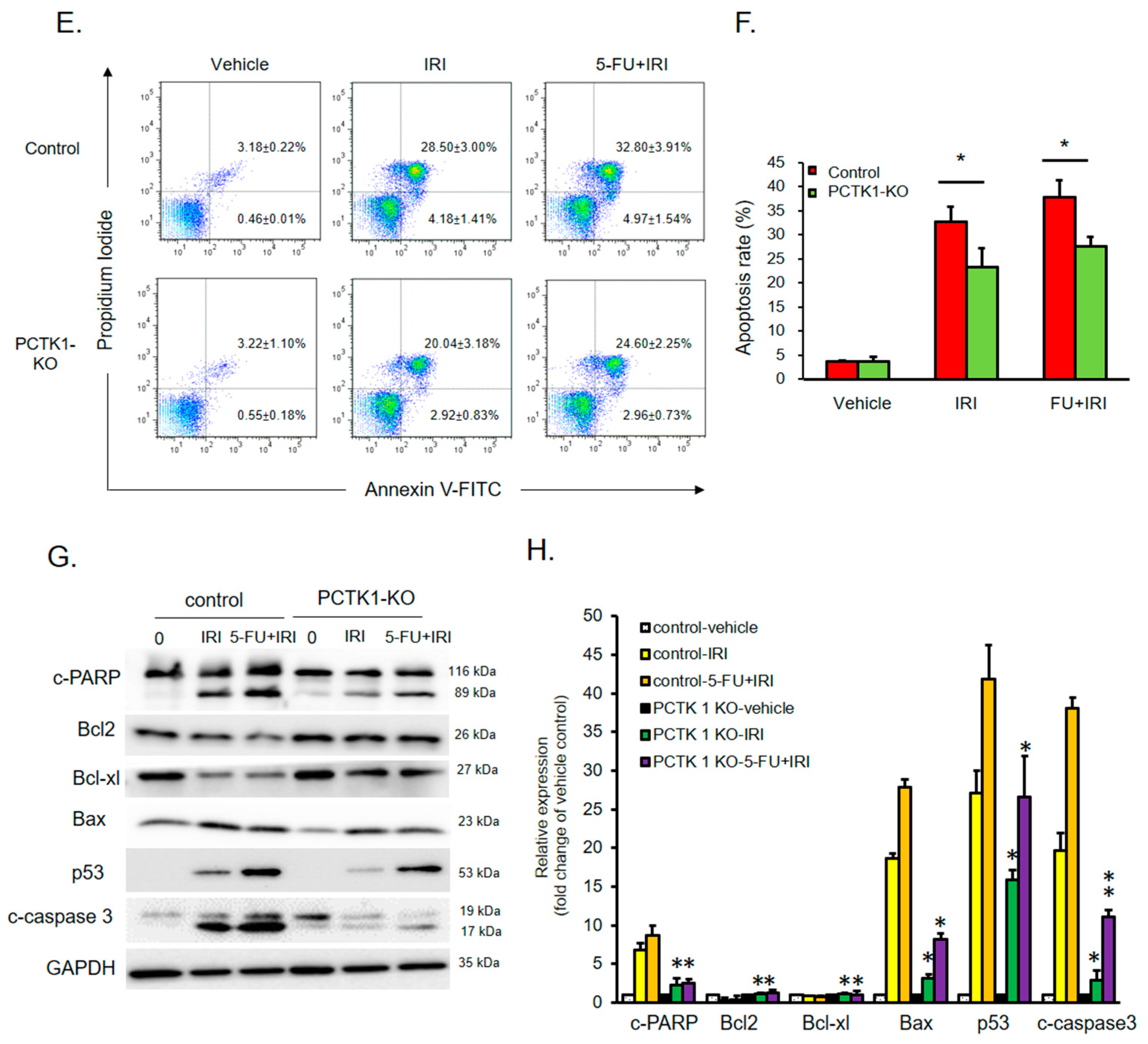
2.4. PCTK1 Negatively Regulated Chemoresistance in CRC
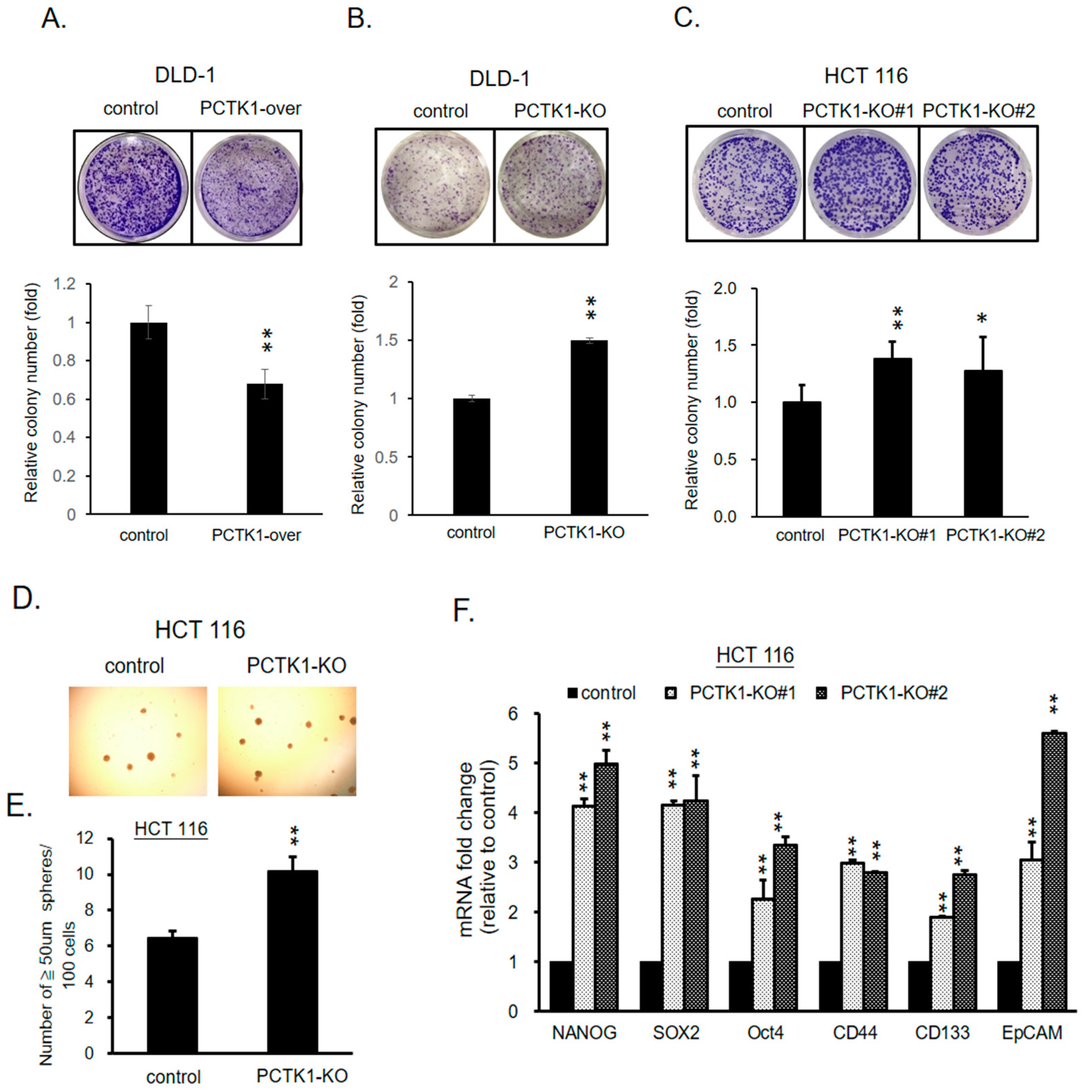
2.5. PCTK1 Expression Altered the Cancer Stem Cell Characteristics of CRC Cells
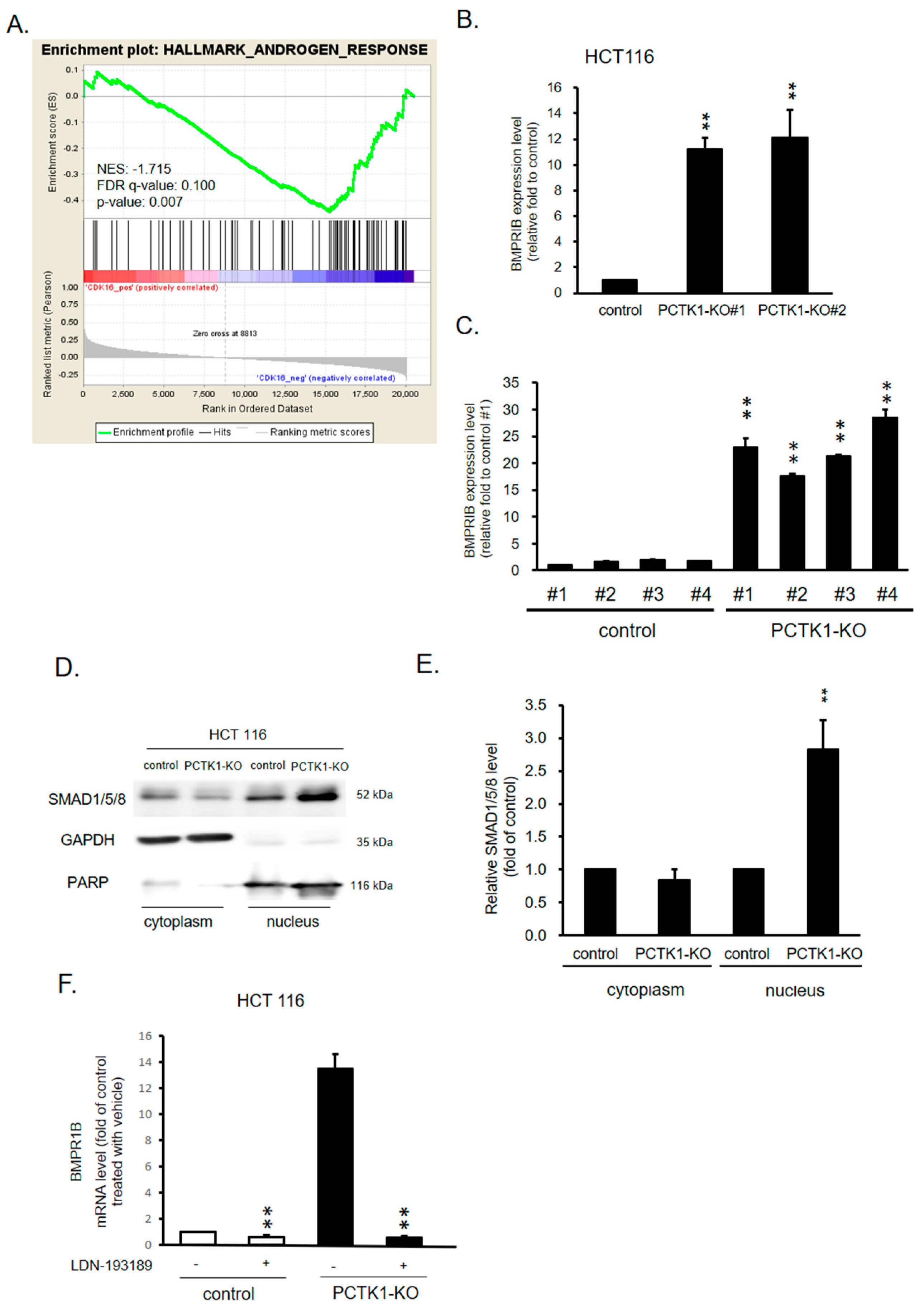
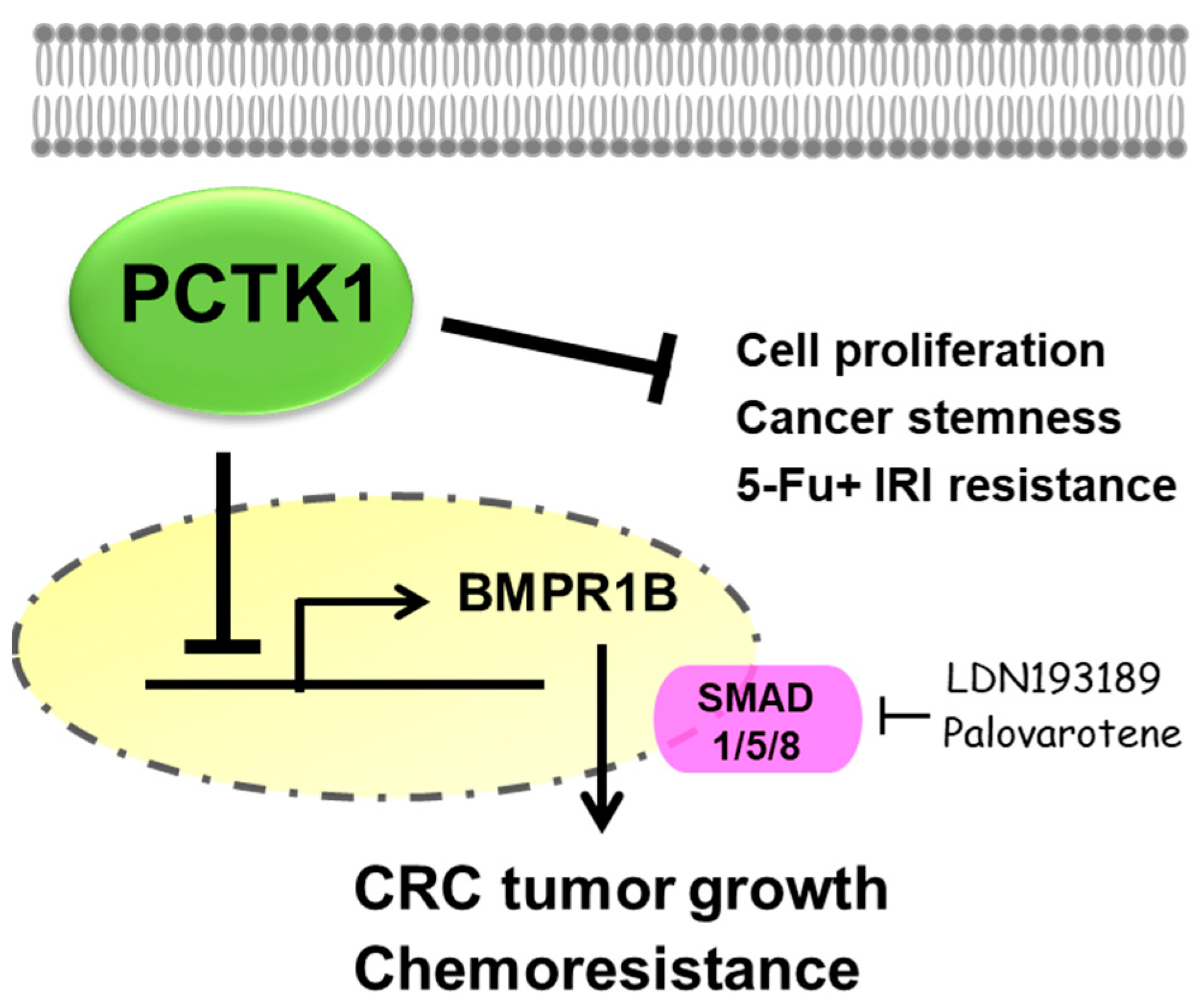
2.6. PCTK1 Suppressed Cell Proliferation, CSC Properties, and Chemoresponse through BMPR1B–Smad Signaling
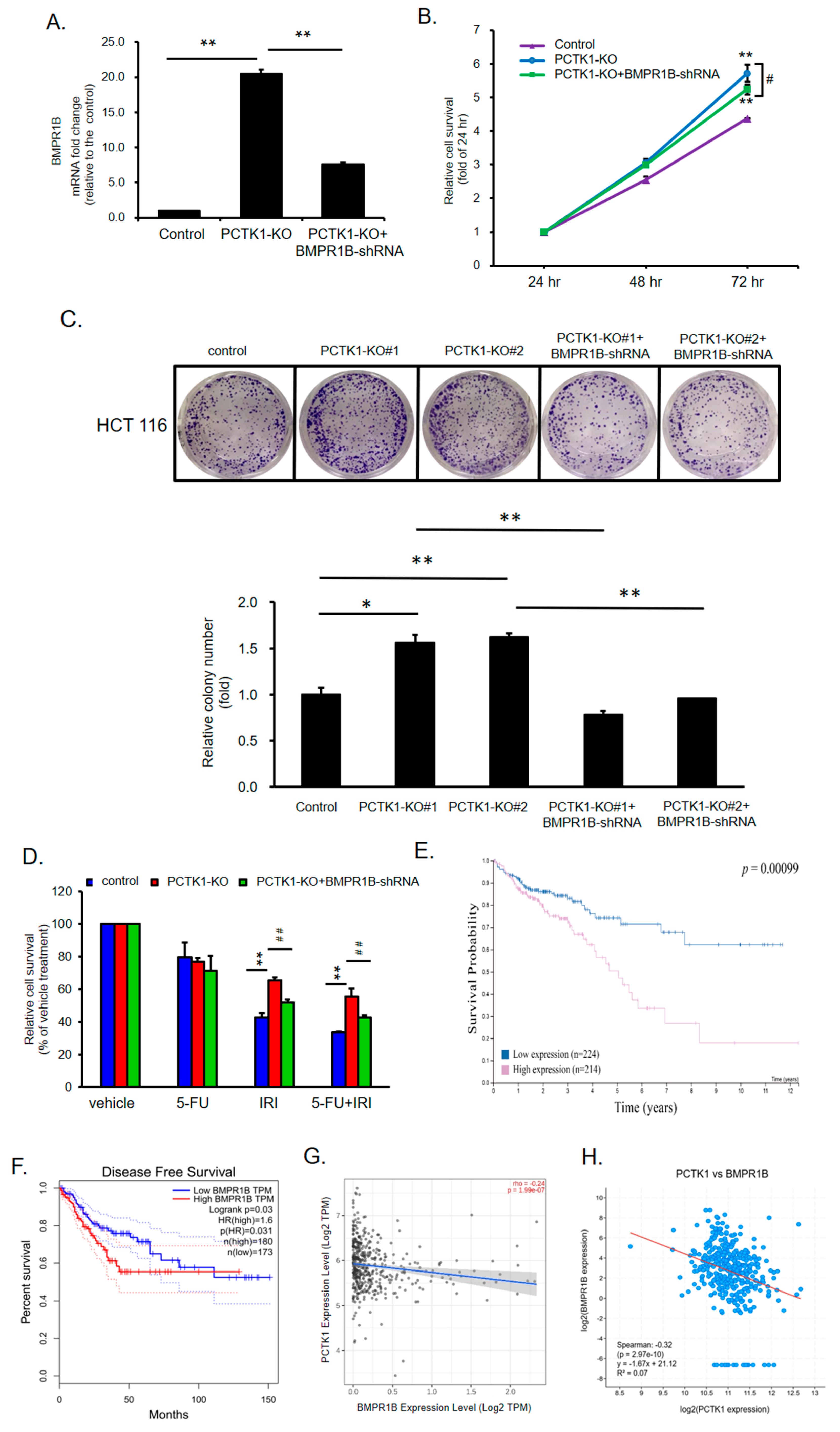
2.7. BMPR1B Knockdown Partially Inhibited the Promotion of PCTK1 Knockout on CRC Cell Malignant Phenotype and Chemoresisitance
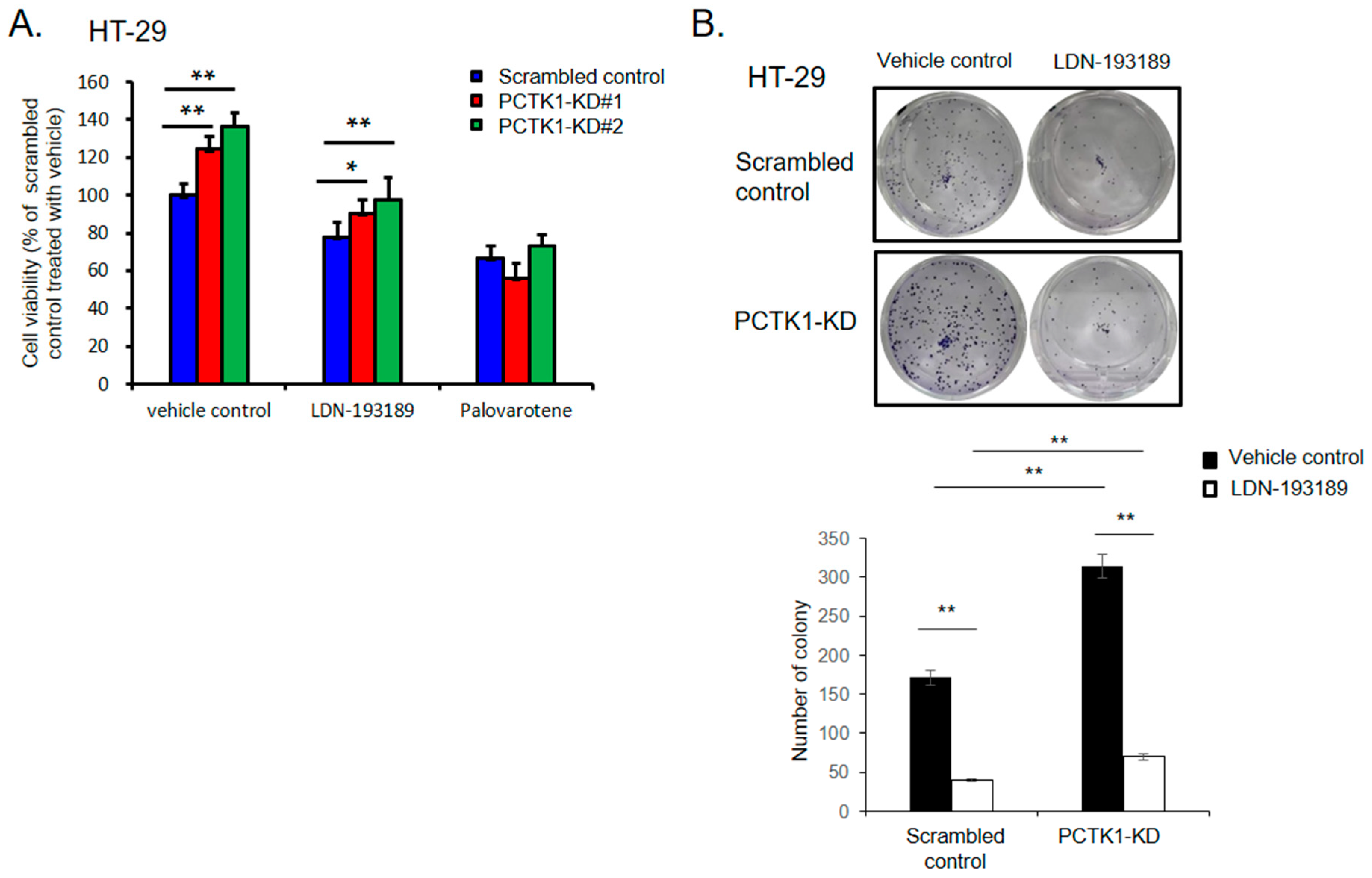
2.8. Pharmacological Targeting BMPR1B-SMAD1/5/8 Signaling with Small Molecules Inhibited the Promotion of PCTK1 Knockout on CRC Cell Malignant Phenotype and Chemoresisitance
3. Discussion
4. Materials and Methods
4.1. Transfection and Generation of Stable Clones
4.2. Generation of PCTK1 Knockout Cell Lines by Using the CRISPR/Cas9 Technology
4.3. Cell Proliferation/Viability
4.4. Evaluation of Cell Proliferation Using the x-CELLigence Biosensor System
4.5. Colony Formation Assay
4.6. Sphere Formation Assay
4.7. In Vivo Tumor Xenograft Experiments
4.8. Annexin V/Propidium Iodide Double Staining Assay
4.9. RT-qPCR
4.10. Western Blotting
4.11. Bioinformatics Data Resources
4.12. Gene Set Enrichment Analysis
4.13. cBioPortal
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef]
- Marin, J.J.; Sanchez de Medina, F.; Castano, B.; Bujanda, L.; Romero, M.R.; Martinez-Augustin, O.; Moral-Avila, R.D.; Briz, O. Chemoprevention, chemotherapy, and chemoresistance in colorectal cancer. Drug Metab. Rev. 2012, 44, 148–172. [Google Scholar] [CrossRef] [PubMed]
- Iwano, S.; Satou, A.; Matsumura, S.; Sugiyama, N.; Ishihama, Y.; Toyoshima, F. PCTK1 regulates integrin-dependent spindle orientation via protein kinase A regulatory subunit KAP0 and myosin X. Mol. Cell Biol. 2015, 35, 1197–1208. [Google Scholar] [CrossRef] [PubMed]
- Meyerson, M.; Enders, G.H.; Wu, C.L.; Su, L.K.; Gorka, C.; Nelson, C.; Harlow, E.; Tsai, L.H. A family of human cdc2-related protein kinases. EMBO J. 1992, 11, 2909–2917. [Google Scholar] [CrossRef] [PubMed]
- Besset, V.; Rhee, K.; Wolgemuth, D.J. The cellular distribution and kinase activity of the Cdk family member Pctaire1 in the adult mouse brain and testis suggest functions in differentiation. Cell Growth Differ. 1999, 10, 173–181. [Google Scholar] [PubMed]
- Charrasse, S.; Carena, I.; Hagmann, J.; Woods-Cook, K.; Ferrari, S. PCTAIRE-1: Characterization, subcellular distribution, and cell cycle-dependent kinase activity. Cell Growth Differ. 1999, 10, 611–620. [Google Scholar]
- Cole, A.R. PCTK proteins: The forgotten brain kinases? Neurosignals 2009, 17, 288–297. [Google Scholar] [CrossRef]
- Mikolcevic, P.; Sigl, R.; Rauch, V.; Hess, M.W.; Pfaller, K.; Barisic, M.; Pelliniemi, L.J.; Boesl, M.; Geley, S. Cyclin-dependent kinase 16/PCTAIRE kinase 1 is activated by cyclin Y and is essential for spermatogenesis. Mol. Cell Biol. 2012, 32, 868–879. [Google Scholar] [CrossRef]
- Yanagi, T.; Krajewska, M.; Matsuzawa, S.; Reed, J.C. PCTAIRE1 phosphorylates p27 and regulates mitosis in cancer cells. Cancer Res. 2014, 74, 5795–5807. [Google Scholar] [CrossRef]
- Yanagi, T.; Reed, J.C.; Matsuzawa, S. PCTAIRE1 regulates p27 stability, apoptosis and tumor growth in malignant melanoma. Oncoscience 2014, 1, 624–633. [Google Scholar] [CrossRef]
- Yanagi, T.; Matsuzawa, S. PCTAIRE1/PCTK1/CDK16: A new oncotarget? Cell Cycle 2015, 14, 463–464. [Google Scholar] [CrossRef]
- Yanagi, T.; Shi, R.; Aza-Blanc, P.; Reed, J.C.; Matsuzawa, S. PCTAIRE1-knockdown sensitizes cancer cells to TNF family cytokines. PLoS ONE 2015, 10, e0119404. [Google Scholar] [CrossRef] [PubMed]
- Cwiek, P.; Leni, Z.; Salm, F.; Dimitrova, V.; Styp-Rekowska, B.; Chiriano, G.; Carroll, M.; Holand, K.; Djonov, V.; Scapozza, L.; et al. RNA interference screening identifies a novel role for PCTK1/CDK16 in medulloblastoma with c-Myc amplification. Oncotarget 2015, 6, 116–129. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.N.; Green, J.; Wang, Z.; Deng, Y.; Qiao, M.; Peabody, M.; Zhang, Q.; Ye, J.; Yan, Z.; Denduluri, S.; et al. Bone Morphogenetic Protein (BMP) signaling in development and human diseases. Genes Dis. 2014, 1, 87–105. [Google Scholar] [CrossRef]
- Dasgupta, Y.; Golovine, K.; Nieborowska-Skorska, M.; Luo, L.; Matlawska-Wasowska, K.; Mullighan, C.G.; Skorski, T. Drugging DNA repair to target T-ALL cells. Leuk. Lymphoma 2018, 59, 1746–1749. [Google Scholar] [CrossRef]
- Hardwick, J.C.; Kodach, L.L.; Offerhaus, G.J.; van den Brink, G.R. Bone morphogenetic protein signalling in colorectal cancer. Nat. Rev. Cancer 2008, 8, 806–812. [Google Scholar] [CrossRef]
- Kodach, L.L.; Wiercinska, E.; de Miranda, N.F.; Bleuming, S.A.; Musler, A.R.; Peppelenbosch, M.P.; Dekker, E.; van den Brink, G.R.; van Noesel, C.J.; Morreau, H.; et al. The bone morphogenetic protein pathway is inactivated in the majority of sporadic colorectal cancers. Gastroenterology 2008, 134, 1332–1341. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.S.; Hutter, C.M.; Potter, J.D.; Liu, Y.; Prentice, R.L.; Peters, U.; Hsu, L. Insights into colon cancer etiology via a regularized approach to gene set analysis of GWAS data. Am. J. Hum. Genet. 2010, 86, 860–871. [Google Scholar] [CrossRef]
- Xie, Y.H.; Chen, Y.X.; Fang, J.Y. Comprehensive review of targeted therapy for colorectal cancer. Signal. Transduct. Target. Ther. 2020, 5, 22. [Google Scholar] [CrossRef]
- Pardini, B.; Kumar, R.; Naccarati, A.; Novotny, J.; Prasad, R.B.; Forsti, A.; Hemminki, K.; Vodicka, P.; Lorenzo Bermejo, J. 5-Fluorouracil-based chemotherapy for colorectal cancer and MTHFR/MTRR genotypes. Br. J. Clin. Pharmacol. 2011, 72, 162–163. [Google Scholar] [CrossRef]
- Cunningham, D.; Pyrhonen, S.; James, R.D.; Punt, C.J.; Hickish, T.F.; Heikkila, R.; Johannesen, T.B.; Starkhammar, H.; Topham, C.A.; Awad, L.; et al. Randomised trial of irinotecan plus supportive care versus supportive care alone after fluorouracil failure for patients with metastatic colorectal cancer. Lancet 1998, 352, 1413–1418. [Google Scholar] [CrossRef]
- Maindrault-Goebel, F.; Louvet, C.; Andre, T.; Carola, E.; Lotz, J.P.; Molitor, J.L.; Garcia, M.L.; Gilles-Amar, V.; Izrael, V.; Krulik, M.; et al. Oxaliplatin added to the simplified bimonthly leucovorin and 5-fluorouracil regimen as second-line therapy for metastatic colorectal cancer (FOLFOX6). Eur. J. Cancer 1999, 35, 1338–1342. [Google Scholar] [CrossRef] [PubMed]
- Rougier, P.; Van Cutsem, E.; Bajetta, E.; Niederle, N.; Possinger, K.; Labianca, R.; Navarro, M.; Morant, R.; Bleiberg, H.; Wils, J.; et al. Randomised trial of irinotecan versus fluorouracil by continuous infusion after fluorouracil failure in patients with metastatic colorectal cancer. Lancet 1998, 352, 1407–1412. [Google Scholar] [CrossRef]
- Prager, B.C.; Xie, Q.; Bao, S.; Rich, J.N. Cancer Stem Cells: The Architects of the Tumor Ecosystem. Cell Stem Cell 2019, 24, 41–53. [Google Scholar] [CrossRef]
- Fang, D.; Nguyen, T.K.; Leishear, K.; Finko, R.; Kulp, A.N.; Hotz, S.; Van Belle, P.A.; Xu, X.; Elder, D.E.; Herlyn, M. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005, 65, 9328–9337. [Google Scholar] [CrossRef] [PubMed]
- Walcher, L.; Kistenmacher, A.K.; Suo, H.; Kitte, R.; Dluczek, S.; Strauss, A.; Blaudszun, A.R.; Yevsa, T.; Fricke, S.; Kossatz-Boehlert, U. Cancer Stem Cells-Origins and Biomarkers: Perspectives for Targeted Personalized Therapies. Front. Immunol. 2020, 11, 1280. [Google Scholar] [CrossRef]
- Kodach, L.L.; Bleuming, S.A.; Musler, A.R.; Peppelenbosch, M.P.; Hommes, D.W.; van den Brink, G.R.; van Noesel, C.J.; Offerhaus, G.J.; Hardwick, J.C. The bone morphogenetic protein pathway is active in human colon adenomas and inactivated in colorectal cancer. Cancer 2008, 112, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Abdurahman, A.; Du, X.; Yao, Y.; Sulaiman, Y.; Aniwashi, J.; Li, Q. Smad4 Feedback Enhances BMPR1B Transcription in Ovine Granulosa Cells. Int. J. Mol. Sci. 2019, 20, 2732. [Google Scholar] [CrossRef]
- Shen, T.; Sun, C.; Zhang, Z.; Xu, N.; Duan, X.; Feng, X.H.; Lin, X. Specific control of BMP signaling and mesenchymal differentiation by cytoplasmic phosphatase PPM1H. Cell Res. 2014, 24, 727–741. [Google Scholar] [CrossRef]
- Graeser, R.; Gannon, J.; Poon, R.Y.; Dubois, T.; Aitken, A.; Hunt, T. Regulation of the CDK-related protein kinase PCTAIRE-1 and its possible role in neurite outgrowth in Neuro-2A cells. J. Cell Sci. 2002, 115, 3479–3490. [Google Scholar] [CrossRef]
- Wang, H.; Liu, H.; Min, S.; Shen, Y.; Li, W.; Chen, Y.; Wang, X. CDK16 overexpressed in non-small cell lung cancer and regulates cancer cell growth and apoptosis via a p27-dependent mechanism. Biomed. Pharmacother. 2018, 103, 399–405. [Google Scholar] [CrossRef]
- Chang, J.W.; Shih, C.L.; Wang, C.L.; Luo, J.D.; Wang, C.W.; Hsieh, J.J.; Yu, C.J.; Chiou, C.C. Transcriptomic Analysis in Liquid Biopsy Identifies Circulating PCTAIRE-1 mRNA as a Biomarker in NSCLC. Cancer Genom. Proteom. 2020, 17, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Yanagi, T.; Tachikawa, K.; Wilkie-Grantham, R.; Hishiki, A.; Nagai, K.; Toyonaga, E.; Chivukula, P.; Matsuzawa, S. Lipid Nanoparticle-mediated siRNA Transfer Against PCTAIRE1/PCTK1/Cdk16 Inhibits In Vivo Cancer Growth. Mol. Ther. Nucleic Acids 2016, 5, e327. [Google Scholar] [CrossRef]
- Miyazono, K.; Kamiya, Y.; Morikawa, M. Bone morphogenetic protein receptors and signal transduction. J. Biochem. 2010, 147, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Pickup, M.W.; Hover, L.D.; Guo, Y.; Gorska, A.E.; Chytil, A.; Novitskiy, S.V.; Moses, H.L.; Owens, P. Deletion of the BMP receptor BMPR1a impairs mammary tumor formation and metastasis. Oncotarget 2015, 6, 22890–22904. [Google Scholar] [CrossRef] [PubMed]
- Saetrom, P.; Biesinger, J.; Li, S.M.; Smith, D.; Thomas, L.F.; Majzoub, K.; Rivas, G.E.; Alluin, J.; Rossi, J.J.; Krontiris, T.G.; et al. A risk variant in an miR-125b binding site in BMPR1B is associated with breast cancer pathogenesis. Cancer Res. 2009, 69, 7459–7465. [Google Scholar] [CrossRef] [PubMed]
- Jeanpierre, S.; Arizkane, K.; Thongjuea, S.; Grockowiak, E.; Geistlich, K.; Barral, L.; Voeltzel, T.; Guillemin, A.; Gonin-Giraud, S.; Gandrillon, O.; et al. The quiescent fraction of chronic myeloid leukemic stem cells depends on BMPR1B, Stat3 and BMP4-niche signals to persist in patients in remission. Haematologica 2021, 106, 111–122. [Google Scholar] [CrossRef]
- Dai, K.; Qin, F.; Zhang, H.; Liu, X.; Guo, C.; Zhang, M.; Gu, F.; Fu, L.; Ma, Y. Low expression of BMPRIB indicates poor prognosis of breast cancer and is insensitive to taxane-anthracycline chemotherapy. Oncotarget 2016, 7, 4770–4784. [Google Scholar] [CrossRef]
- Fukuda, T.; Fukuda, R.; Tanabe, R.; Koinuma, D.; Koyama, H.; Hashizume, Y.; Moustakas, A.; Miyazono, K.; Heldin, C.-H. BMP signaling is a therapeutic target in ovarian cancer. Cell Death Discov. 2020, 6, 139. [Google Scholar] [CrossRef]
- Teufel, A.; Steinmann, S.; Siebler, J.; Zanke, C.; Hohl, H.; Adami, B.; Schroeder, M.; Klein, O.; Hohler, T.; Galle, P.R.; et al. Irinotecan plus folinic acid/continuous 5-fluorouracil as simplified bimonthly FOLFIRI regimen for first-line therapy of metastatic colorectal cancer. BMC Cancer 2004, 4, 38. [Google Scholar] [CrossRef]
- Saltz, L.B.; Cox, J.V.; Blanke, C.; Rosen, L.S.; Fehrenbacher, L.; Moore, M.J.; Maroun, J.A.; Ackland, S.P.; Locker, P.K.; Pirotta, N.; et al. Irinotecan plus fluorouracil and leucovorin for metastatic colorectal cancer. Irinotecan Study Group. N. Engl. J. Med. 2000, 343, 905–914. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.J.; Ren, S.N.; Liu, Y.T.; Yan, H.W.; Chen, X.B. Targeting EGFR sensitizes 5-Fu-resistant colon cancer cells through modification of the lncRNA-FGD5-AS1-miR-330-3p-Hexokinase 2 axis. Mol. Ther. Oncolytics 2021, 23, 14–25. [Google Scholar] [CrossRef]
- Maliekal, T.T.; Antony, M.L.; Nair, A.; Paulmurugan, R.; Karunagaran, D. Loss of expression, and mutations of Smad 2 and Smad 4 in human cervical cancer. Oncogene 2003, 22, 4889–4897. [Google Scholar] [CrossRef] [PubMed]
- Levy, L.; Hill, C.S. Alterations in components of the TGF-beta superfamily signaling pathways in human cancer. Cytokine Growth Factor. Rev. 2006, 17, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Grady, W.M.; Myeroff, L.L.; Swinler, S.E.; Rajput, A.; Thiagalingam, S.; Lutterbaugh, J.D.; Neumann, A.; Brattain, M.G.; Chang, J.; Kim, S.J.; et al. Mutational inactivation of transforming growth factor beta receptor type II in microsatellite stable colon cancers. Cancer Res. 1999, 59, 320–324. [Google Scholar] [PubMed]
- Samanta, D.; Datta, P.K. Alterations in the Smad pathway in human cancers. Front. Biosci. 2012, 17, 1281–1293. [Google Scholar] [CrossRef]
- Shi, Y.; Massague, J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 2003, 113, 685–700. [Google Scholar] [CrossRef]
- Itoh, S.; Itoh, F.; Goumans, M.J.; Ten Dijke, P. Signaling of transforming growth factor-beta family members through Smad proteins. Eur. J. Biochem. 2000, 267, 6954–6967. [Google Scholar] [CrossRef]
- Ali, J.L.; Lagasse, B.J.; Minuk, A.J.; Love, A.J.; Moraya, A.I.; Lam, L.; Arthur, G.; Gibson, S.B.; Morrison, L.C.; Werbowetski-Ogilvie, T.E.; et al. Differential cellular responses induced by dorsomorphin and LDN-193189 in chemotherapy-sensitive and chemotherapy-resistant human epithelial ovarian cancer cells. Int. J. Cancer 2015, 136, E455–E469. [Google Scholar] [CrossRef]
- Hind, M.; Stinchcombe, S. Palovarotene, a novel retinoic acid receptor gamma agonist for the treatment of emphysema. Curr. Opin. Investig. Drugs 2009, 10, 1243–1250. [Google Scholar]
- Kalal, B.S.; Modi, P.K.; Upadhya, D.; Saha, P.; Prasad, T.S.K.; Pai, V.R. Inhibition of bone morphogenetic proteins signaling suppresses metastasis melanoma: A proteomics approach. Am. J. Transl. Res. 2021, 13, 11081–11093. [Google Scholar] [PubMed]
- Sharma, R.; Gogoi, G.; Saikia, S.; Sharma, A.; Kalita, D.J.; Sarma, A.; Limaye, A.M.; Gaur, M.K.; Bhattacharyya, J.; Jaganathan, B.G. BMP4 enhances anoikis resistance and chemoresistance of breast cancer cells through canonical BMP signaling. J. Cell Commun. Signal. 2022, 16, 191–205. [Google Scholar] [CrossRef]
- Chen, Z.; Yuan, L.; Li, X.; Yu, J.; Xu, Z. BMP2 inhibits cell proliferation by downregulating EZH2 in gastric cancer. Cell Cycle 2022, 21, 2298–2308. [Google Scholar] [CrossRef]
- Fukuda, T.; Fukuda, R.; Miyazono, K.; Heldin, C.H. Tumor Promoting Effect of BMP Signaling in Endometrial Cancer. Int. J. Mol. Sci. 2021, 22, 7882. [Google Scholar] [CrossRef]
- Liang, Z.; Wu, B.; Ji, Z.; Liu, W.; Shi, D.; Chen, X.; Wei, Y.; Jiang, J. The binding of LDN193189 to CD133 C-terminus suppresses the tumorigenesis and immune escape of liver tumor-initiating cells. Cancer Lett. 2021, 513, 90–100. [Google Scholar] [CrossRef]
- Thomas, L.; Smith, N.; Saunders, D.; Zalles, M.; Gulej, R.; Lerner, M.; Fung, K.M.; Carcaboso, A.M.; Towner, R.A. OKlahoma Nitrone-007: Novel treatment for diffuse intrinsic pontine glioma. J. Transl. Med. 2020, 18, 424. [Google Scholar] [CrossRef]
- Hoyer-Kuhn, H.; Schonau, E. Pharmacotherapy in Rare Skeletal Diseases. Handb. Exp. Pharmacol. 2020, 261, 87–104. [Google Scholar] [CrossRef] [PubMed]
- Pignolo, R.J.; Pacifici, M. Retinoid Agonists in the Targeting of Heterotopic Ossification. Cells 2021, 10, 3245. [Google Scholar] [CrossRef]
- Wei, P.L.; Lin, J.C.; Hung, C.S.; Makondi, P.T.; Batzorig, U.; Chang, T.C.; Huang, C.Y.; Chang, Y.J. Human alpha-defensin 6 (HD6) suppresses CRC proliferation and metastasis through abolished EGF/EGFR signaling pathway. Int. J. Med. Sci. 2022, 19, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.L.; Prince, G.M.S.H.; Batzorig, U.; Huang, C.Y.; Chang, Y.J. ALDH2 promotes cancer stemness and metastasis in colorectal cancer through activating beta-catenin signaling. J. Cell Biochem. 2023. [Google Scholar] [CrossRef]
- Barrett, T.; Troup, D.B.; Wilhite, S.E.; Ledoux, P.; Rudnev, D.; Evangelista, C.; Kim, I.F.; Soboleva, A.; Tomashevsky, M.; Edgar, R. NCBI GEO: Mining tens of millions of expression profiles--database and tools update. Nucleic Acids Res. 2007, 35, D760–D765. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, P.-L.; Huang, C.-Y.; Chang, T.-C.; Lin, J.-C.; Lee, C.-C.; Prince, G.M.S.H.; Makondi, P.T.; Chui, A.W.-Y.; Chang, Y.-J. PCTAIRE Protein Kinase 1 (PCTK1) Suppresses Proliferation, Stemness, and Chemoresistance in Colorectal Cancer through the BMPR1B-Smad1/5/8 Signaling Pathway. Int. J. Mol. Sci. 2023, 24, 10008. https://doi.org/10.3390/ijms241210008
Wei P-L, Huang C-Y, Chang T-C, Lin J-C, Lee C-C, Prince GMSH, Makondi PT, Chui AW-Y, Chang Y-J. PCTAIRE Protein Kinase 1 (PCTK1) Suppresses Proliferation, Stemness, and Chemoresistance in Colorectal Cancer through the BMPR1B-Smad1/5/8 Signaling Pathway. International Journal of Molecular Sciences. 2023; 24(12):10008. https://doi.org/10.3390/ijms241210008
Chicago/Turabian StyleWei, Po-Li, Chien-Yu Huang, Tung-Cheng Chang, Jang-Chun Lin, Cheng-Chin Lee, G. M. Shazzad Hossain Prince, Precious Takondwa Makondi, Angelina Wong-Ying Chui, and Yu-Jia Chang. 2023. "PCTAIRE Protein Kinase 1 (PCTK1) Suppresses Proliferation, Stemness, and Chemoresistance in Colorectal Cancer through the BMPR1B-Smad1/5/8 Signaling Pathway" International Journal of Molecular Sciences 24, no. 12: 10008. https://doi.org/10.3390/ijms241210008
APA StyleWei, P.-L., Huang, C.-Y., Chang, T.-C., Lin, J.-C., Lee, C.-C., Prince, G. M. S. H., Makondi, P. T., Chui, A. W.-Y., & Chang, Y.-J. (2023). PCTAIRE Protein Kinase 1 (PCTK1) Suppresses Proliferation, Stemness, and Chemoresistance in Colorectal Cancer through the BMPR1B-Smad1/5/8 Signaling Pathway. International Journal of Molecular Sciences, 24(12), 10008. https://doi.org/10.3390/ijms241210008





