Expression of Ceramide-Metabolizing Enzymes in the Heart Adipose Tissue of Cardiovascular Disease Patients
Abstract
1. Introduction
2. Results
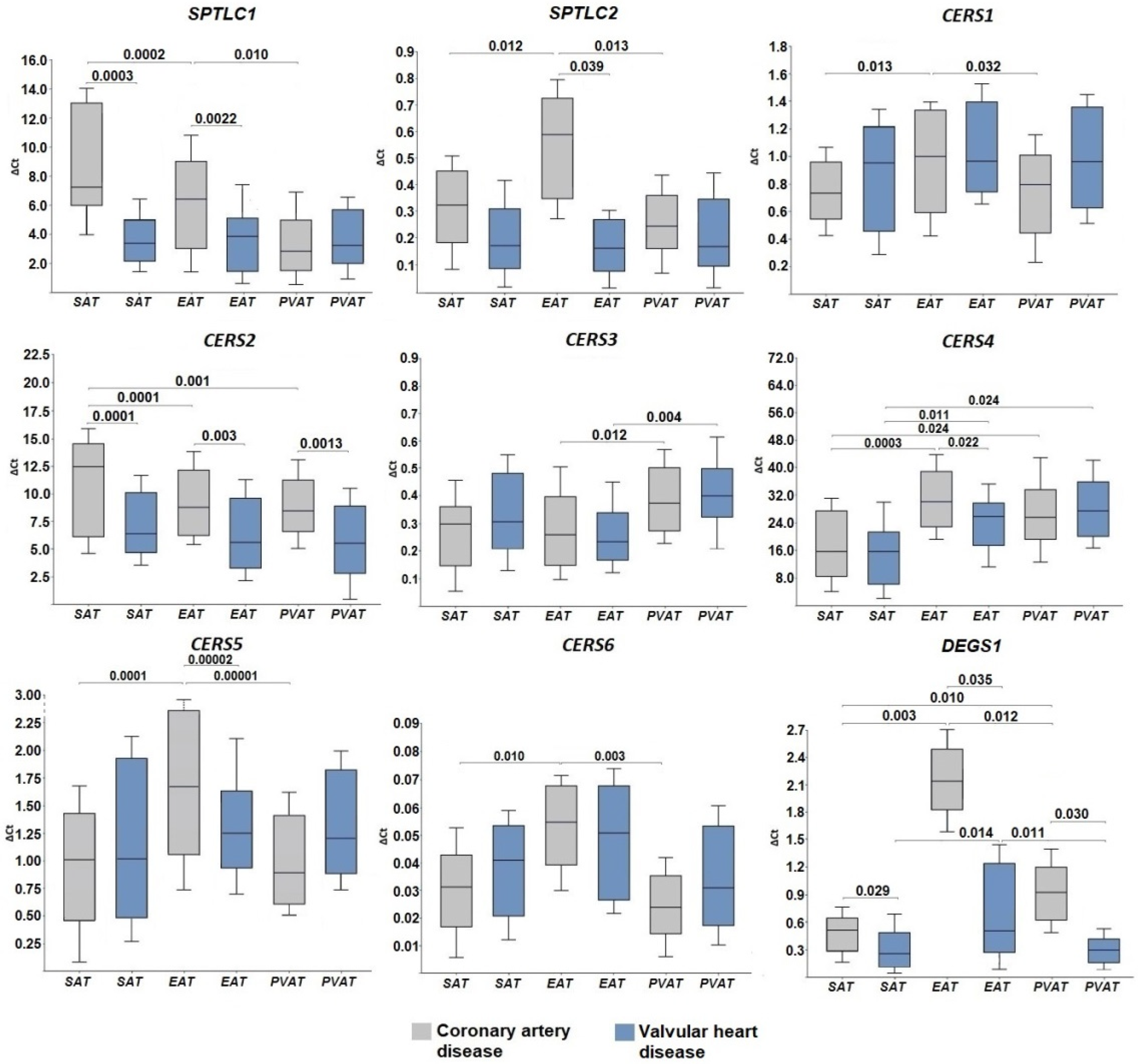
2.1. Expression of Genes for De Novo Ceramide Biosynthesis Enzymes
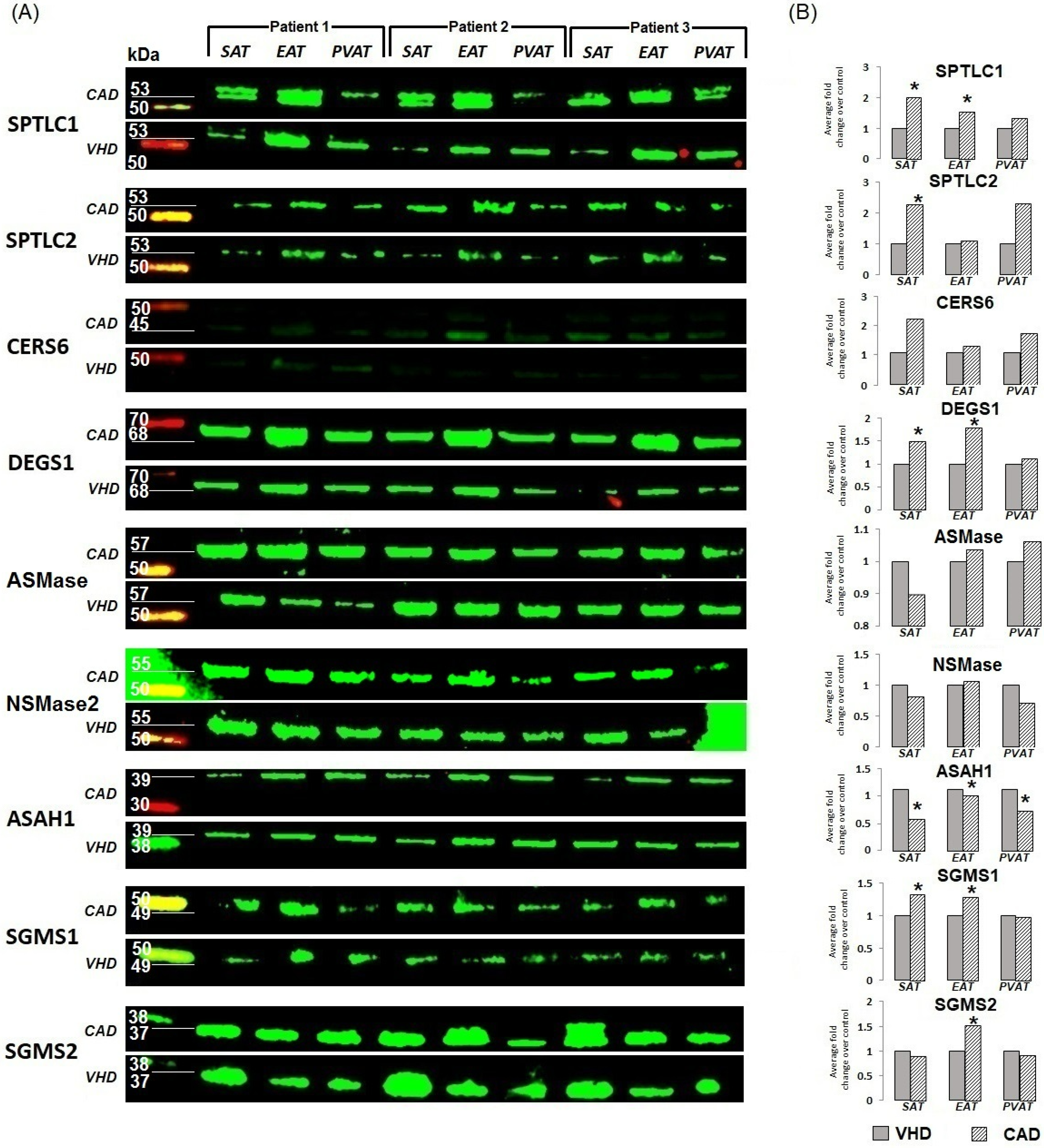
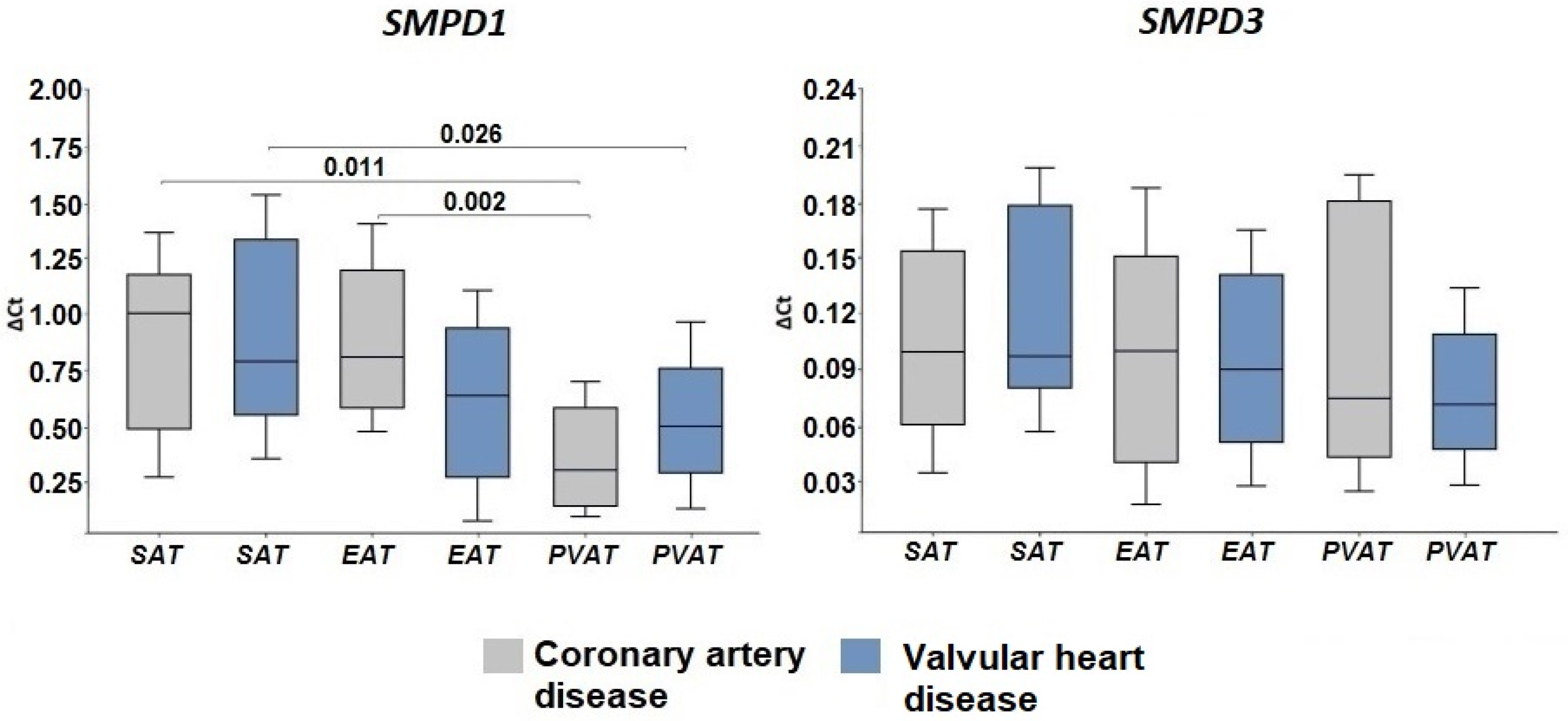
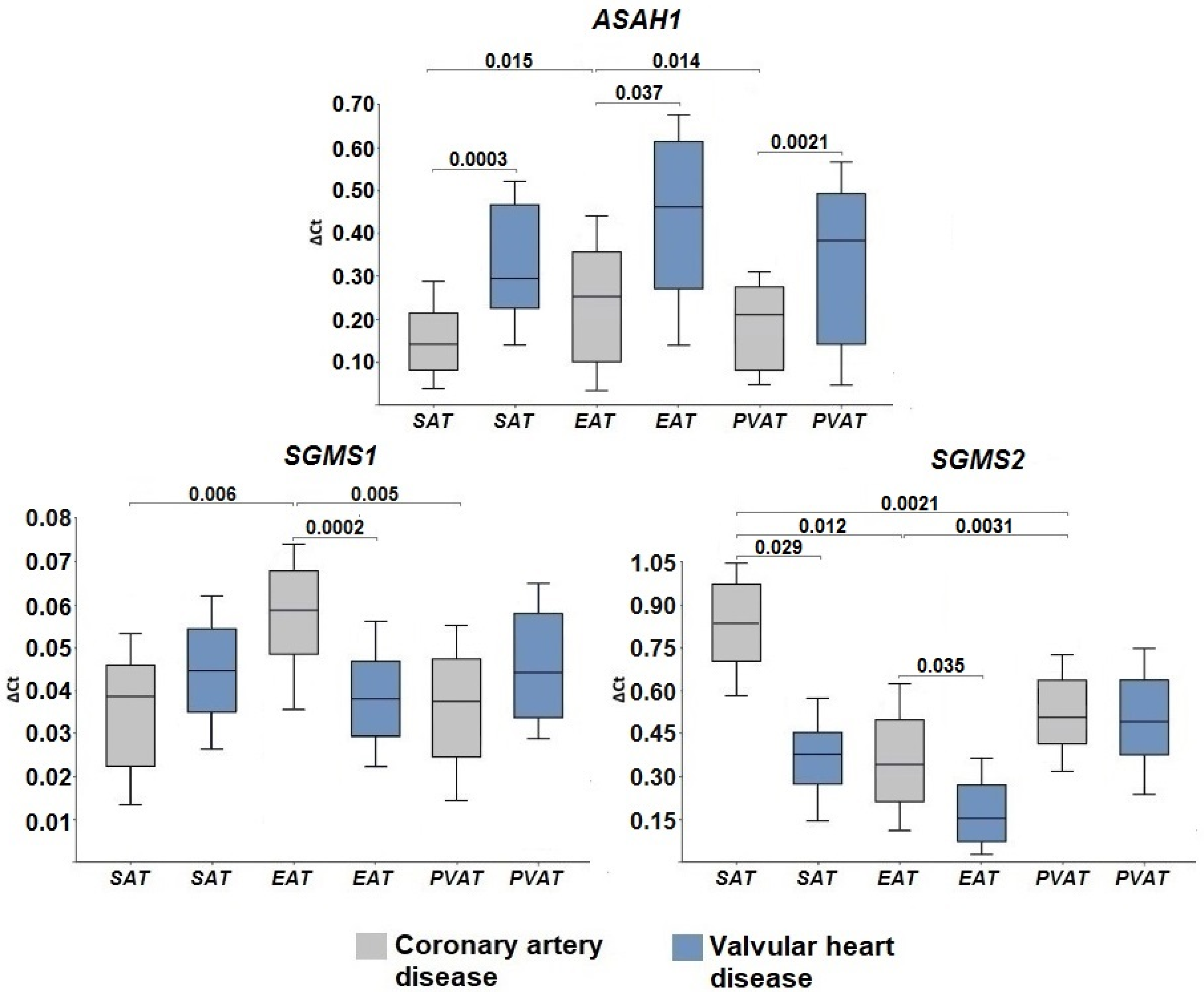
2.2. Expression of Genes Encoding Enzymes of the Sphingomyelinase Pathway for Ceramide Synthesis
2.3. Gene Expression of Ceramide Degradation Enzymes
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Sample Collection
4.3. RNA Extraction and Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
4.4. Fluorescent Western Blotting
4.5. Statistical Analysis
5. Conclusions
Study Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Eckel, R.; Kahn, R.; Robertson, R.; Rizza, R. Preventing Cardiovascular Disease and Diabetes: A Call to Action from the American Diabetes Association and the American Heart Association. Diabetes Care 2006, 29, 1697–1699. [Google Scholar] [CrossRef]
- Iacobellis, G. Epicardial adipose tissue in contemporary cardiology. Nat. Rev. Cardiol. 2022, 19, 593–606. [Google Scholar] [CrossRef] [PubMed]
- Gruzdeva, O.V.; Dyleva, Y.A.; Belik, E.V.; Sinitsky, M.Y.; Stasev, A.N.; Kokov, A.N.; Brel, N.K.; Krivkina, E.O.; Bychkova, E.E.; Tarasov, R.S.; et al. Relationship between Epicardial and Coronary Adipose Tissue and the Expression of Adiponectin, Leptin, and Interleukin 6 in Patients with Coronary Artery Disease. J. Pers. Med. 2022, 12, 129. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-S.; Ahn, S.-G.; Hwang, J.-W.; Lim, H.S.; Choi, B.J.; Choi, S.Y.; Yoon, M.H.; Hwang, G.S.; Tahk, S.J.; Shin, J.H.; et al. Impact of body mass index on the relationship of epicardial adipose tissue to metabolic syndrome and coronary artery disease in an Asian population. Cardiovasc. Diabetol. 2010, 9, 29. [Google Scholar] [CrossRef]
- Park, S.S.; Jung, J.; Mintz, G.S.; Jin, U.; Park, J.-S.; Park, B.; Shin, H.-B.; Seo, K.-W.; Yang, H.-M.; Lim, H.-S.; et al. Epicardial adipose tissue thickness is related to plaque composition in coronary artery disease. Diagnostics 2022, 12, 2836. [Google Scholar] [CrossRef] [PubMed]
- Hogea, T.; Suciu, B.A.; Ivănescu, A.D.; Carașca, C.; Chinezu, L.; Arbănași, E.M.; Russu, E.; Kaller, R.; Arbănași, E.M.; Mureșan, A.V.; et al. Increased Epicardial Adipose Tissue (EAT), Left Coronary Artery Plaque Morphology, and Valvular Atherosclerosis as Risks Factors for Sudden Cardiac Death from a Forensic Perspective. Diagnostics 2023, 13, 142. [Google Scholar] [CrossRef] [PubMed]
- Summers, S.A.; Chaurasia, B.; Holland, W.L. Metabolic Messengers: Ceramides. Nat. Metab. 2019, 1, 1051–1058. [Google Scholar] [CrossRef] [PubMed]
- Mielke, M.M.; Bandaru, V.V.; Han, D.; Resnick, S.M.; Ferrucci, L.; Haughey, N.J. Factors affecting longitudinal trajectories of plasma sphingomyelins: The Baltimore Longitudinal Study of Aging. Aging Cell. 2015, 14, 112–121. [Google Scholar] [CrossRef]
- Park, T.S.; Rosebury, W.; Kindt, E.K.; Kowala, M.C.; Panek, R.L. Serine palmitoyltransferase inhibitor myriocin induces the regression of atherosclerotic plaques in hyperlipidemic ApoE-deficient mice. Pharmacol. Res. 2008, 58, 45–51. [Google Scholar] [CrossRef]
- McGurk, K.A.; Keavney, B.D.; Nicolaou, A. Circulating ceramides as biomarkers of cardiovascular disease: Evidence from phenotypic and genomic studies. Atherosclerosis 2021, 327, 18–30. [Google Scholar] [CrossRef]
- Li, Y.; Talbot, C.L.; Chaurasia, B. Ceramides in Adipose Tissue. Front. Endocrinol. 2020, 11, 407. [Google Scholar] [CrossRef] [PubMed]
- Tidhar, R.; Zelnik, I.D.; Volpert, G.; Ben-Dor, S.; Kelly, S.; Merrill, A.H., Jr.; Futerman, A.H. Eleven residues determine the acyl chain specificity of ceramide synthases. J. Biol. Chem. 2018, 287, 3197–3206. [Google Scholar] [CrossRef]
- Chaurasia, B.; Kaddai, V.A.; Lancaster, G.I.; Henstridge, D.C.; Sriram, S.; Galam, D.L.A.; Gopalan, V.; Prakash, K.N.B.; Velan, S.S.; Bulchand, S.; et al. Adipocyte Ceramides Regulate Subcutaneous Adipose Browning, Inflammation, and Metabolism. Cell Metab. 2016, 24, 820–834. [Google Scholar] [CrossRef] [PubMed]
- Choi, R.H.; Tatum, S.M.; Symons, J.D.; Summers, S.A.; Holland, W.L. Ceramides and other sphingolipids as drivers of cardiovascular disease. Nat. Rev. Cardiol. 2021, 18, 701–711. [Google Scholar] [CrossRef]
- Bandet, C.L.; Tan-Chen, S.; Bourron, O.; Le, S.H.; Hajduch, E. Sphingolipid Metabolism: New Insight into Ceramide-Induced Lipotoxicity in Muscle Cells. Int. J. Mol. Sci. 2019, 20, 479. [Google Scholar] [CrossRef] [PubMed]
- Chaurasia, B.; Talbot, C.L.; Summers, S.A. Adipocyte Ceramides-The Nexus of Inflammation and Metabolic Disease. Front. Immunol. 2020, 11, 576347. [Google Scholar] [CrossRef]
- Hammerschmidt, P.; Brüning, J.C. Contribution of specific ceramides to obesity-associated metabolic diseases. Cell. Mol. Life Sci. 2022, 79, 395. [Google Scholar] [CrossRef]
- Błachnio-Zabielska, A.U.; Baranowski, M.; Hirnle, T.; Zabielski, P.; Lewczuk, A.; Dmitruk, I.; Górski, J. Increased bioactive lipids content in human subcutaneous and epicardial fat tissue correlates with insulin resistance. Lipids 2012, 47, 1131–1141. [Google Scholar] [CrossRef]
- Shen, Y.; Zhao, Z.; Zhang, L.; Shi, L.; Shahriar, S.; Chan, R.B.; Di Paolo, G.; Min, W. Metabolic activity induces membrane phase separation in endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2017, 114, 13394–13399. [Google Scholar] [CrossRef]
- Holland, W.L.; Bikman, B.T.; Wang, L.P.; Yuguang, G.; Sargent, K.M.; Bulchand, S.; Knotts, T.A.; Shui, G.; Clegg, D.J.; Wenk, M.R.; et al. Lipid-induced insulin resistance mediated by the proinflammatory receptor TLR4 requires saturated fatty acid-induced ceramide biosynthesis in mice. J. Clin. Investig. 2011, 121, 1858–1870. [Google Scholar] [CrossRef]
- Gill, J.M.; Sattar, N. Ceramides a new player in the inflammation-insulin resistance paradigm? Diabetologia 2009, 52, 2475–2477. [Google Scholar] [CrossRef] [PubMed]
- Meyer, S.G.; de Groot, H. Cycloserine and threo-dihydrosphingosine inhibit TNF-alpha-induced cytotoxicity: Evidence for the importance of de novo ceramide synthesis in TNF-alpha signaling. Biochim. Biophys. Acta. 2003, 1643, 1–4. [Google Scholar] [CrossRef]
- Mahabadi, A.A.; Berg, M.H.; Lehmann, N.; Kälsch, H.; Bauer, M.; Kara, K.; Dragano, N.; Moebus, S.; Jöckel, K.H.; Erbel, R.; et al. Association of epicardial fat with cardiovascular risk factors and incident myocardial infarction in the general population: The Heinz Nixdorf Recall Study. J. Am. Coll. Cardiol. 2013, 61, 1388–1395. [Google Scholar] [CrossRef] [PubMed]
- Goossens, G.H. The role of adipose tissue dysfunction in the pathogenesis of obesity-related insulin resistance. Physiol. Behav. 2008, 94, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Ginkel, C.; Hartmann, D.; vom Dorp, K.; Zlomuzica, A.; Farwanah, H.; Eckhardt, M.; Sandhoff, R.; Degen, J.; Rabionet, M.; Dere, E.; et al. Ablation of neuronal ceramide synthase 1 in mice decreases ganglioside levels and expression of myelin-associated glycoprotein in oligodendrocytes. J. Biol. Chem. 2012, 287, 41888–41902. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.R.; Lee, E.J.; Shin, K.O.; Kim, M.H.; Pewzner-Jung, Y.; Lee, Y.M.; Park, J.W.; Futerman, A.H.; Park, W.J. Hepatic triglyceride accumulation via endoplasmic reticulum stress-induced SREBP-1 activation is regulated by ceramide synthases. Exp. Mol. Med. 2019, 51, 1–16. [Google Scholar] [CrossRef]
- Goldenberg, J.R.; Carley, A.N.; Ji, R.; Zhang, X.; Fasano, M.; Schulze, P.C.; Lewandowski, E.D. Preservation of Acyl Coenzyme A Attenuates Pathological and Metabolic Cardiac Remodeling Through Selective Lipid Trafficking. Circulation 2019, 139, 2765–2777. [Google Scholar] [CrossRef]
- Laviad, E.L.; Albee, L.; Pankova-Kholmyansky, I.; Park, H.; Merrill, A.H., Jr.; Futerman, A.H. Characterization of ceramide synthase 2: Tissue distribution, substrate specificity, and inhibition by sphingosine 1-phosphate. J. Biol. Chem. 2008, 283, 5677–5684. [Google Scholar] [CrossRef]
- Atilla-Gokcumen, G.E.; Muro, E.; Relat-Goberna, J.; Sasse, S.; Bedigian, A.; Coughlin, M.L.; Garcia-Manyes, S.; Eggert, U.S. Dividing cells regulate their lipid composition and localization. Cell 2014, 156, 428–439. [Google Scholar] [CrossRef]
- Law, B.A.; Liao, X.; Moore, K.S.; Southard, A.; Roddy, P.; Ji, R.; Szulc, Z.; Bielawska, A.; Schulze, P.C.; Cowart, L.A. Lipotoxic, very-long-chain ceramides cause mitochondrial dysfunction, oxidative stress, and cell death in cardiomyocytes. FASEB J. 2018, 32, 1403–1416. [Google Scholar] [CrossRef]
- Gosejacob, D.; Jäger, P.S.; Vom Dorp, K.; Frejno, M.; Carstensen, A.C.; Köhnke, M.; Degen, J.; Dörmann, P.; Hoch, M. Ceramide Synthase 5 Is Essential to Maintain C16:0-Ceramide Pools and Contributes to the Development of Diet-induced Obesity. J. Biol. Chem. 2016, 291, 6989–7003. [Google Scholar] [CrossRef]
- Kolak, M.; Gertow, J.; Westerbacka, J.; Summers, S.A.; Liska, J.; Franco-Cereceda, A.; Orešič, M.; Yki-Järvinen, H.; Eriksson, P.; Fisher, R.M. Expression of ceramide-metabolising enzymes in subcutaneous and intra-abdominal human adipose tissue. Lipids Health Dis. 2012, 11, 115. [Google Scholar] [CrossRef] [PubMed]
- Devlin, C.M.; Lahm, T.; Hubbard, W.C.; Van Demark, M.; Wang, K.C.; Wu, X.; Bielawska, A.; Obeid, L.M.; Petrache, I.M. Dihydroceramide-based response to hypoxia. J. Biol. Chem. 2011, 286, 38069–38078. [Google Scholar] [CrossRef] [PubMed]
- Cataldi, S.; Borrelli, A.; Ceccarini, M.R.; Nakashidze, I.; Codini, M.; Belov, O.; Ivanov, A.; Krasavin, E.; Ferri, I.; Conte, C.; et al. Acid and Neutral Sphingomyelinase Behavior in Radiation-Induced Liver Pyroptosis and in the Protective/Preventive Role of RMnSOD. Int. J. Mol. Sci. 2020, 21, 3281. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, K.; Erlich, S.; Perl, D.P.; Ferlinz, K.; Bisgaier, C.L.; Sandhoff, K.; Desnick, R.J.; Stewart, C.L.; Schuchman, E.H. Acid sphingomyelinase deficient mice: A model of types A and B Niemann-Pick disease. Nat. Genet. 1995, 10, 288–293. [Google Scholar] [CrossRef]
- Argaud, L.; Prigent, A.F.; Chalabreysse, L.; Loufouat, J.; Lagarde, M.; Ovize, M. Ceramide in the antiapoptotic effect of ischemic preconditioning. Am. J. Physiol. Heart Circ. Physiol. 2004, 286, H246–H251. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Yu, J.; Shi, R.; Yan, L.; Yang, T.; Li, Y.; Zhang, Z.; Yu, G.; Bai, Y.; Schuchman, E.H.; et al. Elevation of ceramide and activation of secretory acid sphingomyelinase in patients with acute coronary syndromes. Coron. Artery Dis. 2014, 25, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Pavoine, C.; Pecker, F. Sphingomyelinases: Their regulation and roles in cardiovascular pathophysiology. Cardiovasc. Res. 2009, 82, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Cuschieri, J.; Bulger, E.; Billgrin, J.; Garcia, I.; Maier, R.V. Acid sphingomyelinase is required for lipid Raft TLR4 complex formation. Surg. Infect. 2007, 8, 91–106. [Google Scholar] [CrossRef]
- Wang, P.; Zeng, G.; Yan, Y.; Zhang, S.Y.; Dong, Y.; Zhang, Y.; Zhang, X.; Liu, H.; Zhang, Z.; Jiang, C.; et al. Disruption of adipocyte HIF-1α improves atherosclerosis through the inhibition of ceramide generation. Acta Pharm. Sin. B 2022, 12, 1899–1912. [Google Scholar] [CrossRef]
- Parveen, F.; Bender, D.; Law, S.H. Role of Ceramidases in Sphingolipid Metabolism and Human Diseases. Cells 2019, 8, 1573. [Google Scholar] [CrossRef]
- Gault, C.R.; Obeid, L.M.; Hannun, Y.A. An overview of sphingolipid metabolism: From synthesis to breakdown. Adv. Exp. Med. Biol. 2010, 688, 1–23. [Google Scholar] [CrossRef]
- Zietzer, A.; Düsing, P.; Reese, L.; Nickenig, G.; Jansen, F. Ceramide Metabolism in Cardiovascular Disease: A Network With High Therapeutic Potential. Arterioscler. Thromb. Vasc. Biol. 2022, 42, 1220–1228. [Google Scholar] [CrossRef] [PubMed]
- Hanada, K. Intracellular trafficking of ceramide by ceramide transfer protein. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2010, 86, 426–437. [Google Scholar] [CrossRef]
- Li, Z.; Chiang, Y.P.; He, M.; Zhang, K.; Zheng, J.; Wu, W.; Cai, J.; Chen, Y.; Chen, G.; Chen, Y.; et al. Effect of liver total sphingomyelin synthase deficiency on plasma lipid metabolism. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2021, 1866, 158898. [Google Scholar] [CrossRef] [PubMed]
- Schlitt, A.; Blankenberg, S.; Yan, D.; von Gizycki, H.; Buerke, M.; Werdan, K.; Bickel, C.; Lackner, K.J.; Meyer, J.; Rupprecht, H.J.; et al. Further evaluation of plasma sphingomyelin levels as a risk factor for coronary artery disease. Nutr. Metab. 2006, 3, 5. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Lee, S.H.; Shin, M.J.; Hwang, G.S. Alteration in metabolic signature and lipid metabolism in patients with angina pectoris and myocardial infarction. PLoS ONE 2015, 10, e0135228. [Google Scholar] [CrossRef]
| Parameter | Patients with CAD, n = 30 | Patients with VHD, n = 30 | p |
|---|---|---|---|
| Male gender, n (%) | 18 (60.0) | 17 (56.7) | 0.056 |
| Age, median (range), years | 64.9 (47.8; 69.5) | 59.3 (43.7; 62.1) | 0.071 |
| Body mass index, median (range), kg/m2 | 26.4 (22.5; 30.2) | 27.3 (23.4; 31.2) | 0.062 |
| Arterial hypertension, n (%) | 17 (56.7) | 7 (23.3) | 0.002 |
| Hypercholesterolemia, n (%) | 13 (43.3) | 3 (10.0) | 0.001 |
| Smoking, n (%) | 15 (50.0) | 5 (16.7) | 0.0001 |
| Family history of CAD, n (%) | 18 (60.0) | 11 (36.7) | 0.038 |
| Past medical history of myocardial infarction, n (%) | 21 (70.0) | 0 | |
| Past medical history of stroke, n (%) | 3 (10.0) | 0 | |
| Extracoronary atherosclerosis, n (%) | 5 (16.7) | 0 | |
| No chronic coronary syndrome, n (%) | 1 (3.3) | 30 (100.0) | 0.0001 |
| Chronic coronary syndrome, FC I, n (%) | 0 | 0 | |
| Chronic coronary syndrome, FC II, n (%) | 14 (46.7) | 0 | |
| Chronic coronary syndrome, FC III, n (%) | 15 (50.0) | 0 | |
| Chronic coronary syndrome, FC IV, n (%) | 0 | 0 | |
| Chronic heart failure, NYHA I FC, n (%) | 12 (40.0) | 7 (23.3) | 0.055 |
| Chronic heart failure, CHF NYHA II FC, n (%) | 4 (13.3) | 23 (76.7) | 0.002 |
| Chronic heart failure, NYHA III FC, n (%) | 0 | 0 | |
| Chronic heart failure, NYHA IV FC, n (%) | 0 | 0 | |
| Atherosclerosis of 1 coronary artery, n (%) | 4 (13.3) | 0 | |
| Atherosclerosis of 2 coronary arteries, n (%) | 1 (3.3) | 0 | |
| Atherosclerosis of ≥3 coronary arteries, n (%) | 24 (80.0) | 0 | |
| Ejection fraction, median (range), % | 53.6 (46.3; 58.9) | 51.6 (42.5; 55.8) | 0.046 |
| Treatment strategy (hospital period) | |||
| Aspirin, n (%) | 28 (93.3) | 0 | |
| Clopidogrel, n (%) | 4 (13.3) | 0 | |
| Warfarin, n (%) | 0 | 25 (83.3) | |
| β-blockers, n (%) | 27 (90.0) | 26 (86.7) | 0.091 |
| Angiotensin-converting enzyme inhibitors, n (%) | 23 (76.7) | 24 (80.0) | 0.247 |
| Statins, n (%) | 30 (100.0) | 22 (73.3) | 0.059 |
| Calcium channel blockers, n (%) | 23 (76.7) | 21 (70.0) | 0.166 |
| Nitrates, n (%) | 1 (3.3) | 2 (6.7) | 0.107 |
| Diuretics, n (%) | 24 (80.0) | 25 (83.3) | 0.087 |
| Gene | Orientation | Sequence (5′ -> 3′) | Primer Length |
|---|---|---|---|
| Serine palmitoyltransferase subunit C1 gene (SPTLC1) | Forward primer | aggaagcggctaactatggc | 20 |
| Reverse primer | ccagaggatcagaatcccttcc | 22 | |
| Serine palmitoyltransferase subunit C1 gene (SPTLC2) | Forward primer | cgcctgaaagagatgggcttc | 21 |
| Reverse primer | ccgatgttccgcttcagcat | 20 | |
| Ceramide synthase 1 genes (CERS1) | Forward primer | gcgtttgcagccaaggtgtt | 20 |
| Reverse primer | ttcaccaggccgttcctcag | 20 | |
| Ceramide synthase 2 genes (CERS2) | Forward primer | ggacgtgtctacgccaaagc | 20 |
| Reverse primer | atgttcaagagggcagccagt | 21 | |
| Ceramide synthase 3 genes (CERS3) | Forward primer | ctcgcacagatggtgtcctg | 20 |
| Reverse primer | cctgatgggatgttgcttcctg | 22 | |
| Ceramide synthase 4 genes (CERS4) | Forward primer | caggacttgttggcagccct | 20 |
| Reverse primer | cgttgggcttcacttgcctc | 20 | |
| Ceramide synthase 5 genes (CERS5) | Forward primer | ctcaatggcctgctgctgac | 20 |
| Reverse primer | tgctctccacatcactgcga | 20 | |
| Ceramide synthase 6 genes (CERS6) | Forward primer | cggacctgaagaacacggagga | 22 |
| Reverse primer | atggcgcacggtttggctac | 20 | |
| Dihydroceramide desaturase 1 gene (DEGS1) | Forward primer | ccactgagctggagtttcct | 20 |
| Reverse primer | caggaattgtagtgagggaggt | 22 | |
| Acid sphingomyelinase gene (SMPD1) | Forward primer | ccgctggctctatgaagcgat | 21 |
| Reverse primer | cggggtatggggaaagagcat | 21 | |
| Neutral sphingomyelinase gene (SMPD3) | Forward primer | gaaggacaacaaggtcccagt | 21 |
| Reverse primer | caactccggctggtcaatgg | 20 | |
| Acid ceramidase gene (ASAH1) | Forward primer | ctgaaccgcaccagccaaga | 20 |
| Reverse primer | ggcagtcccgcaggtaagttt | 21 | |
| Sphingomyelin synthase 1 gene (SGMS1) | Forward primer | ccagtgcaacgtgacgacag | 20 |
| Reverse primer | agtccacactccttcagtcgct | 22 | |
| Sphingomyelin synthase 2 gene (SGMS2) | ccgctggctctatgaagcgat | actctacctgtgcctggaatgc | 22 |
| cggggtatggggaaagagcat | |||
| Reverse primer | tcagtgtcagcgtaaccgtgt | 21 | |
| Beta-actin (ACTB) | Forward primer | catcgagcacggcatcgtca | 20 |
| Reverse primer | tagcacagcctggacagcaac | 21 | |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | Forward primer | agccacatcgctcagacac | 19 |
| Reverse primer | gcccaatacgaccaaatcc | 19 | |
| Beta-2-microglobulin (B2M) | Forward primer | tccatccgacattgaagttg | 20 |
| Reverse primer | cggcaggcatactcatctt | 19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gruzdeva, O.; Dyleva, Y.; Belik, E.; Uchasova, E.; Ponasenko, A.; Ivanov, S.; Zinets, M.; Stasev, A.; Kutikhin, A.; Markova, V.; et al. Expression of Ceramide-Metabolizing Enzymes in the Heart Adipose Tissue of Cardiovascular Disease Patients. Int. J. Mol. Sci. 2023, 24, 9494. https://doi.org/10.3390/ijms24119494
Gruzdeva O, Dyleva Y, Belik E, Uchasova E, Ponasenko A, Ivanov S, Zinets M, Stasev A, Kutikhin A, Markova V, et al. Expression of Ceramide-Metabolizing Enzymes in the Heart Adipose Tissue of Cardiovascular Disease Patients. International Journal of Molecular Sciences. 2023; 24(11):9494. https://doi.org/10.3390/ijms24119494
Chicago/Turabian StyleGruzdeva, Olga, Yulia Dyleva, Ekaterina Belik, Evgenia Uchasova, Anastasia Ponasenko, Sergey Ivanov, Maxim Zinets, Alexander Stasev, Anton Kutikhin, Victoria Markova, and et al. 2023. "Expression of Ceramide-Metabolizing Enzymes in the Heart Adipose Tissue of Cardiovascular Disease Patients" International Journal of Molecular Sciences 24, no. 11: 9494. https://doi.org/10.3390/ijms24119494
APA StyleGruzdeva, O., Dyleva, Y., Belik, E., Uchasova, E., Ponasenko, A., Ivanov, S., Zinets, M., Stasev, A., Kutikhin, A., Markova, V., Poddubnyak, A., Gorbatovskaya, E., Fanaskova, E., & Barbarash, O. (2023). Expression of Ceramide-Metabolizing Enzymes in the Heart Adipose Tissue of Cardiovascular Disease Patients. International Journal of Molecular Sciences, 24(11), 9494. https://doi.org/10.3390/ijms24119494





