A Novel Approach for Screening Sericin-Derived Therapeutic Peptides Using Transcriptomics and Immunoprecipitation
Abstract
1. Introduction
2. Results
2.1. Construction of Sericin Peptide Library and Its Promotion of Extracellular Matrix Synthesis in HaCaT Cells
2.2. Sericin Peptide Library Promotes Transcription of the Extracellular Matrix by Regulating the TGF-β Signaling Pathway
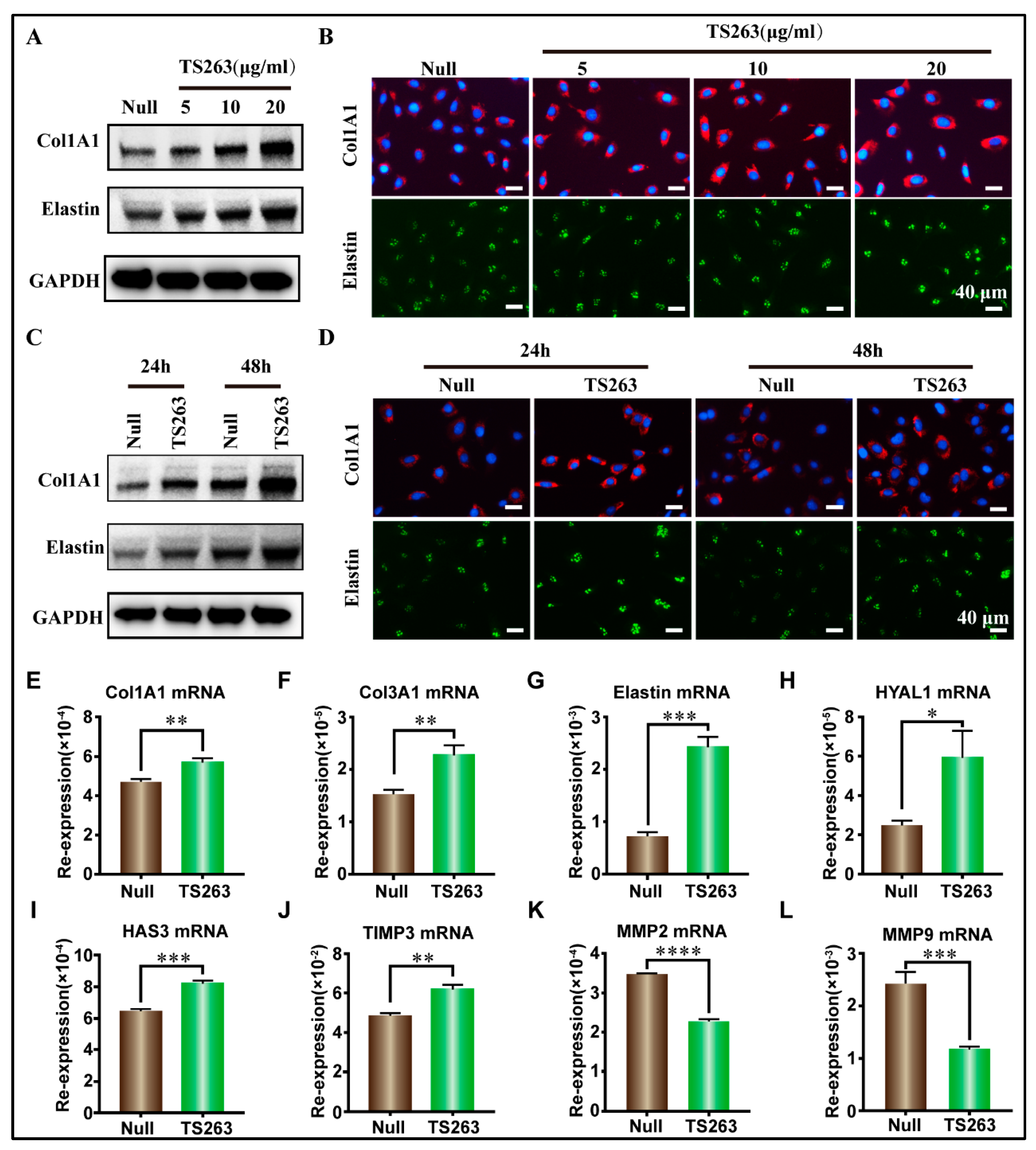
2.3. Targeted Screening of TTPs Based on CoIP Technology
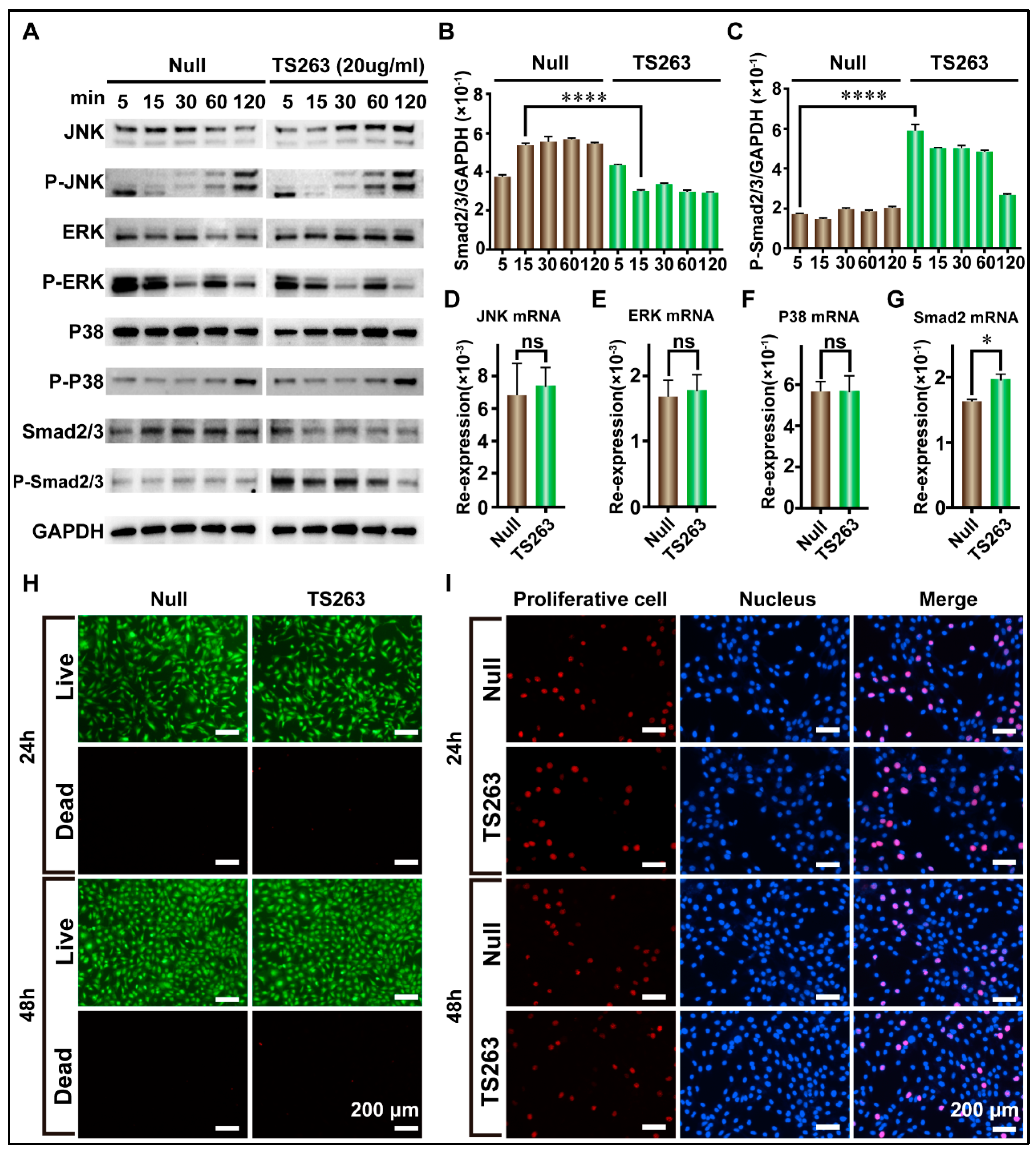
2.4. TTP Regulates the TGF-β Signaling Pathway to Promote Extracellular Matrix Synthesis in HaCaT Cells
3. Discussion
4. Methods and Materials
4.1. Cell Culture
4.2. Preparation of Sericin Peptide Library
4.3. Sericin Peptide Library Promotes Transcription and Synthesis of Extracellular Matrix Proteins
4.4. SDS-PAGE and Western Blotting
4.5. Transcriptome Analysis and Reverse Transcription Polymerase Chain Reaction (RT-PCR)
4.6. Immunofluorescence Staining
4.7. Co-Immunoprecipitation (CoIP)
4.8. LC-MS Analysis
4.9. Molecular Docking, Molecular Dynamics, and Calculation of Molecular Free Energy
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fosgerau, K.; Hoffmann, T. Peptide therapeutics: Current status and future directions. Drug Discov. Today 2015, 20, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, S.; Guha, S.; Majumder, K. Food-Derived Bioactive Peptides in Human Health: Challenges and Opportunities. Nutrients 2018, 10, 1738. [Google Scholar] [CrossRef]
- Henninot, A.; Collins, J.C.; Nuss, J.M. The Current State of Peptide Drug Discovery: Back to the Future? J. Med. Chem. 2018, 61, 1382–1414. [Google Scholar] [CrossRef] [PubMed]
- Machado, E.S.; Passoni, L.F.; Sidi, L.C.; Andrade, H.B.; De Menezes, J.A. Successful desensitization of enfuvirtide after a first attempt failure. AIDS 2006, 20, 2130–2131. [Google Scholar] [CrossRef] [PubMed]
- Drucker, D.J. Advances in oral peptide therapeutics. Nat. Rev. Drug Discov. 2020, 19, 277–289. [Google Scholar] [CrossRef] [PubMed]
- Lien, S.; Lowman, H.B. Therapeutic peptides. Trends Biotechnol. 2003, 21, 556–562. [Google Scholar] [CrossRef] [PubMed]
- Nong, N.T.P.; Chen, Y.K.; Shih, W.L.; Hsu, J.L. Characterization of Novel Dipeptidyl Peptidase-IV Inhibitory Peptides from Soft-Shelled Turtle Yolk Hydrolysate Using Orthogonal Bioassay-Guided Fractionations Coupled with In Vitro and In Silico Study. Pharmaceuticals 2020, 13, 308. [Google Scholar] [CrossRef]
- Han, Z.; Zhou, Z.; Shi, X.; Wang, J.; Wu, X.; Sun, D.; Chen, Y.; Zhu, H.; Magi-Galluzzi, C.; Lu, Z.R. EDB Fibronectin Specific Peptide for Prostate Cancer Targeting. Bioconjugate Chem. 2015, 26, 830–838. [Google Scholar] [CrossRef]
- Tejano, L.A.; Peralta, J.P.; Yap, E.E.S.; Panjaitan, F.C.A.; Chang, Y.W. Prediction of Bioactive Peptides from Chlorella sorokiniana Proteins Using Proteomic Techniques in Combination with Bioinformatics Analyses. Int. J. Mol. Sci. 2019, 20, 1786. [Google Scholar] [CrossRef]
- Reed, J.; Bain, S.; Kanamarlapudi, V. Recent advances in understanding the role of glucagon-like peptide 1. F10001Research 2020, 9, 239. [Google Scholar] [CrossRef]
- Kornilova, E.S. Receptor-mediated endocytosis and cytoskeleton. Biochemistry 2014, 79, 865–878. [Google Scholar] [CrossRef]
- Wu, Q.; Chen, S.; Zhu, H.; Xu, N.; Yang, Q.; Yao, W.; Gao, X. Biased agonists with less glucagon-like peptide-1 receptor-mediated endocytosis prolong hypoglycaemic effects. Eur. J. Pharmacol. 2021, 907, 174203. [Google Scholar] [CrossRef]
- Fletcher, M.M.; Halls, M.L.; Christopoulos, A.; Sexton, P.M.; Wootten, D. The complexity of signalling mediated by the glucagon-like peptide-1 receptor. Biochem. Soc. Trans. 2016, 44, 582–588. [Google Scholar] [CrossRef]
- Nooren, I.M.; Thornton, J.M. Structural characterisation and functional significance of transient protein-protein interactions. J. Mol. Biol. 2003, 325, 991–1018. [Google Scholar] [CrossRef]
- Lagundzin, D.; Krieger, K.L.; Law, H.C.; Woods, N.T. An optimized co-immunoprecipitation protocol for the analysis of endogenous protein-protein interactions in cell lines using mass spectrometry. STAR Protoc. 2022, 3, 101234. [Google Scholar] [CrossRef]
- Cao, T.T.; Zhang, Y.Q. Processing and characterization of silk sericin from Bombyx mori and its application in biomaterials and biomedicines. Mater. Sci. Eng. C Mater. Biol. Appl. 2016, 61, 940–952. [Google Scholar] [CrossRef]
- Wang, R.; Wang, Y.; Song, J.; Tian, C.; Jing, X.; Zhao, P.; Xia Term, Q. A novel method for silkworm cocoons self-degumming and its effect on silk fibers. J. Adv. Res. 2022. [Google Scholar] [CrossRef]
- Jo, Y.Y.; Kweon, H.; Kim, D.W.; Baek, K.; Chae, W.S.; Kang, Y.J.; Oh, J.H.; Kim, S.G.; Garagiola, U. Silk sericin application increases bone morphogenic protein-2/4 expression via a toll-like receptor-mediated pathway. Int. J. Biol. Macromol. 2021, 190, 607–617. [Google Scholar] [CrossRef]
- Lamboni, L.; Gauthier, M.; Yang, G.; Wang, Q. Silk sericin: A versatile material for tissue engineering and drug delivery. Biotechnol. Adv. 2015, 33, 1855–1867. [Google Scholar] [CrossRef]
- Xu, S.; Yang, Q.; Wang, R.; Tian, C.; Ji, Y.; Tan, H.; Zhao, P.; Kaplan, D.L.; Wang, F.; Xia, Q. Genetically engineered pH-responsive silk sericin nanospheres with efficient therapeutic effect on ulcerative colitis. Acta Biomater. 2022, 144, 81–95. [Google Scholar] [CrossRef]
- Lau, J.L.; Dunn, M.K. Therapeutic peptides: Historical perspectives, current development trends, and future directions. Bioorganic Med. Chem. 2018, 26, 2700–2707. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, N.X.; Zhang, W.P.; Cheng, X.R.; Yan, Z.B.; Shao, G.; Wang, X.; Wang, R.; Fu, C.Y. Therapeutic peptides: Current applications and future directions. Signal Transduct. Target. Ther. 2022, 7, 48. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.S.; Costa, E.C.; Reis, S.; Spencer, C.; Calhelha, R.C.; Miguel, S.P.; Ribeiro, M.P.; Barros, L.; Vaz, J.A.; Coutinho, P. Silk Sericin: A Promising Sustainable Biomaterial for Biomedical and Pharmaceutical Applications. Polymers 2022, 14, 4931. [Google Scholar] [CrossRef] [PubMed]
- Aramwit, P.; Kanokpanont, S.; Nakpheng, T.; Srichana, T. The Effect of Sericin from Various Extraction Methods on Cell Viability and Collagen Production. Int. J. Mol. Sci. 2010, 11, 2200–2211. [Google Scholar] [CrossRef]
- Bos, J.D.; Meinardi, M.M. The 500 Dalton rule for the skin penetration of chemical compounds and drugs. Exp. Dermatol. 2000, 9, 165–169. [Google Scholar] [CrossRef]
- Mayba, J.N.; Gooderham, M.J. A Guide to Topical Vehicle Formulations. J. Cutan. Med. Surg. 2018, 22, 207–212. [Google Scholar] [CrossRef]
- Trenker, R.; Jura, N. Receptor tyrosine kinase activation: From the ligand perspective. Curr. Opin. Cell Biol. 2020, 63, 174–185. [Google Scholar] [CrossRef]
- Floriddia, E. Transcriptomics predicts activity. Nat. Neurosci. 2022, 25, 977. [Google Scholar] [CrossRef]
- Zhao, J.; Qin, B.; Nikolay, R.; Spahn, C.M.T.; Zhang, G. Translatomics: The Global View of Translation. Int. J. Mol. Sci. 2019, 20, 212. [Google Scholar] [CrossRef]
- Anderson, S.J.; Feye, K.M.; Schmidt-McCormack, G.R.; Malovic, E.; Mlynarczyk, G.S.A.; Izbicki, P.; Arnold, L.F.; Jefferson, M.A.; de la Rosa, B.M.; Wehrman, R.F.; et al. Off-Target drug effects resulting in altered gene expression events with epigenetic and “Quasi-Epigenetic” origins. Pharmacol. Res. 2016, 107, 229–233. [Google Scholar] [CrossRef]
- Kandasamy, M.; Anusuyadevi, M.; Aigner, K.M.; Unger, M.S.; Kniewallner, K.M.; de Sousa, D.M.B.; Altendorfer, B.; Mrowetz, H.; Bogdahn, U.; Aigner, L. TGF-beta Signaling: A Therapeutic Target to Reinstate Regenerative Plasticity in Vascular Dementia? Aging Dis. 2020, 11, 828–850. [Google Scholar] [CrossRef]
- Tzavlaki, K.; Moustakas, A. TGF-beta Signaling. Biomolecules 2020, 10, 487. [Google Scholar] [CrossRef]
- Derynck, R.; Budi, E.H. Specificity, versatility, and control of TGF-beta family signaling. Sci. Signal. 2019, 12, eaav5183. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, F.; Xu, S.; Wang, R.; Chen, W.; Hou, K.; Tian, C.; Wang, F.; Yu, L.; Lu, Z.; et al. Genetically engineered bi-functional silk material with improved cell proliferation and anti-inflammatory activity for medical application. Acta Biomater. 2019, 86, 148–157. [Google Scholar] [CrossRef]
- Sagui, C.; Darden, T.A. Molecular dynamics simulations of biomolecules: Long-range electrostatic effects. Annu. Rev. Biophys. Biomol. Struct. 1999, 28, 155–179. [Google Scholar] [CrossRef]
- Krautler, V.; Van Gunsteren, W.F.; Hunenberger, P.H. A fast SHAKE: Algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 2001, 22, 501–508. [Google Scholar] [CrossRef]
- Larini, L.; Mannella, R.; Leporini, D. Langevin stabilization of molecular-dynamics simulations of polymers by means of quasisymplectic algorithms. J. Chem. Phys. 2007, 126, 104101. [Google Scholar] [CrossRef]
- Hou, T.; Wang, J.; Li, Y.; Wang, W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 2011, 51, 69–82. [Google Scholar] [CrossRef]
- Altaf, A.; Janjua, N.Z.; Hutin, Y. The cost of unsafe injections in pakistan and challenges for prevention program. J. Coll. Physicians Surg. Pak. 2006, 16, 622–624. [Google Scholar]
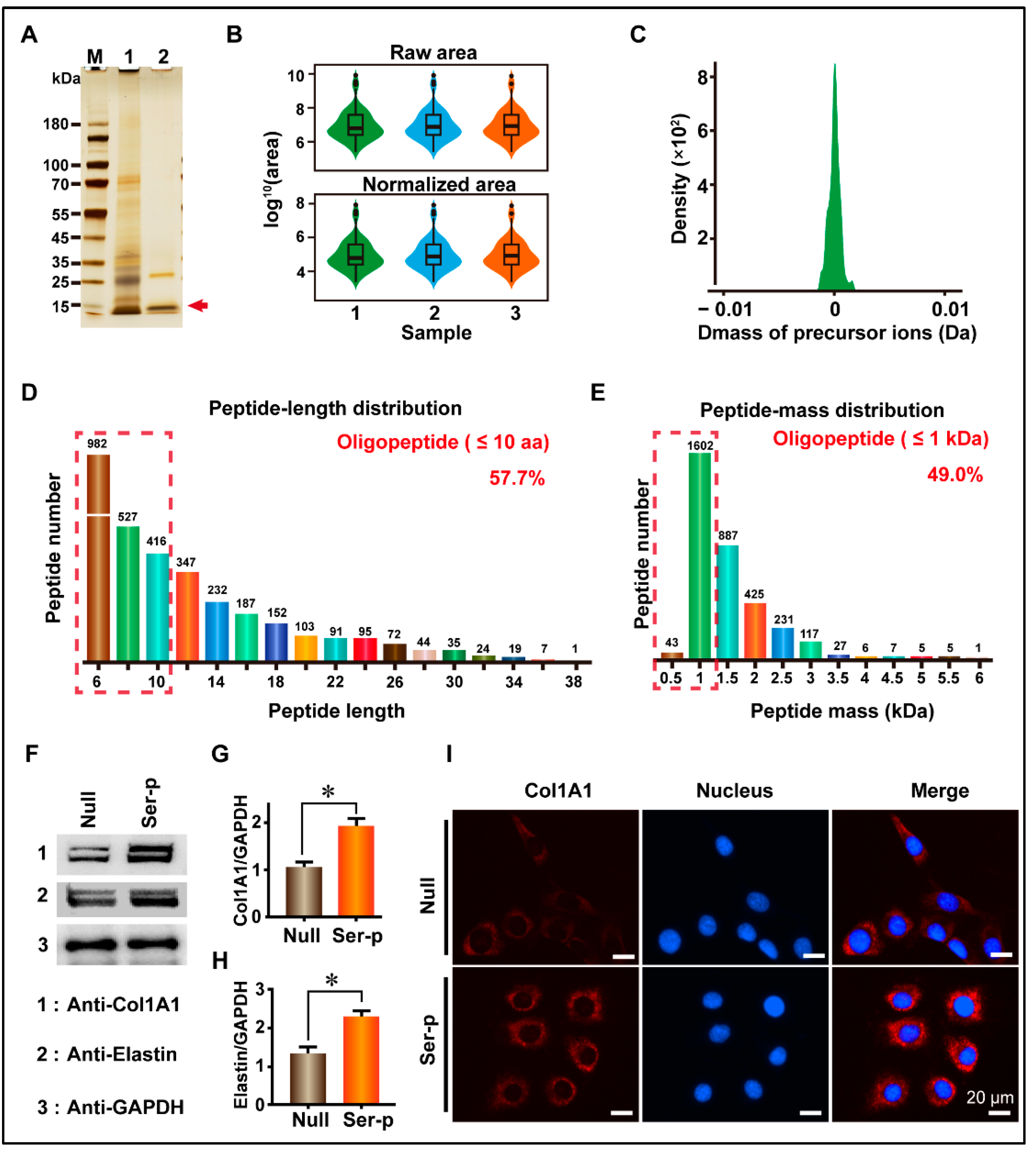
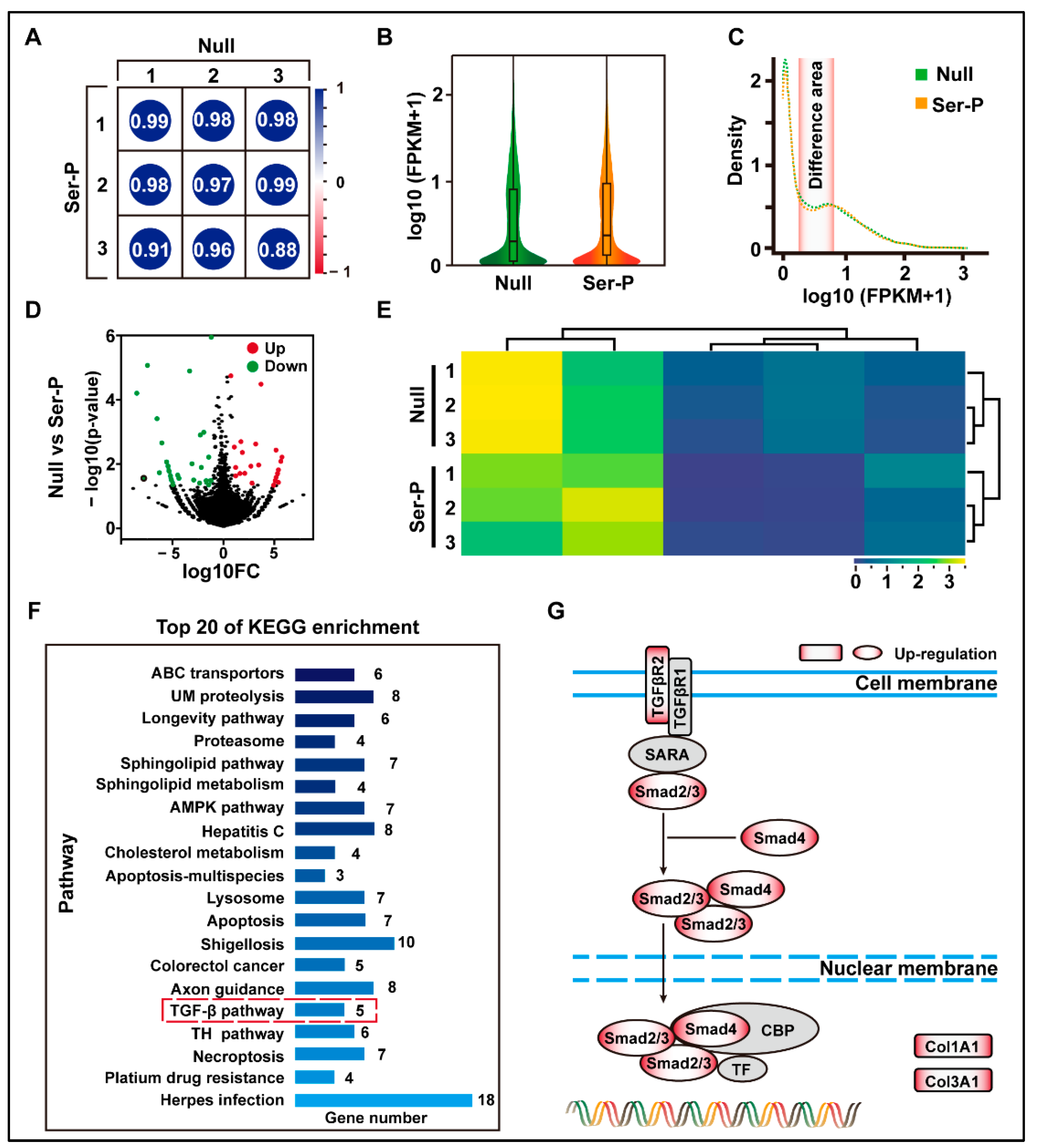
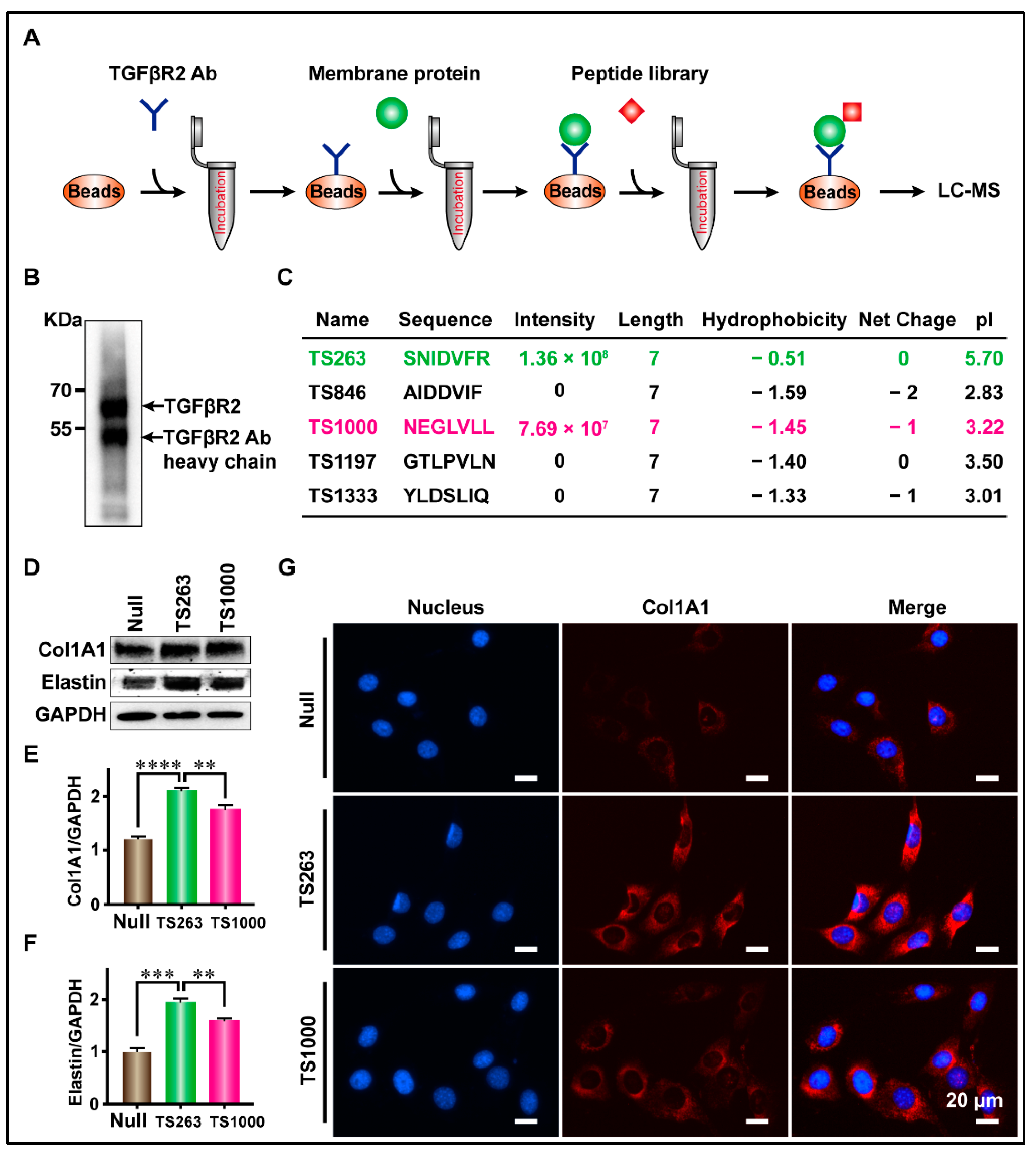
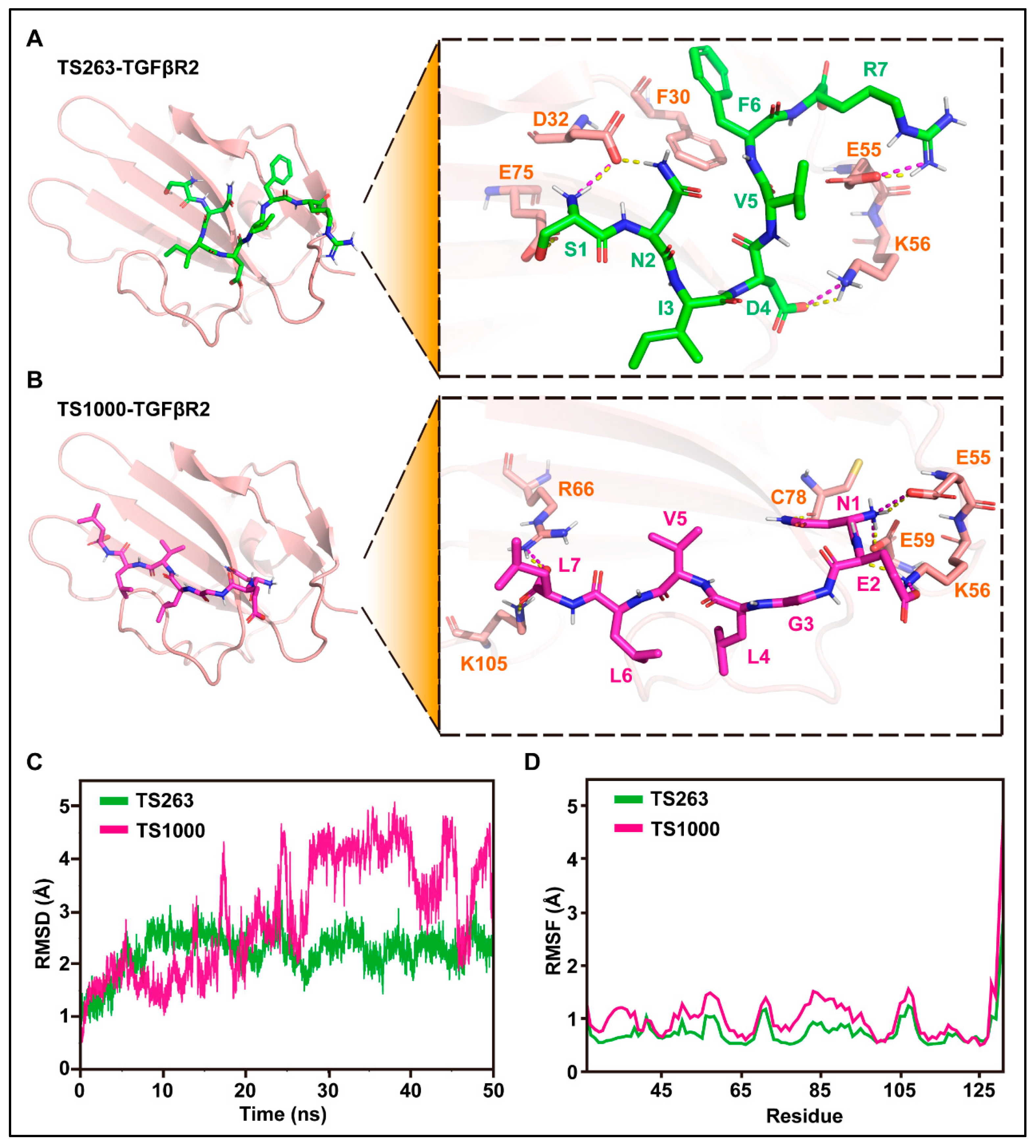
| Name | Col1A1 | Elastin |
|---|---|---|
| Null | 1.11 ± 0.09 | 1.33 ± 0.13 1 |
| Ser-P | 1.91 ± 0.11 | 2.30 ± 0.10 |
| Name | Col1A1 | Elastin |
|---|---|---|
| Null | 1.19 ± 0.04 | 0.99 ± 0.04 |
| TS263 | 2.08 ± 0.03 | 1.94 ± 0.05 |
| TS1000 | 1.73 ± 0.05 | 1.59 ± 0.03 |
| Scheme | TS263_TGFβR2 | TS1000_TGFβR2 |
|---|---|---|
| ΔEvdw | −27.85 ± 3.91 | −24.69 ± 3.69 |
| ΔEelec | −234.13 ± 6.41 | −104.13 ± 7.04 |
| ΔGGB | 245.64 ± 7.42 | 117.90 ± 6.08 |
| ΔGSA | −5.92 ± 0.27 | −4.60 ± 0.06 |
| ΔGbind | −22.26 ± 2.22 | −15.52 ± 2.55 |
| Name | Null | TS263 |
|---|---|---|
| Col1A1 | 4.70 ± 0.08 × 10−1 | 5.68 ± 0.01 × 10−1 |
| Col3A1 | 1.51 ± 0.06 × 10−2 | 2.27 ± 0.11 × 10−2 |
| Elastin | 0.71 ± 0.01 × 100 | 2.41 ± 0.11 × 100 |
| HYAL1 | 2.43 ± 0.17 × 10−2 | 5.92 ± 0.80 × 10−2 |
| HAS3 | 6.40 ± 0.12 × 10−1 | 8.22 ± 0.11 × 10−1 |
| TIMP3 | 4.81 ± 0.10 × 10 | 6.19 ± 0.13 × 10 |
| MMP2 | 3.49 ± 0.01 × 10−1 | 2.28 ± 0.03 × 10−1 |
| MMP9 | 2.40 ± 0.14 × 100 | 1.16 ± 0.03 × 100 |
| Name | Null | TS263 |
|---|---|---|
| JNK | 6.835 ± 1.40 × 10−3 | 7.365 ± 0.69 × 10−3 |
| ERK | 1.694 ± 0.15 × 10−3 | 1.776 ± 0.15 × 10−3 |
| P38 | 5.596 ± 0.31 × 10−1 | 5.633 ± 0.55 × 10−1 |
| Smad2 | 1.632 ± 0.02 × 10−1 | 1.955 ± 0.07 × 10−1 |
| Name. | The Forward Primer | The Reverse Primer |
|---|---|---|
| Col1A1 | 5′-GCAGCCCTGGTGAAAATGGA-3′ | 5′-CAGCACCAGTAGCACCATCA-3′ |
| Col3A1 | 5′-TGGAGGATGGTTGCACGAAA-3′ | 5′-ACAGCCTTGCGTGTTCGATA-3′ |
| Elastin | 5′-GGCCATTCCTGGTGGAGTTCC-3′ | 5′-AACTGGCTTAAGAGGTTTGCCT-3′ |
| HYAL1 | 5′-CTTCAGCCCCAAGGTTGTCC-3′ | 5′-AAGCCTTGGGCCATATCGAG-3′ |
| HAS3 | 5′-TCGGGGAAGAGTGCTCTCAG-3′ | 5′-GTGGATGAACTGGTAGCCCG-3′ |
| TIMP3 | 5′-GTGGCACTCATTGCTTGTGG-3′ | 5′-CAAGGTGACGGGACTGGAAG-3′ |
| MMP2 | 5′-ATCCAGACTTCCTCAGGCGG-3′ | 5′-TCCTGGCAATCCCTTTGTATGT-3′ |
| MMP9 | 5′-GTACTCGACCTGTACCAGCG-3′ | 5′-AGAAGCCCCACTTCTTGTCG-3′ |
| JNK | 5′-TTGGAACACCATGTCCTGAA-3′ | 5′-ATGTACGGGTGTTGGAGAGC-3′ |
| ERK | 5′-CGTGTTTGAGTCAAGCCAGA-3′ | 5′-GCTGCAATGGTATCCACCTT-3′ |
| P38 | 5′-GCCCCAGTAGTCAGAAGCAG-3′ | 5′-TAGGGGCTGAAGAGAGGTGA-3′ |
| Smad2 | 5′-CGAAATGCCACGGTAGAAAT-3′ | 5′-CCAGAAGAGCAGCAAATTCC-3′ |
| GAPDH | 5′-CCATGGGGAAGGTGAAGGTC-3′ | 5′-TGAAGGGGTCATTGATGGCA-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, R.; Wang, Y.; Song, J.; Tan, H.; Tian, C.; Zhao, D.; Xu, S.; Zhao, P.; Xia, Q. A Novel Approach for Screening Sericin-Derived Therapeutic Peptides Using Transcriptomics and Immunoprecipitation. Int. J. Mol. Sci. 2023, 24, 9425. https://doi.org/10.3390/ijms24119425
Wang R, Wang Y, Song J, Tan H, Tian C, Zhao D, Xu S, Zhao P, Xia Q. A Novel Approach for Screening Sericin-Derived Therapeutic Peptides Using Transcriptomics and Immunoprecipitation. International Journal of Molecular Sciences. 2023; 24(11):9425. https://doi.org/10.3390/ijms24119425
Chicago/Turabian StyleWang, Riyuan, Yuancheng Wang, Jianxin Song, Huanhuan Tan, Chi Tian, Dongchao Zhao, Sheng Xu, Ping Zhao, and Qingyou Xia. 2023. "A Novel Approach for Screening Sericin-Derived Therapeutic Peptides Using Transcriptomics and Immunoprecipitation" International Journal of Molecular Sciences 24, no. 11: 9425. https://doi.org/10.3390/ijms24119425
APA StyleWang, R., Wang, Y., Song, J., Tan, H., Tian, C., Zhao, D., Xu, S., Zhao, P., & Xia, Q. (2023). A Novel Approach for Screening Sericin-Derived Therapeutic Peptides Using Transcriptomics and Immunoprecipitation. International Journal of Molecular Sciences, 24(11), 9425. https://doi.org/10.3390/ijms24119425






