Effects of Different Types of Recombinant SARS-CoV-2 Spike Protein on Circulating Monocytes’ Structure
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sampath, S.; Khedr, A.; Qamar, S.; Tekin, A.; Singh, R.; Green, R.; Kashyap, R. Pandemics Throughout the History. Cureus 2021, 13, e18136. [Google Scholar] [CrossRef] [PubMed]
- McVernon, J.; Liberman, J. WHO keeps COVID-19 a public health emergency of international concern. BMJ 2023, 380, 504. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.P.; Liu, S.-L. Role of host factors in SARS-CoV-2 entry. J. Biol. Chem. 2021, 297, 100847. [Google Scholar] [CrossRef]
- Lim, S.; Zhang, M.; Chang, T.L. ACE2-Independent Alternative Receptors for SARS-CoV-2. Viruses 2022, 14, 2535. [Google Scholar] [CrossRef]
- Nocini, R.; Henry, B.M.; Mattiuzzi, C.; Lippi, G. Improving Nasal Protection for Preventing SARS-CoV-2 Infection. Biomedicines 2022, 10, 2966. [Google Scholar] [CrossRef] [PubMed]
- Ligi, D.; Sasso, B.L.; Henry, B.M.; Ciaccio, M.; Lippi, G.; Plebani, M.; Mannello, F. Deciphering the role of monocyte and monocyte distribution width (MDW) in COVID-19: An updated systematic review and meta-analysis. Clin. Chem. Lab. Med. 2023, 61, 960–973. [Google Scholar] [CrossRef] [PubMed]
- Grant, R.A.; Morales-Nebreda, L.; Markov, N.S.; Swaminathan, S.; Querrey, M.; Guzman, E.R.; Abbott, D.A.; Donnelly, H.K.; Donayre, A.; Goldberg, I.A.; et al. Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature 2021, 590, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Lippi, G.; Plebani, M. Cytokine “storm”, cytokine “breeze”, or both in COVID-19? Clin. Chem. Lab. Med. 2020, 59, 637–639. [Google Scholar] [CrossRef]
- Park, J.; Dean, L.S.; Jiyarom, B.; Gangcuangco, L.M.; Shah, P.; Awamura, T.; Ching, L.L.; Nerurkar, V.R.; Chow, D.C.; Igno, F.; et al. Elevated circulating monocytes and monocyte activation in COVID-19 convalescent individuals. Front. Immunol. 2023, 14, 1151780. [Google Scholar] [CrossRef]
- Kilercik, M.; Demirelce, Ö.; Serdar, M.A.; Mikailova, P.; Serteser, M. A new haematocytometric index: Predicting severity and mortality risk value in COVID-19 patients. PLoS ONE 2021, 16, e0254073. [Google Scholar] [CrossRef]
- Ait-Belkacem, I.; García, C.C.; Millet-Wallisky, E.; Izquierdo, N.; Loosveld, M.; Arnoux, I.; Morange, P.-E.; Galland, F.; Lambert, N.; Malergue, F.; et al. SARS-CoV-2 spike protein induces a differential monocyte activation that may contribute to age bias in COVID-19 severity. Sci. Rep. 2022, 12, 20824. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Subedi, K.; Liu, T.; Khalasawi, N.; Pretto-Kernahan, C.D.; Wotring, J.W.; Wang, J.; Yin, C.; Jiang, A.; Fu, C.; et al. Surface translocation of ACE2 and TMPRSS2 upon TLR4/7/8 activation is required for SARS-CoV-2 infection in circulating monocytes. Cell Discov. 2022, 8, 89. [Google Scholar] [CrossRef] [PubMed]
- Jalloh, S.; Olejnik, J.; Berrigan, J.; Nisa, A.; Suder, E.L.; Akiyama, H.; Lei, M.; Ramaswamy, S.; Tyagi, S.; Bushkin, Y.; et al. CD169-mediated restrictive SARS-CoV-2 infection of macrophages induces pro-inflammatory responses. PLOS Pathog. 2022, 18, e1010479. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Tian, Y.; Nguyen, V.; Zhang, Y.; Gao, C.; Yin, R.; Carver, W.; Fan, D.; Albrecht, H.; Cui, T.; et al. Spike protein of SARS-CoV-2 activates macrophages and contributes to induction of acute lung inflammation in male mice. FASEB J. 2021, 35, e21801. [Google Scholar] [CrossRef] [PubMed]
- Palestra, F.; Poto, R.; Ciardi, R.; Opromolla, G.; Secondo, A.; Tedeschi, V.; Ferrara, A.L.; Di Crescenzo, R.M.; Galdiero, M.R.; Cristinziano, L.; et al. SARS-CoV-2 Spike Protein Activates Human Lung Macrophages. Int. J. Mol. Sci. 2023, 24, 3036. [Google Scholar] [CrossRef] [PubMed]
- Sukumaran, P.; Da Conceicao, V.N.; Sun, Y.; Ahamad, N.; Saraiva, L.R.; Selvaraj, S.; Singh, B.B. Calcium Signaling Regulates Autophagy and Apoptosis. Cells 2021, 10, 2125. [Google Scholar] [CrossRef] [PubMed]
- Medina, D.L.; Di Paola, S.; Peluso, I.; Armani, A.; De Stefani, D.; Venditti, R.; Montefusco, S.; Scotto-Rosato, A.; Prezioso, C.; Forrester, A.; et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 2015, 17, 288–299. [Google Scholar] [CrossRef]
- Chandrasekar, A.P.; Maynes, M.; Natesampillai, S.; Shweta, F.N.U.; Badley, A.D.; Cummins, N.W. SARS-CoV-2 spike protein induces monocyte apoptosis and interleukin-8 production. Top Antivir Med. 2021, 29, 61. [Google Scholar]
- Schroeder, J.T.; Bieneman, A.P. The S1 Subunit of the SARS-CoV-2 Spike Protein Activates Human Monocytes to Produce Cytokines Linked to COVID-19: Relevance to Galectin-3. Front. Immunol. 2022, 13, 831763. [Google Scholar] [CrossRef]
- Barhoumi, T.; Alghanem, B.; Shaibah, H.; Mansour, F.A.; Alamri, H.S.; Akiel, M.A.; Alroqi, F.; Boudjelal, M. SARS-CoV-2 Coronavirus Spike Protein-Induced Apoptosis, Inflammatory, and Oxidative Stress Responses in THP-1-Like-Macrophages: Potential Role of Angiotensin-Converting Enzyme Inhibitor (Perindopril). Front. Immunol. 2021, 12, 728896. [Google Scholar] [CrossRef]
- Cao, B.; Jing, X.; Liu, Y.; Wen, R.; Wang, C. Comparison of laboratory parameters in mild vs. severe cases and died vs. survived patients with COVID-19: Systematic review and meta-analysis. J. Thorac. Dis. 2022, 14, 1478–1487. [Google Scholar] [CrossRef] [PubMed]
- Utrero-Rico, A.; González-Cuadrado, C.; Chivite-Lacaba, M.; Cabrera-Marante, O.; Laguna-Goya, R.; Almendro-Vazquez, P.; Díaz-Pedroche, C.; Ruiz-Ruigómez, M.; Lalueza, A.; Folgueira, M.D.; et al. Alterations in Circulating Monocytes Predict COVID-19 Severity and Include Chromatin Modifications Still Detectable Six Months after Recovery. Biomedicines 2021, 9, 1253. [Google Scholar] [CrossRef] [PubMed]
- Kubánková, M.; Hohberger, B.; Hoffmanns, J.; Fürst, J.; Herrmann, M.; Guck, J.; Kräter, M. Physical phenotype of blood cells is altered in COVID-19. Biophys. J. 2021, 120, 2838–2847. [Google Scholar] [CrossRef] [PubMed]
- Ropa, J.; Cooper, S.; Capitano, M.L.; Hof, W.V.; Broxmeyer, H.E. Human Hematopoietic Stem, Progenitor, and Immune Cells Respond Ex Vivo to SARS-CoV-2 Spike Protein. Stem Cell Rev. Rep. 2021, 17, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Guo, R.; Lei, L.; Liu, H.; Wang, Y.; Wang, Y.; Qian, H.; Dai, T.; Zhang, T.; Lai, Y.; et al. Frontline Science: COVID-19 infection induces readily detectable morphologic and inflammation-related phenotypic changes in peripheral blood monocytes. J. Leukoc. Biol. 2021, 109, 13–22. [Google Scholar] [CrossRef]
- Malinovska, A.; Hernried, B.; Lin, A.; Badaki-Makun, O.; Fenstermacher, K.; Ervin, A.M.; Ehrhardt, S.; Levin, S.; Hinson, J.S. Monocyte Distribution Width as a Diagnostic Marker for Infection: A Systematic Review and Meta-analysis. Chest 2023. [Google Scholar] [CrossRef]
- Bajić, D.; Matijašević, J.; Andrijević, L.; Zarić, B.; Lalić-Popović, M.; Andrijević, I.; Todorović, N.; Mihajlović, A.; Tapavički, B.; Ostojić, J. Prognostic Role of Monocyte Distribution Width, CRP, Procalcitonin and Lactate as Sepsis Biomarkers in Critically Ill COVID-19 Patients. J. Clin. Med. 2023, 12, 1197. [Google Scholar] [CrossRef]
- Lorubbio, M.; Tacconi, D.; Iannelli, G.; Feri, M.; Scala, R.; Montemerani, S.; Mandò, M.; Ognibene, A. The role of Monocyte Distribution Width (MDW) in the prognosis and monitoring of COVID-19 patients. Clin. Biochem. 2022, 103, 29–31. [Google Scholar] [CrossRef]
- Riva, G.; Castellano, S.; Nasillo, V.; Ottomano, A.M.; Bergonzini, G.; Paolini, A.; Lusenti, B.; Milić, J.; De Biasi, S.; Gibellini, L.; et al. Monocyte Distribution Width (MDW) as novel inflammatory marker with prognostic significance in COVID-19 patients. Sci. Rep. 2021, 11, 12716. [Google Scholar] [CrossRef]
- Lippi, G.; Sanchis-Gomar, F.; Henry, B.M. Pooled analysis of monocyte distribution width in subjects with SARS-CoV-2 infection. Int. J. Lab. Hematol. 2021, 43, O161–O163. [Google Scholar] [CrossRef]
- Khedmati, M.E.; Sharifi, M.J. Platelet phagocytosis by monocytes. Clin. Chem. Lab. Med. 2022, 60, e204–e206. [Google Scholar] [CrossRef] [PubMed]
- Fadlallah, M.M.; Salman, S.M.; Fadlallah, M.M.; Rahal, H. Hemophagocytic Syndrome and COVID-19: A Comprehensive Review. Cureus 2023, 15, e36140. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, W.; Liu, Z.; Kang, Y.; Zhang, X.; Xu, Z.; Gao, Y.; Qi, Y. A comparative study of spike protein of SARS-CoV-2 and its variant Omicron (B.1.1.529) on some immune characteristics. Sci. Rep. 2022, 12, 17058. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Geng, B.; Marcon, E.; Pu, S.; Tang, H.; Merluza, J.; Bello, A.; Snider, J.; Lu, P.; Wood, H.; et al. Omicron Spike Protein is Vulnerable to Reduction. J. Mol. Biol. 2023, 435, 168128. [Google Scholar] [CrossRef]
- Hu, F.-H.; Jia, Y.-J.; Zhao, D.-Y.; Fu, X.-L.; Zhang, W.-Q.; Tang, W.; Hu, S.-Q.; Wu, H.; Ge, M.-W.; Du, W.; et al. Clinical outcomes of the severe acute respiratory syndrome coronavirus 2 Omicron and Delta variant: Systematic review and meta-analysis of 33 studies covering 6 037 144 coronavirus disease 2019–positive patients. Clin. Microbiol. Infect. 2023. Epub ahead of print. [Google Scholar] [CrossRef]
- Esper, F.P.; Adhikari, T.M.; Tu, Z.J.; Cheng, Y.-W.; El-Haddad, K.; Farkas, D.H.; Bosler, D.; Rhoads, D.; Procop, G.W.; Ko, J.S.; et al. Alpha to Omicron: Disease Severity and Clinical Outcomes of Major SARS-CoV-2 Variants. J. Infect. Dis. 2023, 227, 344–352. [Google Scholar] [CrossRef]
- Varea-Jiménez, E.; Cano, E.A.; Vega-Piris, L.; Sánchez, E.V.M.; Mazagatos, C.; Rodríguez-Alarcón, L.G.S.M.; Casas, I.; Moros, M.J.S.; Iglesias-Caballero, M.; Vazquez-Morón, S.; et al. Comparative severity of COVID-19 cases caused by Alpha, Delta or Omicron SARS-CoV-2 variants and its association with vaccination. Enfermedades Infecc. y Microbiol. Clin. (Engl. ed.) 2023. Epub ahead of print. [Google Scholar] [CrossRef]
- Korobova, Z.R.; Arsentieva, N.A.; Liubimova, N.E.; Batsunov, O.K.; Dedkov, V.G.; Gladkikh, A.S.; Sharova, A.A.; Adish, Z.; Chernykh, E.I.; Kaschenko, V.A.; et al. Cytokine Profiling in Different SARS-CoV-2 Genetic Variants. Int. J. Mol. Sci. 2022, 23, 14146. [Google Scholar] [CrossRef]
- Barh, D.; Tiwari, S.; Gomes, L.G.R.; Pinto, C.H.R.; Andrade, B.S.; Ahmad, S.; Aljabali, A.A.A.; Alzahrani, K.J.; Banjer, H.J.; Hassan, S.S.; et al. SARS-CoV-2 Variants Show a Gradual Declining Pathogenicity and Pro-Inflammatory Cytokine Stimulation, an Increasing Antigenic and Anti-Inflammatory Cytokine Induction, and Rising Structural Protein Instability: A Minimal Number Genome-Based Approach. Inflammation 2023, 46, 297–312. [Google Scholar] [CrossRef]
- Park, C.; Tavakoli-Tabasi, S.; Sharafkhaneh, A.; Seligman, B.J.; Hicken, B.; Amos, C.I.; Chou, A.; Razjouyan, J. Inflammatory Biomarkers Differ among Hospitalized Veterans Infected with Alpha, Delta, and Omicron SARS-CoV-2 Variants. Int. J. Environ. Res. Public Heal. 2023, 20, 2987. [Google Scholar] [CrossRef]
- McCrindle, B.W.; Harahsheh, A.S.; Handoko, R.; Raghuveer, G.; Portman, M.A.; Khoury, M.; Newburger, J.W.; Lee, S.; Jain, S.S.; Khare, M.; et al. SARS-CoV-2 Variants and Multisystem Inflammatory Syndrome in Children. N. Engl. J. Med. 2023, 388, 1624–1626. [Google Scholar] [CrossRef] [PubMed]
- Craddock, V.; Mahajan, A.; Spikes, L.; Krishnamachary, B.; Ram, A.K.; Kumar, A.; Chen, L.; Chalise, P.; Dhillon, N.K. Persistent circulation of soluble and extracellular vesicle-linked Spike protein in individuals with postacute sequelae of COVID-19. J. Med Virol. 2023, 95, e28568. [Google Scholar] [CrossRef] [PubMed]
- Lippi, G.; Salvagno, G.L.; Montagnana, M.; Lima-Oliveira, G.; Guidi, G.C.; Favaloro, E.J. Quality Standards for Sample Collection in Coagulation Testing. Semin. Thromb. Hemost. 2012, 38, 565–575. [Google Scholar] [CrossRef]
- Buoro, S.; Seghezzi, M.; Vavassori, M.; Dominoni, P.; Esposito, S.A.; Manenti, B.; Mecca, T.; Marchesi, G.; Castellucci, E.; Azzarà, G.; et al. Clinical significance of cell population data (CPD) on Sysmex XN-9000 in septic patients with our without liver impairment. Ann. Transl. Med. 2016, 4, 418. [Google Scholar] [CrossRef] [PubMed]
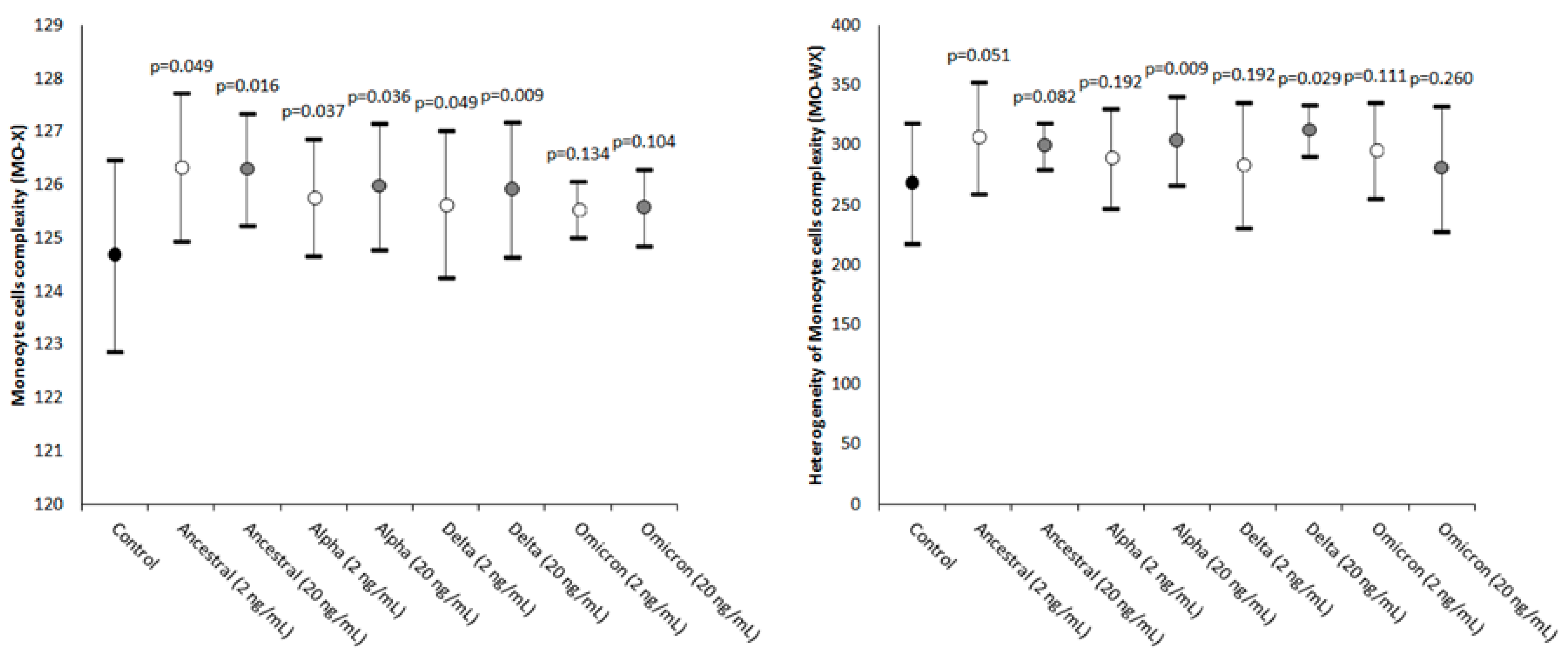
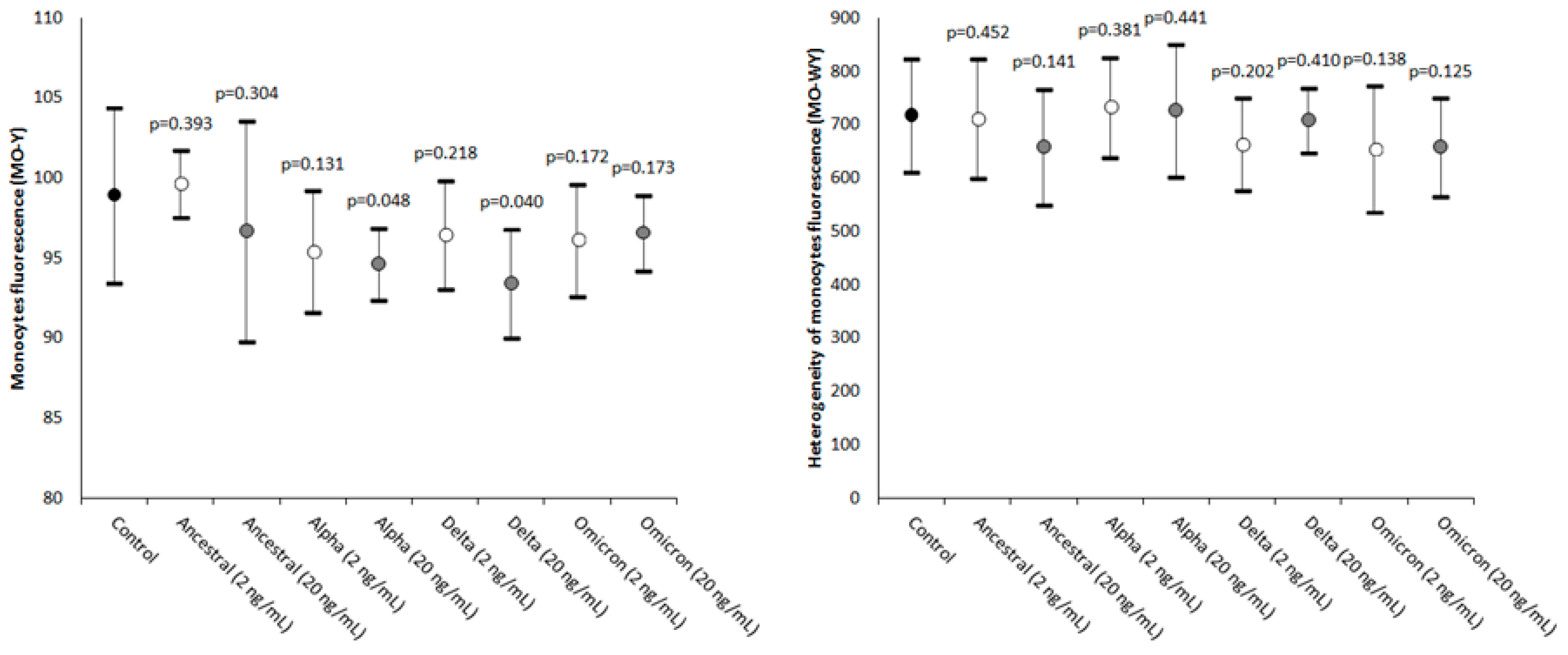
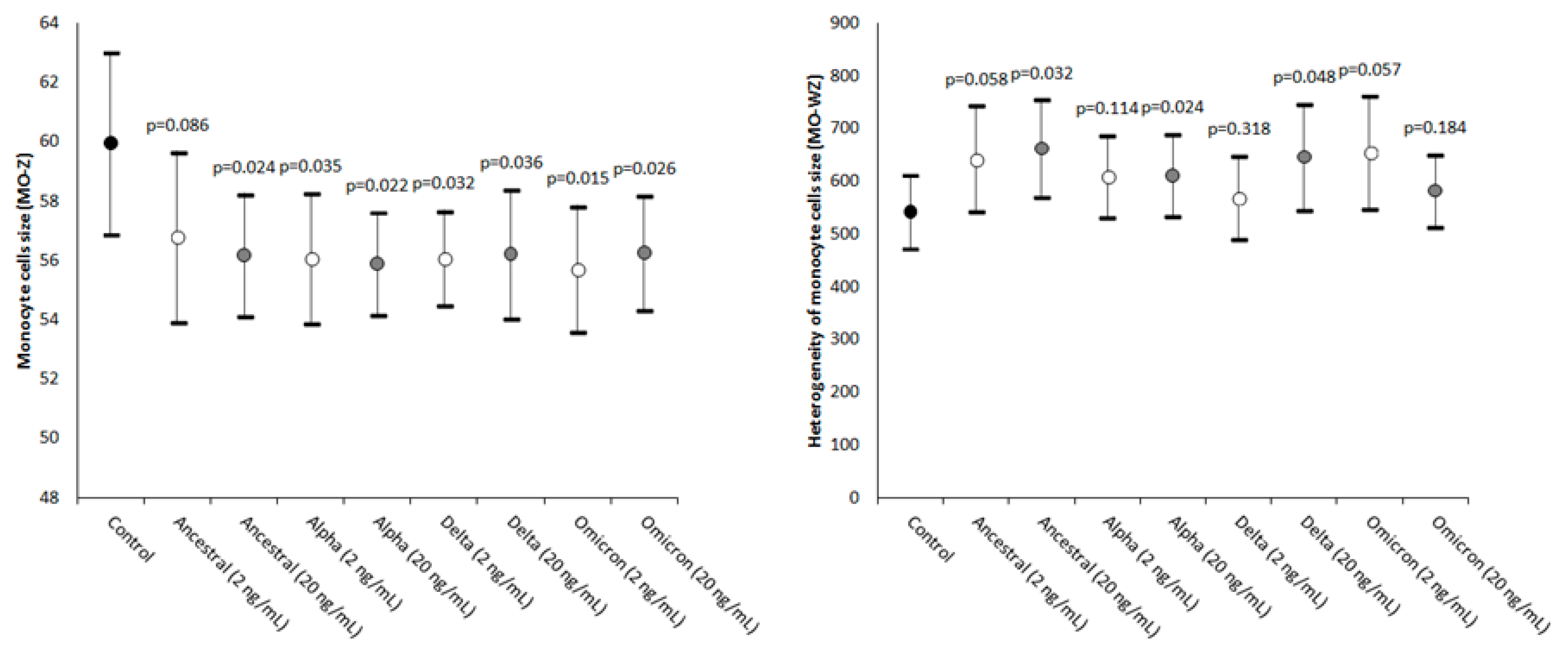
| Parameter | Statistical Significance (20 vs. 2 ng/mL) |
|---|---|
| MO-X | Ancestral: p = 0.467; Alpha: p = 0.188; Delta: p = 0.386; Omicron: p = 0.093 |
| MO-WX | Ancestral: p = 0.374; Alpha: p = 0.453; Delta: p = 0.334; Omicron: p = 0.040 |
| MO-Y | Ancestral: p = 0.156; Alpha: p = 0.008; Delta: p < 0.001; Omicron: p = 0.003 |
| MO-WY | Ancestral: p = 0.049; Alpha: p = 0.417; Delta: p = 0.470; Omicron: p = 0.163 |
| MO-Z | Ancestral: p = 0.256; Alpha: p = 0.204; Delta: p = 0.247; Omicron: p = 0.312 |
| MO-WZ | Ancestral: p = 0.346; Alpha: p = 0.287; Delta: p = 0.430; Omicron: p = 0.136 |
| Spike Protein Variants | Additional Mutations |
|---|---|
| Ancestral | D614G |
| Alpha (B.1.1.7) | AAVal16, Pro1213, HV69-70del, Y144del, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H |
| Delta (B.1.617.2) | AAVal16, Pro 1213, T19R, G142D, EF156-157del, R158G, L452R, T478K, D614G, P681R, D950N |
| Omicron (B.1.1.529) | AAVal16, Pro 1213, A67V, HV69-70del, T95I, G142D, VYY143-145del, N211del, L212I, ins214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F |
| Parameter | Abbreviation | Clinical Significance |
|---|---|---|
| Monocyte cells’ complexity | MO-X | Presence of granules, vacuoles, and other cytoplasmic inclusions |
| Width of dispersion of the monocytes’ complexity | MO-WX | Heterogeneity of monocytes’ complexity |
| Monocytes’ fluorescence intensity | MO-Y | Quantity of cellular DNA and RNA |
| Width of dispersion of the monocytes’ fluorescence intensity | MO-WY | Heterogeneity of monocytes’ fluorescence intensity |
| Monocyte cells’ size | MO-Z | Abnormal sized cells |
| Width of dispersion of the monocytes’ cell size | MO-WZ | Heterogeneity of monocyte cell size |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vettori, M.; Dima, F.; Henry, B.M.; Carpenè, G.; Gelati, M.; Celegon, G.; Salvagno, G.L.; Lippi, G. Effects of Different Types of Recombinant SARS-CoV-2 Spike Protein on Circulating Monocytes’ Structure. Int. J. Mol. Sci. 2023, 24, 9373. https://doi.org/10.3390/ijms24119373
Vettori M, Dima F, Henry BM, Carpenè G, Gelati M, Celegon G, Salvagno GL, Lippi G. Effects of Different Types of Recombinant SARS-CoV-2 Spike Protein on Circulating Monocytes’ Structure. International Journal of Molecular Sciences. 2023; 24(11):9373. https://doi.org/10.3390/ijms24119373
Chicago/Turabian StyleVettori, Marco, Francesco Dima, Brandon Michael Henry, Giovanni Carpenè, Matteo Gelati, Giovanni Celegon, Gian Luca Salvagno, and Giuseppe Lippi. 2023. "Effects of Different Types of Recombinant SARS-CoV-2 Spike Protein on Circulating Monocytes’ Structure" International Journal of Molecular Sciences 24, no. 11: 9373. https://doi.org/10.3390/ijms24119373
APA StyleVettori, M., Dima, F., Henry, B. M., Carpenè, G., Gelati, M., Celegon, G., Salvagno, G. L., & Lippi, G. (2023). Effects of Different Types of Recombinant SARS-CoV-2 Spike Protein on Circulating Monocytes’ Structure. International Journal of Molecular Sciences, 24(11), 9373. https://doi.org/10.3390/ijms24119373










