Markers of NETosis in Patients with Systemic Lupus Erythematosus and Antiphospholipid Syndrome
Abstract
1. Introduction
2. Results
2.1. MPO-DNA Complex in the Studied Groups
2.2. Clinical Characteristics of SLE Patients and the Serum MPO-DNA Complex Levels
2.3. The MPO-DNA Complex Levels Depend on the Disease Activity and Clinical Manifestations of SLE
2.4. MPO-DNA Complex in Antiphospholipid Syndrome
2.5. Nucleosomes in Patients with SLE and APS
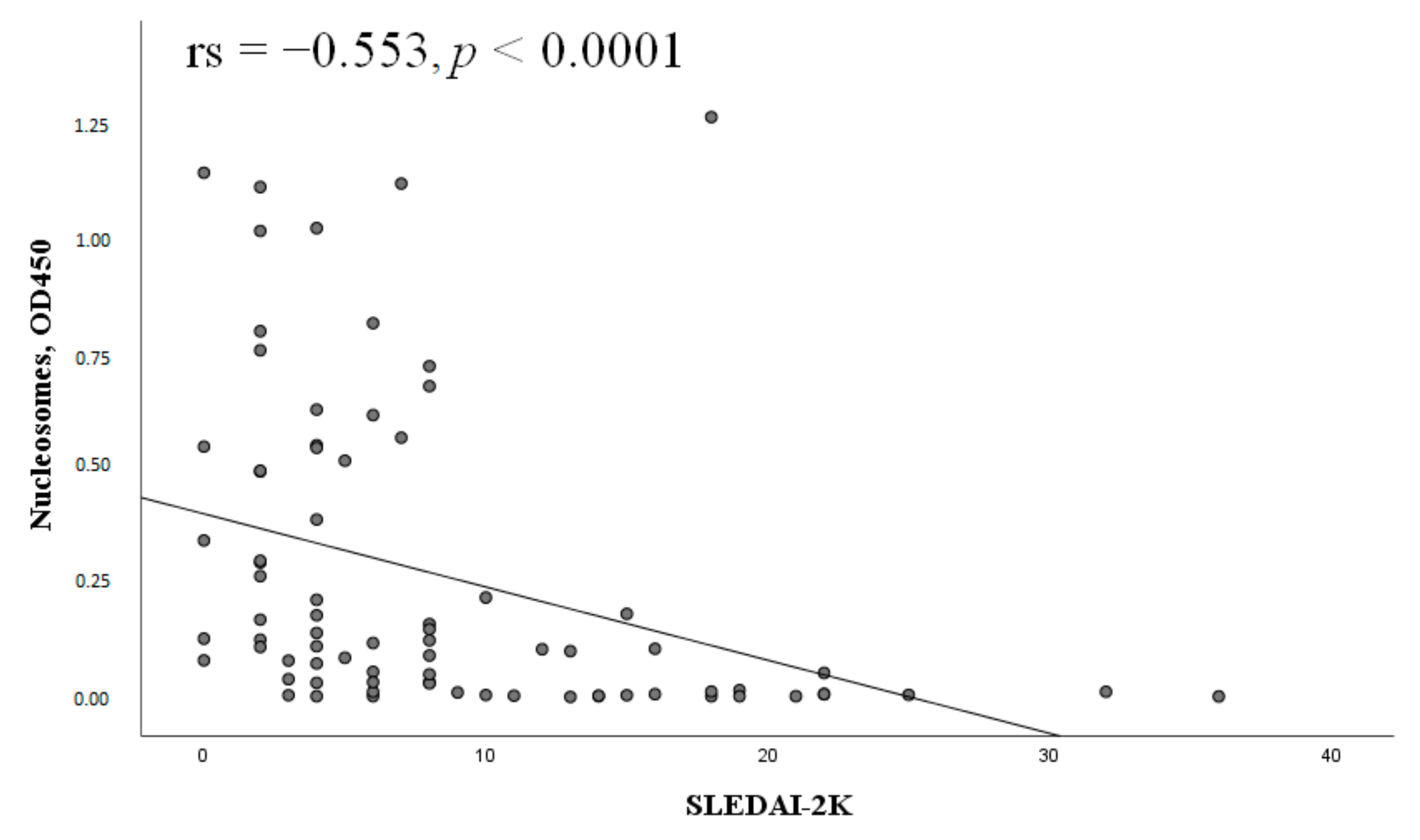
2.6. Clinical Characteristics of SLE Patients and Serum Nucleosomes Levels
2.7. Nucleosomes in Antiphospholipid Syndrome
3. Discussion
4. Materials and Methods
4.1. Serum Histone-Associated-DNA-Fragments Immunoanalysis (Nucleosomes)
4.2. Serum NETs Immunoanalysis (MPO-DNA Complex)
4.3. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nasonov, E.L.; Soloviev, S.K.; Arshinov, A.V. Systemic lupus erythematosus: History and modernity. Rheumatol. Sci. Pract. 2022, 60, 397–412. (In Russian) [Google Scholar] [CrossRef]
- Reshetnyak, T.M.; Cheldieva, F.A.; Nurbaeva, K.S.; Lila, A.M.; Nasonov, E.L. Antiphospholipid syndrome: Diagnosis, mechanism of development, issues of therapy. Thromb. Hemost. Rheol. 2020, 4, 4–21. (In Russian) [Google Scholar] [CrossRef]
- Ibrahim, A.A.G.; Shadi, H.W.E.; Elamin, A.A.Y.; Draz, H.E. Retrospective cohort study of thromboembolic events in systemic lupus erythematosus with or without secondary antiphospholipid syndrome and their correlation to lupus activity and dyslipidemia. Egypt. Rheumatol. Rehabil. 2023, 50, 10. [Google Scholar] [CrossRef]
- Wirestam, L.; Arve, S.; Linge, P.; Bengtsson, A.A. Neutrophils-Important Communicators in Systemic Lupus Erythematosus and Antiphospholipid Syndrome. Front. Immunol. 2019, 10, 2734. [Google Scholar] [CrossRef] [PubMed]
- Fousert, E.; Toes, R.; Desai, J. Neutrophil Extracellular Traps (NETs) Take the Central Stage in Driving Autoimmune Responses. Cells 2020, 9, 915. [Google Scholar] [CrossRef] [PubMed]
- Stark, K.; Massberg, S. Interplay between inflammation and thrombosis in cardiovascular pathology. Nat. Rev. Cardiol. 2021, 18, 666–682. [Google Scholar] [CrossRef] [PubMed]
- Stoimenou, M.; Tzorosm, G.; Skendros, P.; Chrysanthopoulou, A. Methods for the Assessment of NET Formation: From Neutrophil Biology to Translational Research. Int. J. Mol. Sci. 2022, 23, 15823. [Google Scholar] [CrossRef] [PubMed]
- Masuda, S.; Nakazawa, D.; Shida, H.; Miyoshi, A.; Kusunoki, Y.; Tomaru, U.; Ishizu, A. NETosis markers: Quest for specific, objective, and quantitative markers. Clin. Chim. Acta 2016, 459, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, M.A.; Elmofty, S.A.; Elmaghraby, A.A.; Khalifa, R.H. Anti-nucleosome antibodies in systemic lupus erythematosus patients: Relation to anti-double stranded deoxyribonucleic acid and disease activity. Egypt. Rheumatol. 2018, 40, 29–33. [Google Scholar] [CrossRef]
- Ebrahimi, F.; Giaglis, S.; Hahn, S.; Blum, C.A.; Baumgartner, C.; Kutz, A.; van Breda, S.V.; Mueller, B.; Schuetz, P.; Christ-Crain, M.; et al. Markers of neutrophil extracellular traps predict adverse outcome in community-acquired pneumonia: Secondary analysis of a randomised controlled trial. Eur. Respir. J. 2018, 51, 1701389. [Google Scholar] [CrossRef]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil Extracellular Traps Kill Bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef]
- Grayson, P.C.; Kaplan, M.J. At the Bench: Neutrophil extracellular traps (NETs) high-light novel aspects of innate immune system involvement in autoimmune diseases. J. Leukoc. Biol. 2016, 99, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Hanata, N.; Ota, M.; Tsuchida, Y.; Nagafuchi, Y.; Okamura, T.; Shoda, H.; Fujio, K. Serum extracellular traps associate with the activation of myeloid cells in SLE patients with the low level of anti-DNA antibodies. Sci. Rep. 2022, 12, 18397. [Google Scholar] [CrossRef] [PubMed]
- Bruschi, M.; Bonanni, A.; Petretto, A.; Vaglio, A.; Pratesi, F.; Santucci, L.; Migliorini, P.; Bertelli, R.; Galetti, M.; Belletti, S.; et al. Neutrophil Extracellular Traps Profiles in Patients with Incident Systemic Lupus Erythematosus and Lupus Nephritis. J. Rheumatol. 2020, 47, 377–386. [Google Scholar] [CrossRef] [PubMed]
- van der Linden, M.; van den Hoogen, L.L.; Westerlaken, G.H.A.; Fritsch-Stork, R.D.E.; van Roon, J.A.G.; Radstake, T.R.D.J.; Meyaard, L. Neutrophil extracellular trap release is associated with antinuclear antibodies in systemic lupus erythematosus and anti-phospholipid syndrome. Rheumatology 2018, 57, 1228–1234. [Google Scholar] [CrossRef]
- El-Ghoneimy, D.H.; Hesham, M.; Hasan, R.; Tarif, M.; Gouda, S. The behavior of neutrophil extracellular traps and NADPH oxidative activity in pediatric systemic lupus erythematosus: Relation to disease activity and lupus nephritis. Clin. Rheumatol. 2019, 38, 2585–2593. [Google Scholar] [CrossRef] [PubMed]
- Hakkim, A.; Fürnrohr, B.G.; Amann, K.; Laube, B.; Abed, U.A.; Brinkmann, V.; Herrmann, M.; Voll, R.E.; Zychlinsky, A. Impairment of neutrophil extracellular trap degradation is associated with lupus nephritis. Proc. Natl. Acad. Sci. USA 2010, 107, 9813–9818. [Google Scholar] [CrossRef]
- Leffler, J.; Martin, M.; Gullstrand, B.; Tydén, H.; Lood, C.; Truedsson, L.; Bengtsson, A.A.; Blom, A.M. Neutrophil extracellular traps that are not degraded in systemic lupus erythematosus activate complement exacerbating the disease. J. Immunol. 2012, 188, 3522–3531. [Google Scholar] [CrossRef]
- Leffler, J.; Gullstrand, B.; Jönsen, A.; Nilsson, J.; Martin, M.; Blom, A.M.; Bengtsson, A.A. Degradation of neutrophil extracellular traps co-varies with disease activity in patients with systemic lupus erythematosus. Arthritis Res. Ther. 2013, 15, R84. [Google Scholar] [CrossRef]
- Zhang, S.; Lu, X.; Shu, X.; Tian, X.; Yang, H.; Yang, W.; Zhang, Y.; Wang, G. Elevated plasma cfDNA may be associated with active lupus nephritis and partially attributed to abnormal regulation of neutrophil extracellular traps (NETs) in patients with systemic lupus erythematosus. Intern. Med. 2014, 53, 2763–2771. [Google Scholar] [CrossRef]
- Jeremic, I.; Djuric, O.; Nikolic, M.; Vlajnic, M.; Nikolic, A.; Radojkovic, D.; Bonaci-Nikolic, B. Neutrophil extracellular traps-associated markers are elevated in patients with systemic lupus erythematosus. Rheumatol. Int. 2019, 39, 1849–1857. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, E.; Yalavarthi, S.; Berthier, C.C.; Hodgin, J.B.; Khandpur, R.; Li, A.M.; Rubin, C.J.; Zhao, W.; Olsen, S.H.; Klinker, M.; et al. Netting neutrophils induce endothelial damage, infiltrate tissues, and expose immunostimulatory molecules in systemic lupus erythematosus. J. Immunol. 2011, 187, 538–552. [Google Scholar] [CrossRef]
- Wang, Y.; Xiao, S.; Xia, Y.; Wang, H. The Therapeutic Strategies for SLE by Targeting Anti-dsDNA Antibodies. Clinic. Rev. Allergy Immunol. 2022, 63, 152–165. [Google Scholar] [CrossRef] [PubMed]
- Patiño-Trives, A.M.; Pérez-Sánchez, C.; Pérez-Sánchez, L.; Luque-Tévar, M.; Ábalos-Aguilera, M.C.; Alcaide-Ruggiero, L.; Arias-de la Rosa, I.; Román-Rodríguez, C.; Seguí, P.; Espinosa, M.; et al. Anti-dsDNA Antibodies Increase the Cardiovascular Risk in Systemic Lupus Erythematosus Promoting a Distinctive Immune and Vascular Activation. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 2417–2430. [Google Scholar] [CrossRef] [PubMed]
- De Bont, C.M.; Boelens, W.C.; Pruijn, G.J.M. NETosis, complement, and coagulation: A triangular relationship. Cell. Mol. Immunol. 2019, 16, 19–27. [Google Scholar] [CrossRef]
- Mukai, M. Systemic lupus erythematosus and nucleosome. Nihon Rinsho Meneki Gakkai Kaishi 2006, 29, 127–135. (In Japanese) [Google Scholar] [CrossRef] [PubMed]
- Amoura, Z.; Piette, J.C.; Chabre, H.; Cacoub, P.; Papo, T.; Wechsler, B.; Bach, J.F.; Koutouzov, S. Circulating plasma levels of nucleosomes in patients with systemic lupus erythematosus: Correlation with serum antinucleosome antibody titers and absence of clear association with disease activity. Arthritis Rheum. 1997, 40, 2217–2225. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.C., Jr.; Malone, C.C.; Meyers, C.; Decker, P.; Muller, S. Detection of nucleosome particles in serum and plasma from patients with systemic lupus erythematosus using monoclonal antibody 4H7. J. Rheumatol. 2001, 28, 81–94. [Google Scholar]
- Marsman, G.; Stephan, F.; de Leeuw, K.; Bulder, I.; Ruinard, J.T.; de Jong, J.; Westra, J.; Bultink, I.E.; Voskuyl, A.E.; Aarden, L.A.; et al. FSAP-mediated nucleosome release from late apoptotic cells is inhibited by autoantibodies present in SLE. Eur. J. Immunol. 2016, 46, 762–771. [Google Scholar] [CrossRef]
- Yalavarthi, S.; Gould, T.J.; Rao, A.N.; Mazza, L.F.; Morris, A.E.; Núñez-Álvarez, C.; Hernández-Ramírez, D.; Bockenstedt, P.L.; Liaw, P.C.; Cabral, A.R.; et al. Release of neutrophil extracellular traps by neutrophils stimulated with antiphospholipid antibodies: A newly identified mechanism of thrombosis in the antiphospholipid syndrome. Arthritis Rheumatol. 2015, 67, 2990–3003. [Google Scholar] [CrossRef] [PubMed]
- Hell, L.; Lurger, K.; Mauracher, L.-M.; Grilz, E.; Reumiller, C.M.; Schmidt, G.J.; Ercan, H.; Koder, S.; Assinger, A.; Basílio, J.; et al. Altered platelet proteome in lupus anticoagulant (LA)-positive patients-protein disulfide isomerase and NETosis as new players in LA-related thrombosis. Exp. Mol. Med. 2020, 52, 66–78. [Google Scholar] [CrossRef] [PubMed]
- Mazetto, B.D.M.; Hounkpe, B.W.; Saraiva, S.D.S.; Vieira-Damiani, G.; dos Santos, A.P.R.; Jacinto, B.C.; Vaz, C.D.O.; Mesquita, G.T.V.; Annichino-Bizzacchi, J.M.; De Paula, E.V.; et al. Association between neutrophil extracellular traps (NETs) and thrombosis in antiphospholipid syndrome. Thromb. Res. 2022, 214, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zuo, Y.; Zhang, S.; Makris, U.E.; Karp, D.R.; Li, Z. Additional risk factors associated with thrombosis and pregnancy morbidity in a unique cohort of antiphospholipid antibody-positive patients. Chin. Med. J. 2022, 135, 658–664. [Google Scholar] [CrossRef] [PubMed]
- Petri, M.; Orbai, A.M.; Alarcon, G.S.; Gordon, C.; Merrill, J.T.; Fortin, P.R.; Bruce, I.N.; Isenberg, D.; Wallace, D.J.; Nived, O.; et al. Derivation and validation of the systemic lupus international collaborating clinics classification criteria for systemic lupus erythematosus. Arthritis Rheum. 2012, 64, 2677–2686. [Google Scholar] [CrossRef]
- Hochberg, M.C. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997, 40, 1725. [Google Scholar] [CrossRef] [PubMed]
- Touma, Z.; Urowitz, M.B.; Ibanez, D.; Gladman, D.D. Sledai-2k 10 days versus sledai-2k 30 days in a longitudinal evaluation. Lupus 2011, 20, 67–70. [Google Scholar] [CrossRef]
- Gladman, D.D.; Goldsmith, C.H.; Urowitz, M.B.; Bacon, P.; Fortin, P.; Ginzler, E.; Gordon, C.; Hanly, J.G.; Isenberg, D.A.; Petri, M.; et al. The Systemic Lupus International Collaborating Clinics/American College of Rheumatology (SLICC/ACR) Damage Index for Systemic Lupus Erythematosus International Comparison. J. Rheumatol. 2000, 27, 373–376. [Google Scholar]
- Miyakis, S.; Lockshin, M.D.; Atsumi, T.; Branch, D.W.; Brey, R.L.; Cervera, R.; Derksen, R.H.W.M.; De Groot, P.G.; Koike, T.; Meroni, P.L.; et al. International consensus statement on an update of the classification criteria for definite antiphospholipid syndrome (APS). J. Thromb. Haemost. 2006, 4, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Aleksandrova, E.N.; Novikov, A.A.; Reshetnyak, T.M.; Nasonov, E.L. Antibodies to β2-glycoprotein 1 and antibodies to cardiolipin in antiphospholipid syndrome: Sensitivity and specificity analysis. Klin. Meditsina 2003, 81, 25–31. (In Russian) [Google Scholar]
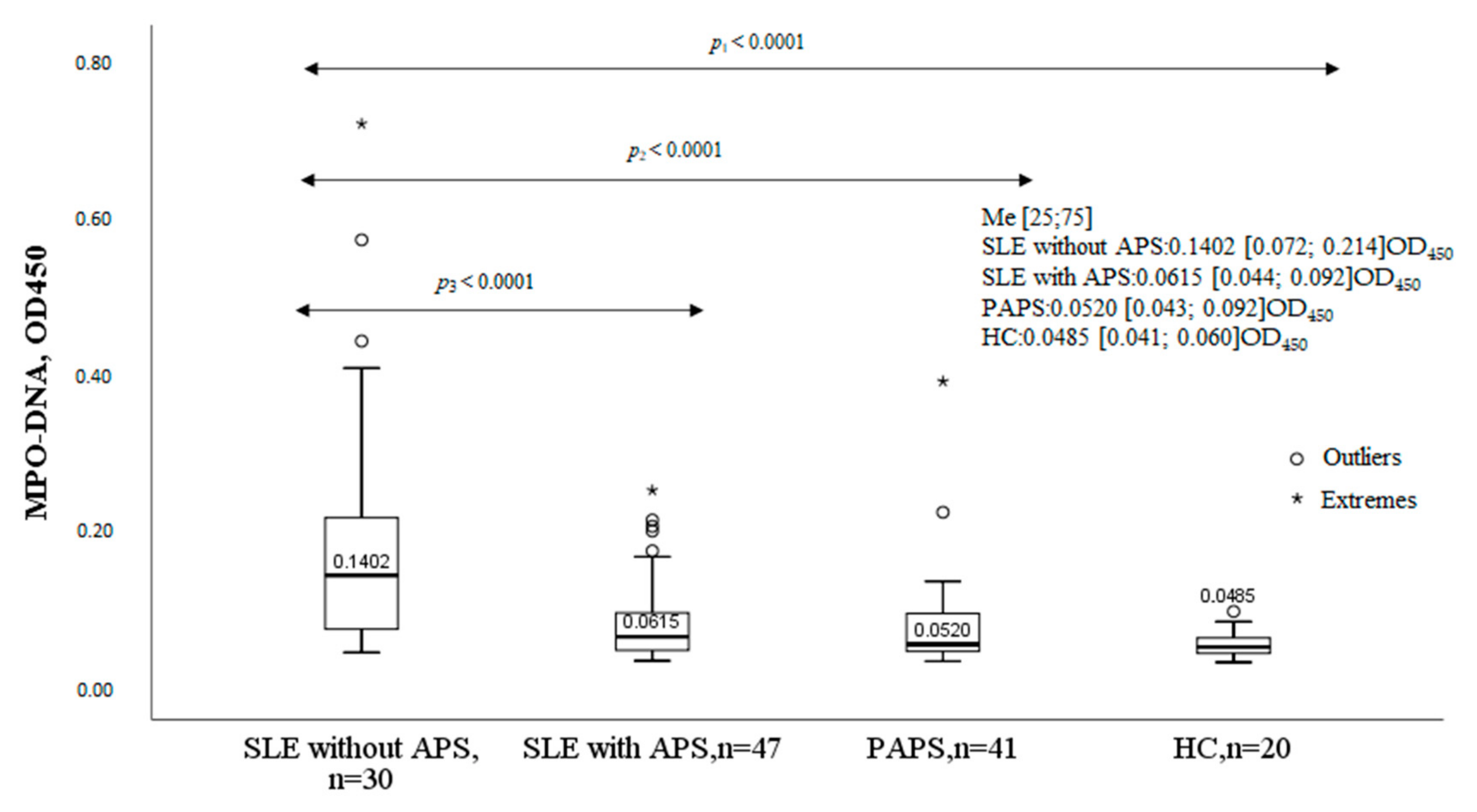
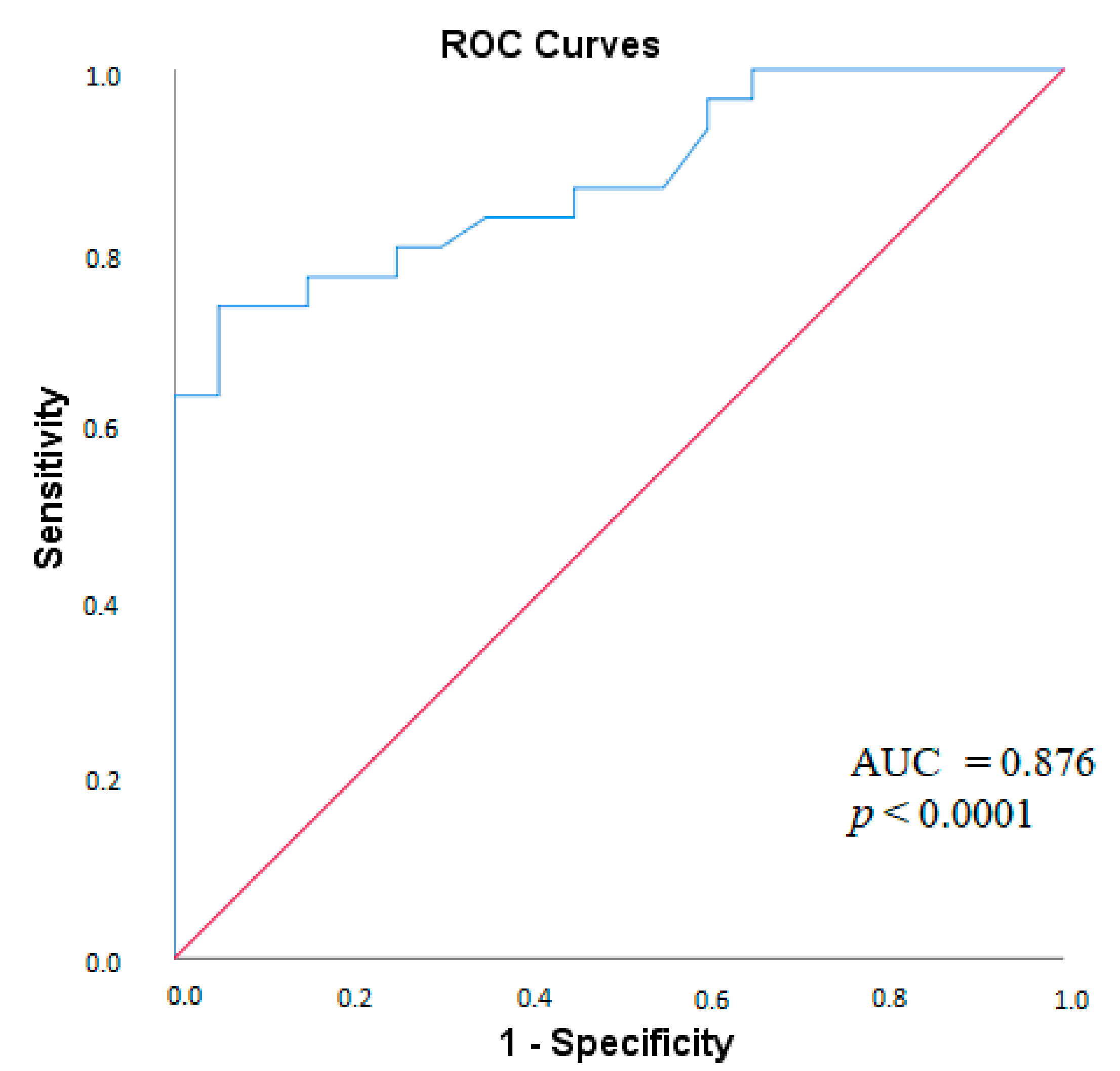
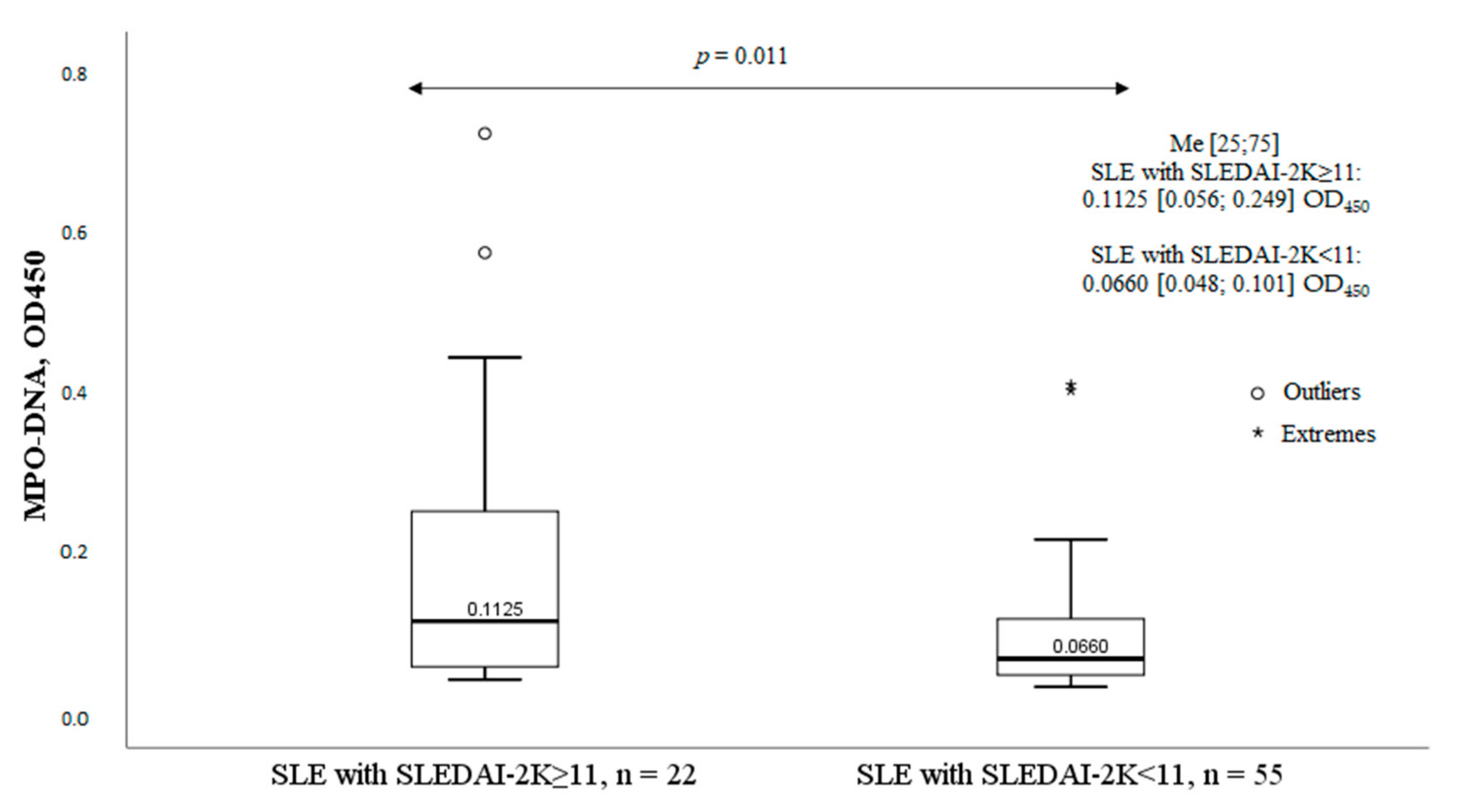
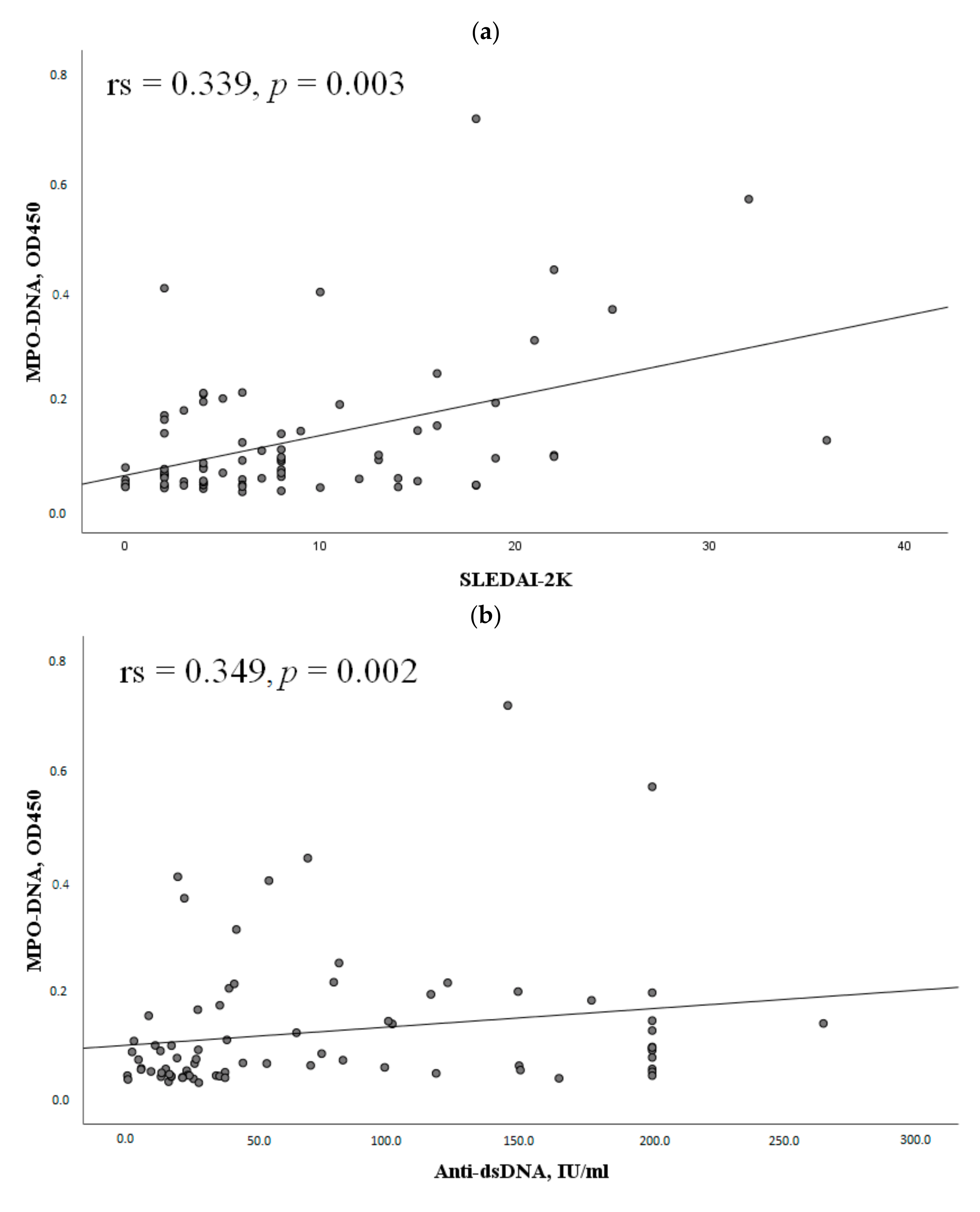
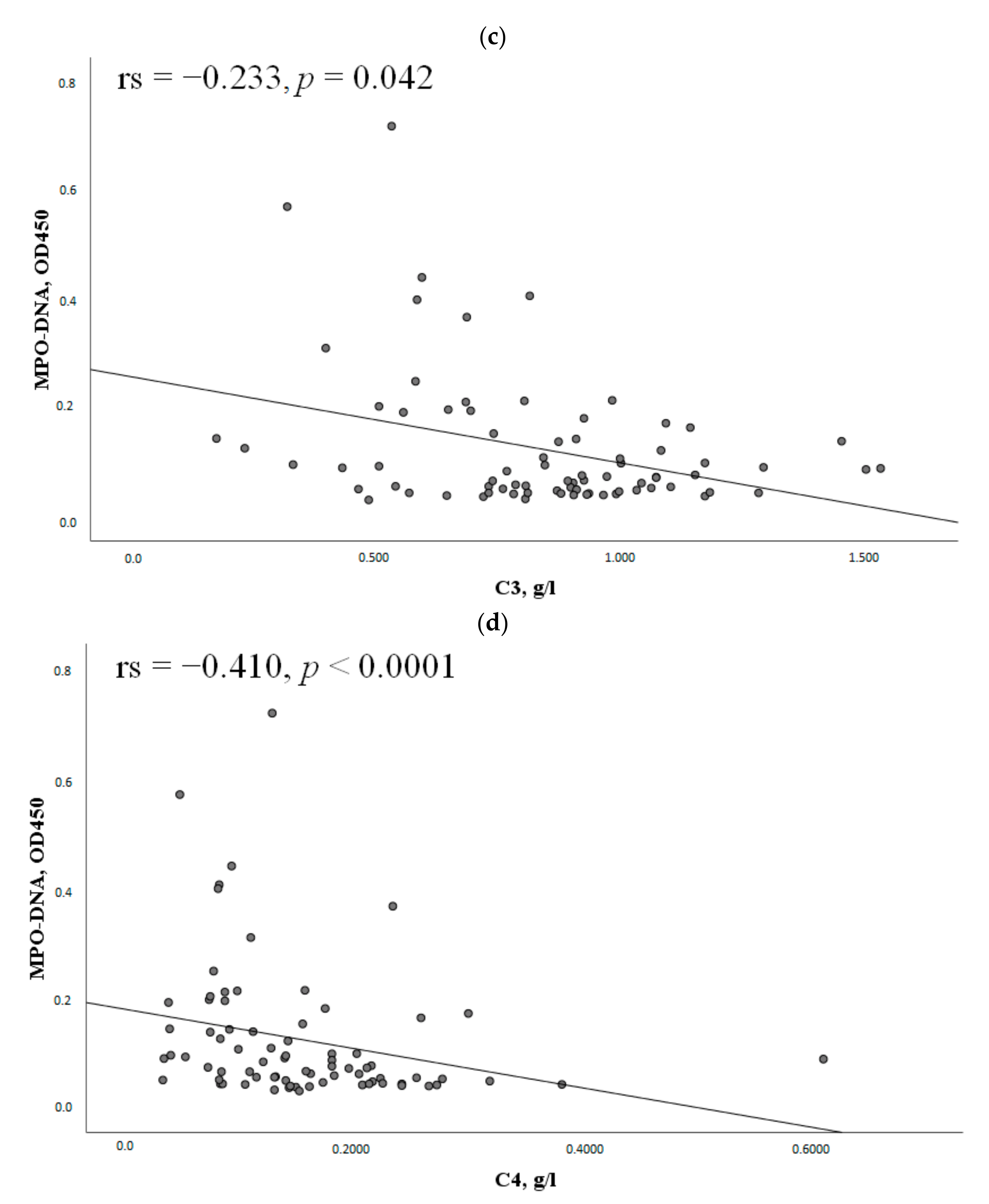

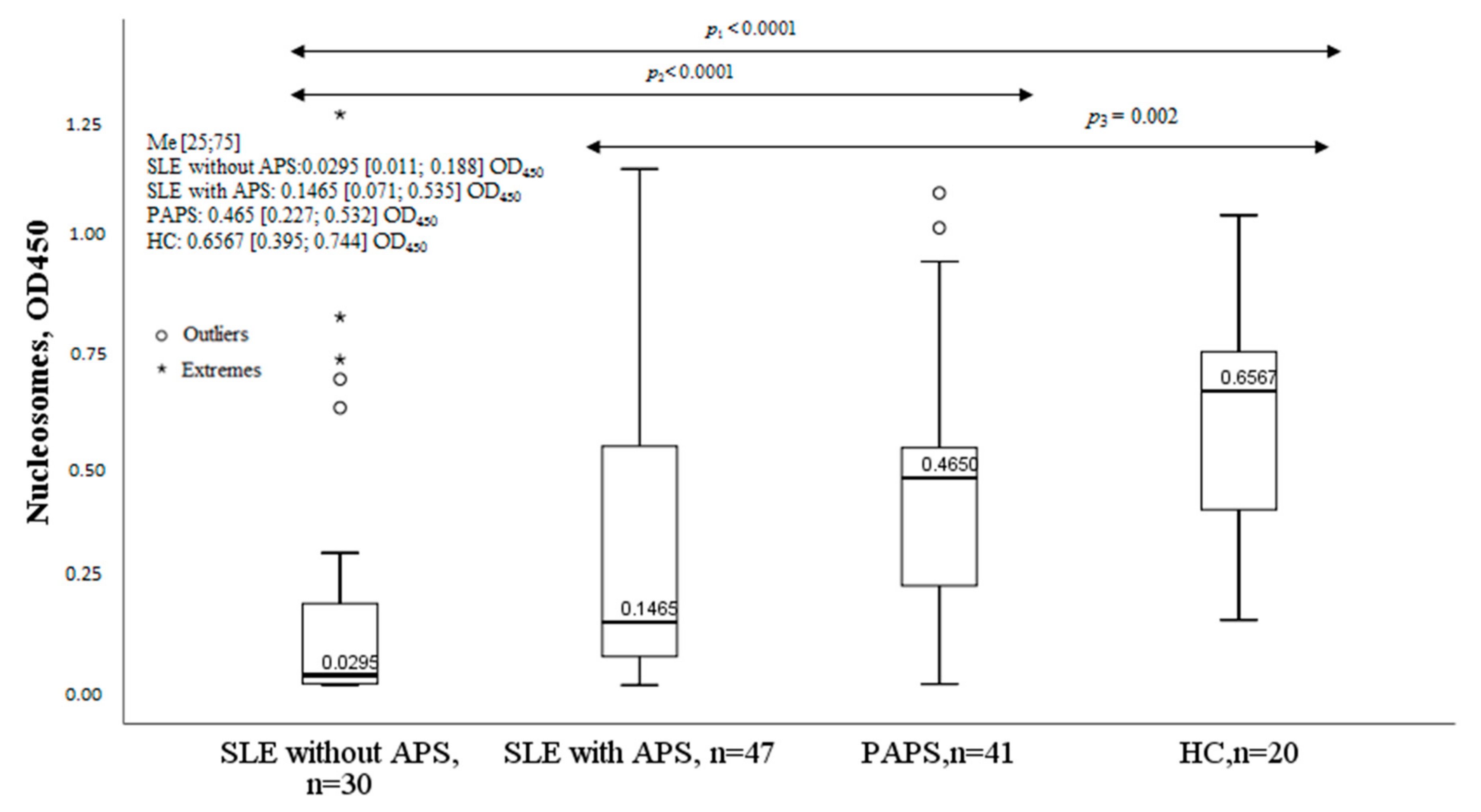
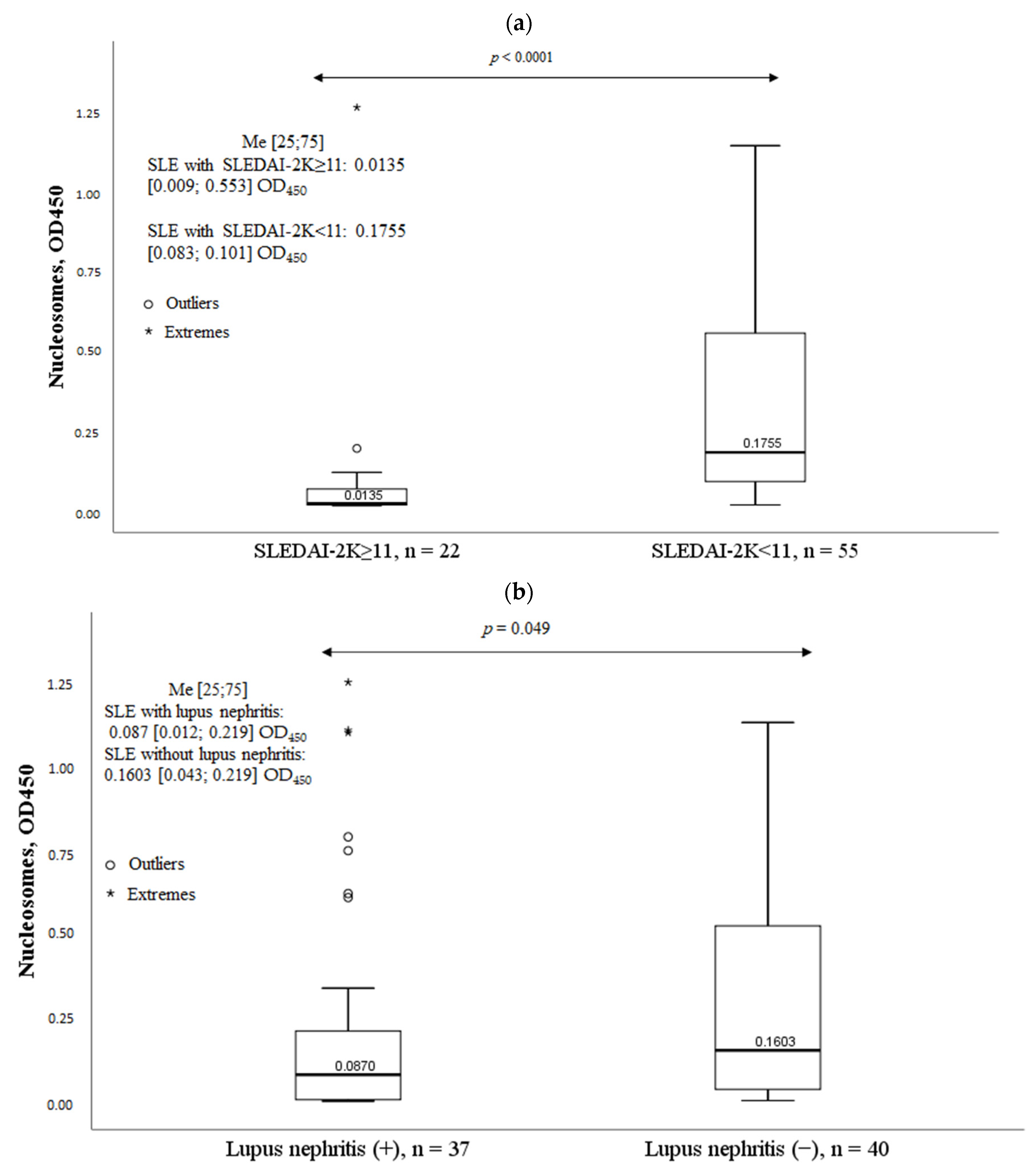
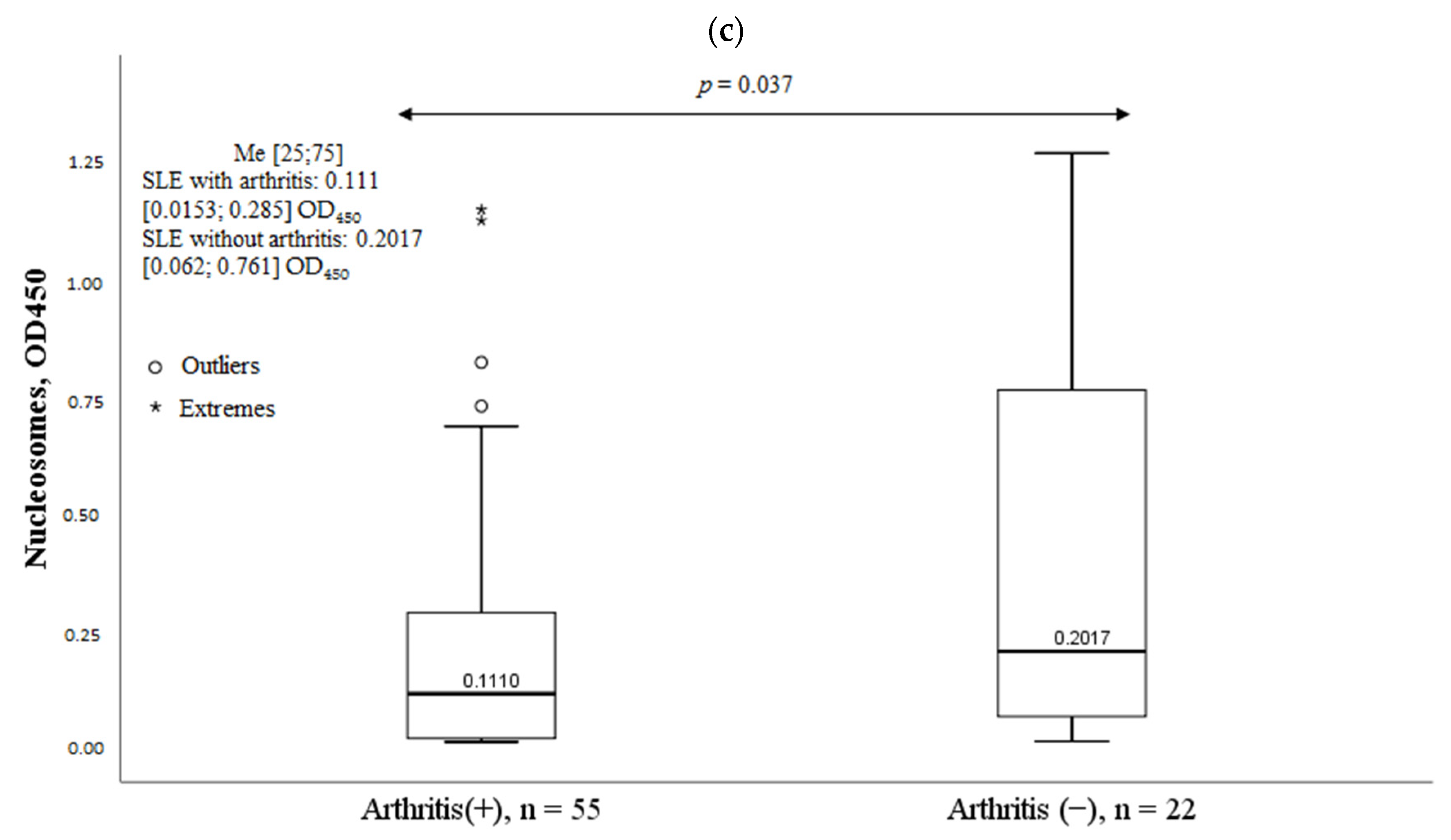


| Parameter | MPO-DNA Complex (+), n = 30, n (%) | MPO-DNA Complex (-), n = 47, n (%) | χ2; p OR and 95% CI | |
|---|---|---|---|---|
| Lupus nephritis | yes | 20 (67) | 17 (36) | 6.82; 0.009 3.53 [1.35–9.26] |
| no | 10 (33) | 30 (64) | ||
| Positive antibodies to dsDNA | yes | 26 (87) | 30 (64) | 4.82; 0.036 3.68 [1.09–12.34] |
| no | 4 (13) | 17 (36) | ||
| Hypocomplementemia | yes | 23 (77) | 22 (47) | 6.72; 0.01 3.73 [1.34–10.37] |
| no | 7 (23) | 25 (53) | ||
| High SLE activity | yes | 13 (43) | 9 (19) | 5.25; 0.037 3.23 [1.16–8.99] |
| no | 17 (57) | 38 (81) | ||
| Parameters | MPO-DNA Complex Levels, OD450 Me, [25;75] | p | |
|---|---|---|---|
| Positive anti-dsDNA | yes (n = 56) | 0.090 [0.0518;0.186] | p = 0.037 |
| no (n = 21) | 0.056 [0.0435;0.089] | ||
| Hypocomplementemia | yes (n = 45) | 0.095 [0.054;0.197] | p = 0.032 |
| no (n = 32) | 0.065 [0.044;0.095] | ||
| Positive anti-Sm antibody | yes (n = 9) | 0.089 [0.0655;0.181] | p = 0.706 |
| no (n = 42) | 0.089 [0.050;0.153] | ||
| Positive aPL | yes (n = 35) | 0.063 [0.044;0.142] | p = 0.142 |
| no (n = 42) | 0.091 [0.055;0.143] | ||
| Parameters | Nucleosomes (−), n = 45, n (%) | Nucleosomes (+), n = 32, n (%) | χ2; p OR and 95% CI | |
|---|---|---|---|---|
| Lupus nephritis | yes | 26 (57.8) | 11 (34.4) | 4.1; 0.043 2.61 [1.02–6.68] |
| no | 19 (42.2) | 21 (65.6) | ||
| Arthritis | yes | 36 (80) | 19 (59.4) | 3.89; 0.048 1.71 [1.03–2.83] |
| no | 9 (20) | 13 (40.6) | ||
| High SLE activity | yes | 20 (44.4) | 2 (6.25) | 13.37; <0.0001 12.0 [2.55–56.39] |
| no | 25 (55.6) | 30 (93.8) | ||
| Positive anti-dsDNA | yes | 36 (80) | 20 (62.5) | 2.89; 0.076 2.4 [0.86–6.67] |
| no | 9 (20) | 12 (37.5) | ||
| Hypocomplementemia | yes | 29 (64.4) | 16 (50) | 1.61; 0.205 1.81 [0.72–4.56] |
| no | 16 (35.6) | 16 (50) | ||
| Parameter | Nucleosomes (−), n = 32, n (%) | Nucleosomes (+), n = 56, n (%) | χ2; p OR and 95% CI | |
|---|---|---|---|---|
| Highly positive levels of IgM aCL | yes | 1 (3) | 11 (20) | 4.52; 0.049 0.14 [0.017–1.112] |
| no | 31 (97) | 45 (80) | ||
| Highly positive levels of IgM aβ2GP1 | yes | 0 | 49 (88) | 4.21; 0.047 0.61 [0.52–0.73] |
| no | 32 (100) | 7 (12) | ||
| Authors | Number of Patients | Methods of NETs Detection | Association of NETs with Lupus Nephritis | Association of NETs with Daily Proteinuria | Association of NETs with Anti-dsDNA | Association of NETs with C3, C4 | Association of NETs with SLE Activity |
|---|---|---|---|---|---|---|---|
| Hakkim et al., 2010 [17] | SLE 61, RA 30, HC 54 | Destruction of NETs by serum | + | ND | + | ND | ND |
| Leffler et al., 2012 [18] | SLE 94, HC 54 | 1. Destruction of NETs by serum 2.DNAase activity | + | ND | + | + | + |
| Leffler et al., 2013 [19] | SLE 69 | Destruction of NETs by serum | + | ND | + | + | + |
| Zhang et al., 2014 [20] | SLE 54, HC 43 | Plasma cfDNA levels | + | + | − | − | − |
| van der Linden et al., 2018 [15] | SLE 55, SLE with APS 38, PAPS 28, HC 27 | NETs production | − | ND | + | − | − |
| El-Ghoneimy et al., 2019 [16] | SLE 50, HC 50 | NETs production | + | + | + | + | + |
| Jeremic et al., 2019 [21] | SLE 111, HC 50 | cfDNA levels | − | ND | + | − | − |
| Bruschi et al., 2020 [14] | SLE 216, SLE with lupus nephritis 103, HC 50 | 1.MPO-DNA complex ELISA 2.NETs production 3.DNAase activity | + | ND | ND | − | − |
| Hanata et al., 2022 [13] | SLE 33, HC 19 | MPO-DNA complex ELISA | − | ND | + | - | − |
| Reshetnyak et al., 2023 (current study) | SLE 30, SLE + APS 47, PAPS 41, HC 20 | MPO-DNA complex ELISA | + | - | + | + | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reshetnyak, T.; Nurbaeva, K.; Ptashnik, I.; Kudriaeva, A.; Belogurov, A., Jr.; Lila, A.; Nasonov, E. Markers of NETosis in Patients with Systemic Lupus Erythematosus and Antiphospholipid Syndrome. Int. J. Mol. Sci. 2023, 24, 9210. https://doi.org/10.3390/ijms24119210
Reshetnyak T, Nurbaeva K, Ptashnik I, Kudriaeva A, Belogurov A Jr., Lila A, Nasonov E. Markers of NETosis in Patients with Systemic Lupus Erythematosus and Antiphospholipid Syndrome. International Journal of Molecular Sciences. 2023; 24(11):9210. https://doi.org/10.3390/ijms24119210
Chicago/Turabian StyleReshetnyak, Tatiana, Kamila Nurbaeva, Ivan Ptashnik, Anna Kudriaeva, Alexey Belogurov, Jr., Aleksandr Lila, and Evgeny Nasonov. 2023. "Markers of NETosis in Patients with Systemic Lupus Erythematosus and Antiphospholipid Syndrome" International Journal of Molecular Sciences 24, no. 11: 9210. https://doi.org/10.3390/ijms24119210
APA StyleReshetnyak, T., Nurbaeva, K., Ptashnik, I., Kudriaeva, A., Belogurov, A., Jr., Lila, A., & Nasonov, E. (2023). Markers of NETosis in Patients with Systemic Lupus Erythematosus and Antiphospholipid Syndrome. International Journal of Molecular Sciences, 24(11), 9210. https://doi.org/10.3390/ijms24119210







