Investigating Mammalian Formins with SMIFH2 Fifteen Years in: Novel Targets and Unexpected Biology
Abstract
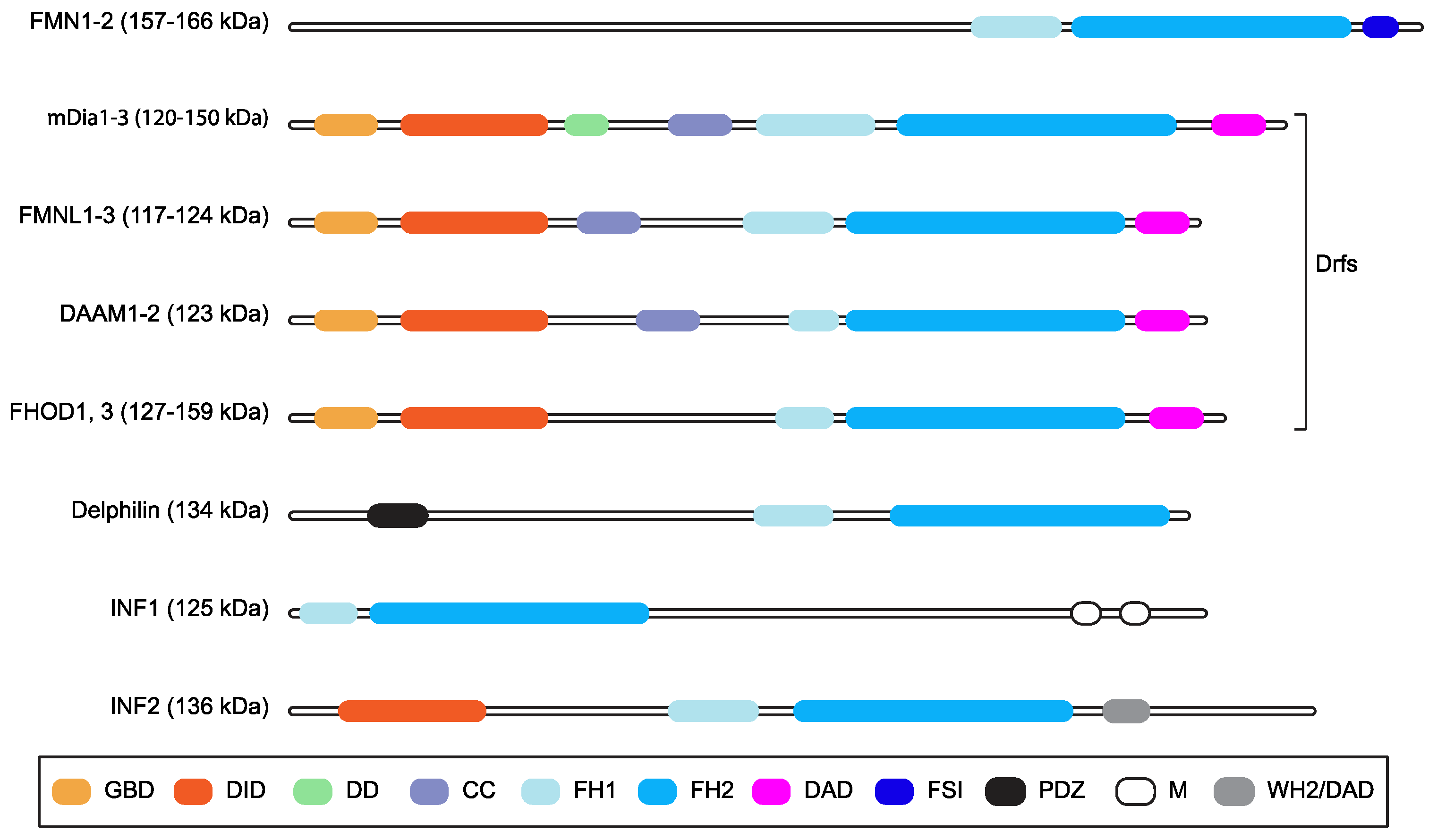
1. Introduction
2. SMIFH2 Discovery and Characterization In Vitro and in Cells
3. Identification of Mammalian Myosins and Interferons as SMIFH2 Targets
3.1. Myosins
3.2. Interferons
4. High-Throughput Bioactivity Assays Reveal Novel SMIFH2 Targets
4.1. Ribonuclease H (RNase H) Activity of the HIV-1 Reverse Transcriptase
4.2. Dual-Specificity Phosphatase 3 and Dual-Specificity Phosphatase 6
4.3. Tyrosine-Protein Phosphatase Non-Receptor Type 7 Isoform 2
4.4. Heat Shock 70kDa Protein 1A
5. Possible Mechanism(s) for Formin Inhibition by SMIFH2
5.1. Non-Covalent Reversible Inhibition of the FH2 Domain
5.2. Covalent Inhibition of Formins
6. Can SMIFH2 Be Modified to Obtain an Isoform-Specific Formin Inhibitor?
7. Conclusions and Future Directions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chesarone, M.A.; DuPage, A.G.; Goode, B.L. Unleashing Formins to Remodel the Actin and Microtubule Cytoskeletons. Nat. Rev. Mol. Cell Biol. 2010, 11, 62–74. [Google Scholar] [CrossRef] [PubMed]
- Isogai, T.; Innocenti, M. New Nuclear and Perinuclear Functions of Formins. Biochem. Soc. Trans. 2016, 44, 1701–1708. [Google Scholar] [CrossRef] [PubMed]
- Valencia, D.A.; Quinlan, M.E. Formins. Curr. Biol. 2021, 31, R517–R522. [Google Scholar] [CrossRef]
- Schonichen, A.; Geyer, M. Fifteen Formins for an Actin Filament: A Molecular View on the Regulation of Human Formins. Biochim. Biophys. Acta 2010, 1803, 152–163. [Google Scholar] [CrossRef][Green Version]
- Peng, J.; Wallar, B.J.; Flanders, A.; Swiatek, P.J.; Alberts, A.S. Disruption of the Diaphanous-Related Formin Drf1 Gene Encoding MDia1 Reveals a Role for Drf3 as an Effector for Cdc42. Curr. Biol. 2003, 13, 534–545. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wallar, B.J.; Stropich, B.N.; Schoenherr, J.A.; Holman, H.A.; Kitchen, S.M.; Alberts, A.S. The Basic Region of the Diaphanous-Autoregulatory Domain (DAD) Is Required for Autoregulatory Interactions with the Diaphanous-Related Formin Inhibitory Domain. J. Biol. Chem. 2006, 281, 4300–4307. [Google Scholar] [CrossRef][Green Version]
- Alberts, A.S. Identification of a Carboxyl-Terminal Diaphanous-Related Formin Homology Protein Autoregulatory Domain. J. Biol. Chem. 2001, 276, 2824–2830. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gould, C.J.; Maiti, S.; Michelot, A.; Graziano, B.R.; Blanchoin, L.; Goode, B.L. The Formin DAD Domain Plays Dual Roles in Autoinhibition and Actin Nucleation. Curr. Biol. 2011, 21, 384–390. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Thurston, S.F.; Kulacz, W.A.; Shaikh, S.; Lee, J.M.; Copeland, J.W. The Ability to Induce Microtubule Acetylation Is a General Feature of Formin Proteins. PLoS ONE 2012, 7, e48041. [Google Scholar] [CrossRef][Green Version]
- Bartolini, F.; Moseley, J.B.; Schmoranzer, J.; Cassimeris, L.; Goode, B.L.; Gundersen, G.G. The Formin MDia2 Stabilizes Microtubules Independently of Its Actin Nucleation Activity. J. Cell Biol. 2008, 181, 523–536. [Google Scholar] [CrossRef][Green Version]
- Gaillard, J.; Ramabhadran, V.; Neumanne, E.; Gurel, P.; Blanchoin, L.; Vantard, M.; Higgs, H.N. Differential Interactions of the Formins INF2, MDia1, and MDia2 with Microtubules. Mol. Biol. Cell 2011, 22, 4575–4587. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Eng, C.H.; Schmoranzer, J.; Cabrera-Poch, N.; Morris, E.J.S.; Chen, M.; Wallar, B.J.; Alberts, A.S.; Gundersen, G.G. EB1 and APC Bind to MDia to Stabilize Microtubules Downstream of Rho and Promote Cell Migration. Nat. Cell Biol. 2004, 6, 820–830. [Google Scholar] [CrossRef]
- Manor, U.; Bartholomew, S.; Golani, G.; Christenson, E.; Kozlov, M.; Higgs, H.; Spudich, J.; Lippincott-Schwartz, J. A Mitochondria-Anchored Isoform of the Actin-Nucleating Spire Protein Regulates Mitochondrial Division. eLife 2015, 4, e08828. [Google Scholar] [CrossRef] [PubMed]
- Cangkrama, M.; Liu, H.; Whipman, J.; Zubair, M.; Matsushita, M.; Di Filippo, M.; Kopf, M.; Innocenti, M.; Werner, S. A Pro-Tumorigenic MDia2-MIRO1 Axis Controls Mitochondrial Positioning and Function in Cancer-Associated Fibroblasts. Cancer Res 2022, 82, 3701–3717. [Google Scholar] [CrossRef] [PubMed]
- Wallar, B.J.; Deward, A.D.; Resau, J.H.; Alberts, A.S. RhoB and the Mammalian Diaphanous-Related Formin MDia2 in Endosome Trafficking. Exp. Cell Res. 2007, 313, 560–571. [Google Scholar] [CrossRef] [PubMed]
- Copeland, J. Actin-Based Regulation of Ciliogenesis–The Long and the Short of It. Semin. Cell Dev. Biol. 2020, 102, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Beli, P.; Mascheroni, D.; Xu, D.; Innocenti, M. WAVE and Arp2/3 Jointly Inhibit Filopodium Formation by Entering into a Complex with MDia2. Nat. Cell Biol. 2008, 10, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Isogai, T.; van der Kammen, R.; Leyton-Puig, D.; Kedziora, K.M.; Jalink, K.; Innocenti, M. Initiation of Lamellipodia and Ruffles Involves Cooperation between MDia1 and the Arp2/3 Complex. J. Cell Sci. 2015, 128, 3796–3810. [Google Scholar] [CrossRef][Green Version]
- Kedziora, K.M.; Isogai, T.; Jalink, K.; Innocenti, M. Invadosomes-Shaping Actin Networks to Follow Mechanical Cues. Front. Biosci. (Landmark Ed) 2016, 21, 1092–1117. [Google Scholar]
- Dhanda, A.S.; Vogl, A.W.; Ness, F.; Innocenti, M.; Guttman, J.A. MDia1 Assembles a Linear F-Actin Coat at Membrane Invaginations To Drive Listeria Monocytogenes Cell-to-Cell Spreading. mBio 2021, 12, e0293921. [Google Scholar] [CrossRef]
- Colucci-Guyon, E.; Niedergang, F.; Wallar, B.J.; Peng, J.; Alberts, A.S.; Chavrier, P. A Role for Mammalian Diaphanous-Related Formins in Complement Receptor (CR3)-Mediated Phagocytosis in Macrophages. Curr. Biol. 2005, 15, 2007–2012. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Damiani, D.; Goffinet, A.M.; Alberts, A.; Tissir, F. Lack of Diaph3 Relaxes the Spindle Checkpoint Causing the Loss of Neural Progenitors. Nat. Commun. 2016, 7, 13509. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lau, E.O.-C.; Damiani, D.; Chehade, G.; Ruiz-Reig, N.; Saade, R.; Jossin, Y.; Aittaleb, M.; Schakman, O.; Tajeddine, N.; Gailly, P.; et al. DIAPH3 Deficiency Links Microtubules to Mitotic Errors, Defective Neurogenesis, and Brain Dysfunction. eLife 2021, 10, e61974. [Google Scholar] [CrossRef]
- Olson, E.N.; Nordheim, A. Linking Actin Dynamics and Gene Transcription to Drive Cellular Motile Functions. Nat. Rev. Mol. Cell Biol. 2010, 11, 353–365. [Google Scholar] [CrossRef][Green Version]
- Hyrskyluoto, A.; Vartiainen, M.K. Regulation of Nuclear Actin Dynamics in Development and Disease. Curr. Opin. Cell Biol. 2020, 64, 18–24. [Google Scholar] [CrossRef]
- Labat-de-Hoz, L.; Alonso, M.A. Formins in Human Disease. Cells 2021, 10, 2554. [Google Scholar] [CrossRef] [PubMed]
- Chiereghin, C.; Robusto, M.; Massa, V.; Castorina, P.; Ambrosetti, U.; Asselta, R.; Soldà, G. Role of Cytoskeletal Diaphanous-Related Formins in Hearing Loss. Cells 2022, 11, 1726. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, S.A.; Neidt, E.M.; Cui, J.; Feiger, Z.; Skau, C.T.; Gardel, M.L.; Kozmin, S.A.; Kovar, D.R. Identification and Characterization of a Small Molecule Inhibitor of Formin-Mediated Actin Assembly. Chem. Biol. 2009, 16, 1158–1168. [Google Scholar] [CrossRef][Green Version]
- Orman, M.; Landis, M.; Oza, A.; Nambiar, D.; Gjeci, J.; Song, K.; Huang, V.; Klestzick, A.; Hachicho, C.; Liu, S.Q.; et al. Alterations to the Broad-Spectrum Formin Inhibitor SMIFH2 Modulate Potency but Not Specificity. Sci. Rep. 2022, 12, 13520. [Google Scholar] [CrossRef]
- Nishimura, Y.; Shi, S.; Zhang, F.; Liu, R.; Takagi, Y.; Bershadsky, A.D.; Viasnoff, V.; Sellers, J.R. The Formin Inhibitor SMIFH2 Inhibits Members of the Myosin Superfamily. J. Cell Sci. 2021, 134, jcs253708. [Google Scholar] [CrossRef]
- Isogai, T.; van der Kammen, R.; Innocenti, M. SMIFH2 Has Effects on Formins and P53 That Perturb the Cell Cytoskeleton. Sci. Rep. 2015, 5, 9802. [Google Scholar] [CrossRef][Green Version]
- Thoidingjam, L.K.; Blouin, C.M.; Gaillet, C.; Brion, A.; Solier, S.; Niyomchon, S.; El Marjou, A.; Mouasni, S.; Sepulveda, F.E.; de Saint Basile, G.; et al. Small Molecule Inhibitors of Interferon-Induced JAK-STAT Signalling. Angew. Chem. Int. Ed. Engl. 2022, 61, e202205231. [Google Scholar] [CrossRef]
- Houdusse, A.; Titus, M.A. The Many Roles of Myosins in Filopodia, Microvilli and Stereocilia. Curr. Biol. 2021, 31, R586–R602. [Google Scholar] [CrossRef] [PubMed]
- Innocenti, M. New Insights into the Formation and the Function of Lamellipodia and Ruffles in Mesenchymal Cell Migration. Cell Adhes. Migr. 2018, 12, 401–416. [Google Scholar] [CrossRef] [PubMed]
- Limouze, J.; Straight, A.F.; Mitchison, T.; Sellers, J.R. Specificity of Blebbistatin, an Inhibitor of Myosin II. J. Muscle Res. Cell Motil. 2004, 25, 337–341. [Google Scholar] [CrossRef]
- Tee, Y.H.; Shemesh, T.; Thiagarajan, V.; Hariadi, R.F.; Anderson, K.L.; Page, C.; Volkmann, N.; Hanein, D.; Sivaramakrishnan, S.; Kozlov, M.M.; et al. Cellular Chirality Arising from the Self-Organization of the Actin Cytoskeleton. Nat. Cell Biol. 2015, 17, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Alieva, N.O.; Efremov, A.K.; Hu, S.; Oh, D.; Chen, Z.; Natarajan, M.; Ong, H.T.; Jégou, A.; Romet-Lemonne, G.; Groves, J.T.; et al. Myosin IIA and Formin Dependent Mechanosensitivity of Filopodia Adhesion. Nat. Commun. 2019, 10, 3593. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jacquemet, G.; Hamidi, H.; Ivaska, J. Filopodia in Cell Adhesion, 3D Migration and Cancer Cell Invasion. Curr. Opin. Cell Biol. 2015, 36, 23–31. [Google Scholar] [CrossRef][Green Version]
- Benci, J.L.; Xu, B.; Qiu, Y.; Wu, T.J.; Dada, H.; Twyman-Saint Victor, C.; Cucolo, L.; Lee, D.S.M.; Pauken, K.E.; Huang, A.C.; et al. Tumor Interferon Signaling Regulates a Multigenic Resistance Program to Immune Checkpoint Blockade. Cell 2016, 167, 1540–1554 e12. [Google Scholar] [CrossRef][Green Version]
- Macarron, R.; Banks, M.N.; Bojanic, D.; Burns, D.J.; Cirovic, D.A.; Garyantes, T.; Green, D.V.; Hertzberg, R.P.; Janzen, W.P.; Paslay, J.W.; et al. Impact of High-Throughput Screening in Biomedical Research. Nat. Rev. Drug Discov. 2011, 10, 188–195. [Google Scholar] [CrossRef]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug Repurposing: Progress, Challenges and Recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Madia, V.N.; Messore, A.; De Leo, A.; Tudino, V.; Pindinello, I.; Saccoliti, F.; De Vita, D.; Scipione, L.; Costi, R.; Di Santo, R. Small-Molecule Inhibitors of HIV-1 Reverse Transcriptase-Associated Ribonuclease H Function: Challenges and Recent Developments. Curr. Med. Chem. 2021, 28, 6146–6178. [Google Scholar] [CrossRef] [PubMed]
- Parniak, M.A.; Min, K.L.; Budihas, S.R.; Le Grice, S.F.; Beutler, J.A. A Fluorescence-Based High-Throughput Screening Assay for Inhibitors of Human Immunodeficiency Virus-1 Reverse Transcriptase-Associated Ribonuclease H Activity. Anal. Biochem. 2003, 322, 33–39. [Google Scholar] [CrossRef] [PubMed]
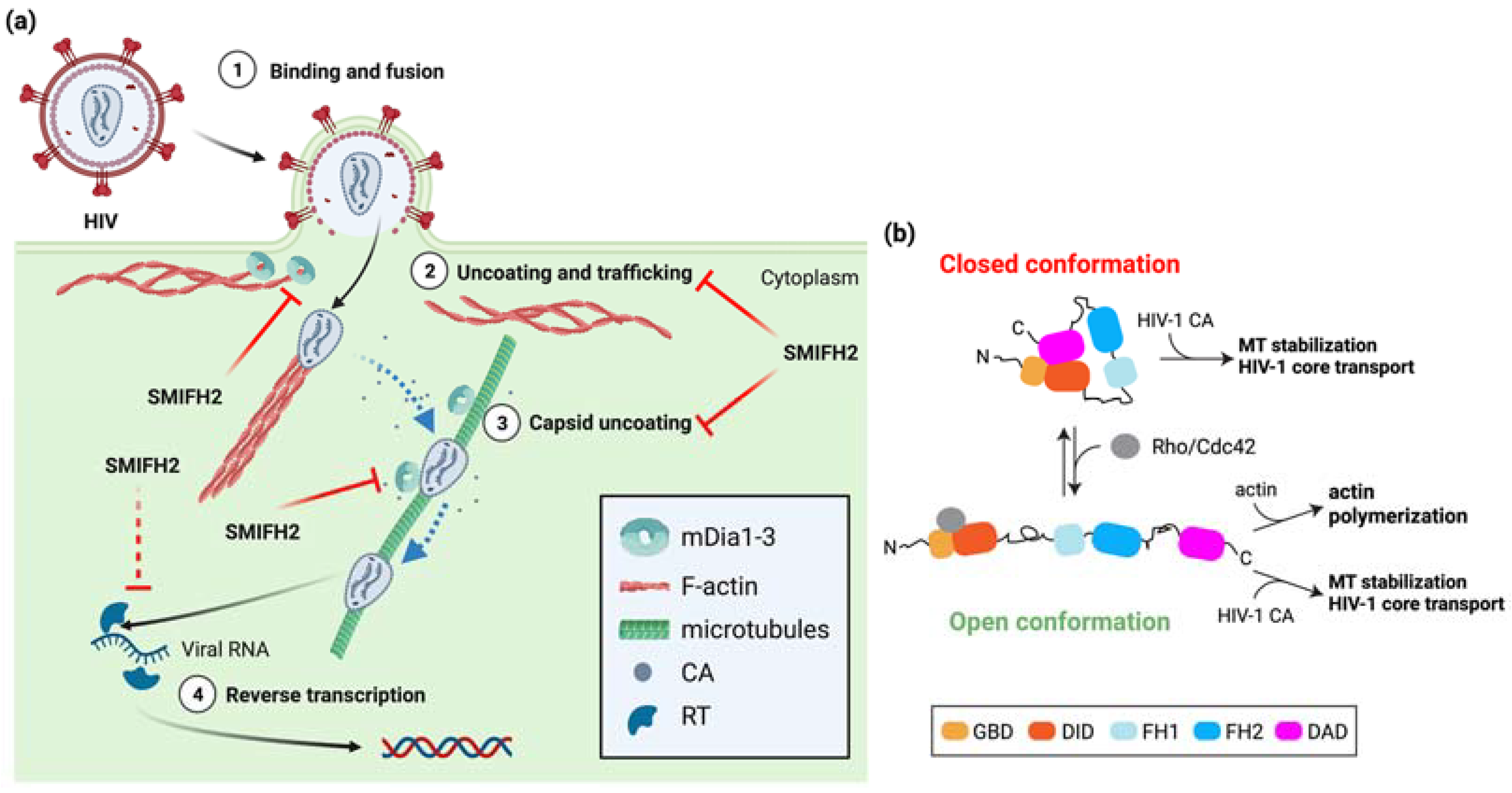
- Rawle, D.J.; Harrich, D. Toward the “Unravelling” of HIV: Host Cell Factors Involved in HIV-1 Core Uncoating. PLoS Pathog. 2018, 14, e1007270. [Google Scholar] [CrossRef][Green Version]
- Naghavi, M.H. HIV-1 Capsid Exploitation of the Host Microtubule Cytoskeleton during Early Infection. Retrovirology 2021, 18, 19. [Google Scholar] [CrossRef]
- Sabo, Y.; Walsh, D.; Barry, D.S.; Tinaztepe, S.; de Los Santos, K.; Goff, S.P.; Gundersen, G.G.; Naghavi, M.H. HIV-1 Induces the Formation of Stable Microtubules to Enhance Early Infection. Cell Host Microbe 2013, 14, 535–546. [Google Scholar] [CrossRef][Green Version]
- Delaney, M.K.; Malikov, V.; Chai, Q.; Zhao, G.; Naghavi, M.H. Distinct Functions of Diaphanous-Related Formins Regulate HIV-1 Uncoating and Transport. Proc. Natl. Acad. Sci. USA 2017, 114, E6932–E6941. [Google Scholar] [CrossRef][Green Version]
- Naranatt, P.P.; Krishnan, H.H.; Smith, M.S.; Chandran, B. Kaposi’s Sarcoma-Associated Herpesvirus Modulates Microtubule Dynamics via RhoA-GTP-Diaphanous 2 Signaling and Utilizes the Dynein Motors to Deliver Its DNA to the Nucleus. J. Virol. 2005, 79, 1191–1206. [Google Scholar] [CrossRef][Green Version]
- del Real, G.; Jimenez-Baranda, S.; Mira, E.; Lacalle, R.A.; Lucas, P.; Gomez-Mouton, C.; Alegret, M.; Pena, J.M.; Rodriguez-Zapata, M.; Alvarez-Mon, M.; et al. Statins Inhibit HIV-1 Infection by down-Regulating Rho Activity. J. Exp. Med. 2004, 200, 541–547. [Google Scholar] [CrossRef][Green Version]
- Lucera, M.B.; Fleissner, Z.; Tabler, C.O.; Schlatzer, D.M.; Troyer, Z.; Tilton, J.C. HIV Signaling through CD4 and CCR5 Activates Rho Family GTPases That Are Required for Optimal Infection of Primary CD4+ T Cells. Retrovirology 2017, 14, 4. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bartolini, F.; Ramalingam, N.; Gundersen, G.G. Actin-Capping Protein Promotes Microtubule Stability by Antagonizing the Actin Activity of MDia1. Mol. Biol. Cell 2012, 23, 4032–4040. [Google Scholar] [CrossRef]
- Destaing, O.; Saltel, F.; Gilquin, B.; Chabadel, A.; Khochbin, S.; Ory, S.; Jurdic, P. A Novel Rho-MDia2-HDAC6 Pathway Controls Podosome Patterning through Microtubule Acetylation in Osteoclasts. J. Cell Sci. 2005, 118, 2901–2911. [Google Scholar] [CrossRef] [PubMed][Green Version]
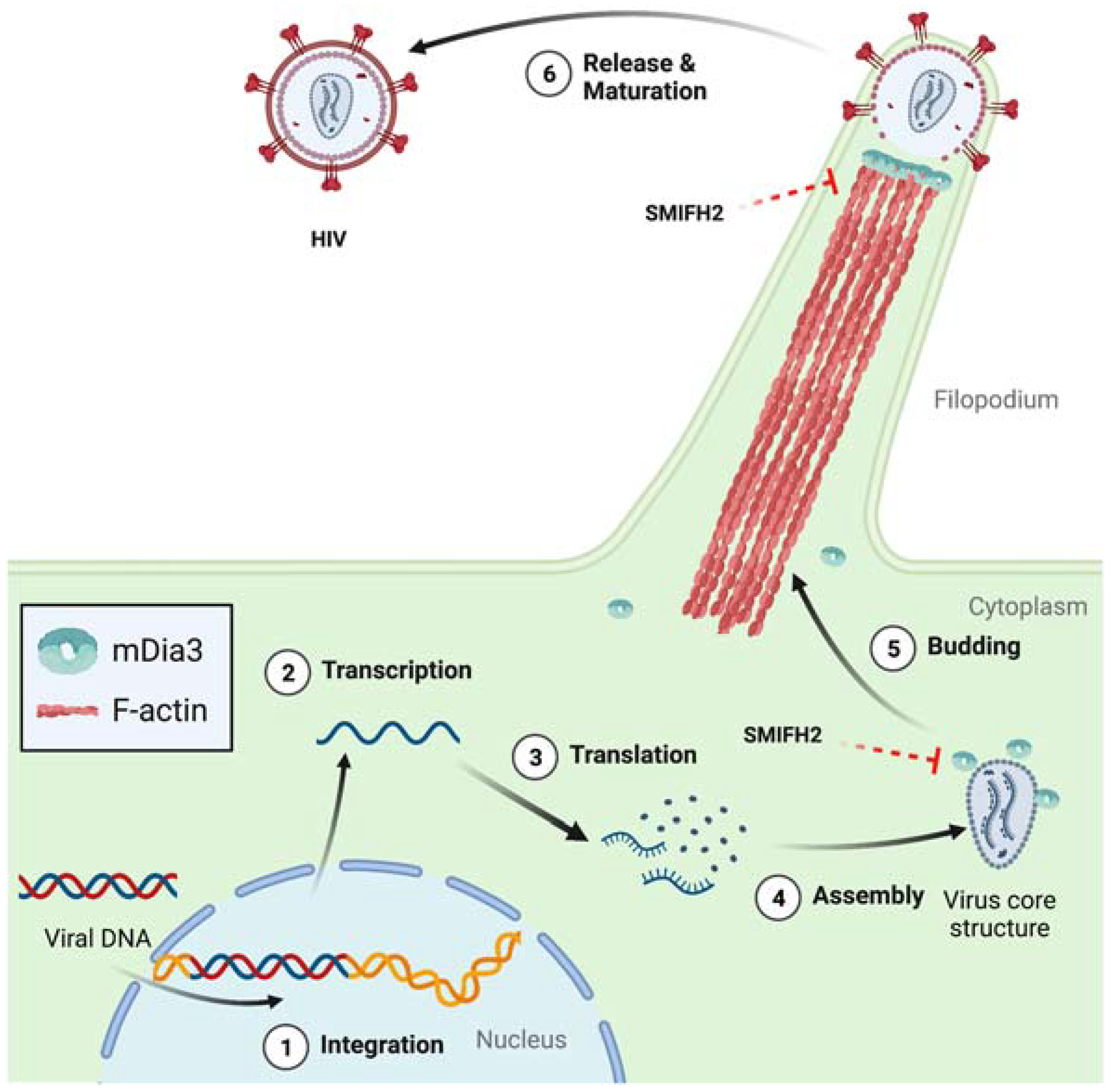
- Aggarwal, A.; Iemma, T.L.; Shih, I.; Newsome, T.P.; McAllery, S.; Cunningham, A.L.; Turville, S.G. Mobilization of HIV Spread by Diaphanous 2 Dependent Filopodia in Infected Dendritic Cells. PLoS Pathog. 2012, 8, e1002762. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sowinski, S.; Jolly, C.; Berninghausen, O.; Purbhoo, M.A.; Chauveau, A.; Kohler, K.; Oddos, S.; Eissmann, P.; Brodsky, F.M.; Hopkins, C.; et al. Membrane Nanotubes Physically Connect T Cells over Long Distances Presenting a Novel Route for HIV-1 Transmission. Nat. Cell Biol. 2008, 10, 211–219. [Google Scholar] [CrossRef]
- Zurzolo, C. Tunneling Nanotubes: Reshaping Connectivity. Curr. Opin. Cell Biol. 2021, 71, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Argenzio, E.; Innocenti, M. The Chloride Intracellular Channel Protein CLIC4 Inhibits Filopodium Formation Induced by Constitutively Active Mutants of Formin MDia2. FEBS Lett. 2020, 594, 1750–1758. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Argenzio, E.; Klarenbeek, J.; Kedziora, K.M.; Nahidiazar, L.; Isogai, T.; Perrakis, A.; Jalink, K.; Moolenaar, W.H.; Innocenti, M. Profilin Binding Couples Chloride Intracellular Channel Protein CLIC4 to RhoA-MDia2 Signaling and Filopodium Formation. J. Biol. Chem. 2018, 293, 19161–19176. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mattila, P.K.; Lappalainen, P. Filopodia: Molecular Architecture and Cellular Functions. Nat. Rev. Mol. Cell Biol. 2008, 9, 446–454. [Google Scholar] [CrossRef]
- Zhong, L.; Li, Y.; Xiong, L.; Wang, W.; Wu, M.; Yuan, T.; Yang, W.; Tian, C.; Miao, Z.; Wang, T.; et al. Small Molecules in Targeted Cancer Therapy: Advances, Challenges, and Future Perspectives. Signal Transduct. Target. Ther. 2021, 6, 201. [Google Scholar] [CrossRef]
- Lang, R.; Raffi, F.A.M. Dual-Specificity Phosphatases in Immunity and Infection: An Update. Int. J. Mol. Sci. 2019, 20, 2710. [Google Scholar] [CrossRef][Green Version]
- Pavic, K.; Duan, G.; Kohn, M. VHR/DUSP3 Phosphatase: Structure, Function and Regulation. FEBS J. 2015, 282, 1871–1890. [Google Scholar] [CrossRef]
- Amand, M.; Erpicum, C.; Bajou, K.; Cerignoli, F.; Blacher, S.; Martin, M.; Dequiedt, F.; Drion, P.; Singh, P.; Zurashvili, T.; et al. DUSP3/VHR Is a pro-Angiogenic Atypical Dual-Specificity Phosphatase. Mol. Cancer 2014, 13, 108. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Maillet, M.; Purcell, N.H.; Sargent, M.A.; York, A.J.; Bueno, O.F.; Molkentin, J.D. DUSP6 (MKP3) Null Mice Show Enhanced ERK1/2 Phosphorylation at Baseline and Increased Myocyte Proliferation in the Heart Affecting Disease Susceptibility. J. Biol. Chem. 2008, 283, 31246–31255. [Google Scholar] [CrossRef][Green Version]
- Francis, D.M.; Rozycki, B.; Koveal, D.; Hummer, G.; Page, R.; Peti, W. Structural Basis of P38alpha Regulation by Hematopoietic Tyrosine Phosphatase. Nat. Chem. Biol. 2011, 7, 916–924. [Google Scholar] [CrossRef]
- Saxena, M.; Williams, S.; Tasken, K.; Mustelin, T. Crosstalk between CAMP-Dependent Kinase and MAP Kinase through a Protein Tyrosine Phosphatase. Nat. Cell Biol. 1999, 1, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Sergienko, E.; Xu, J.; Liu, W.H.; Dahl, R.; Critton, D.A.; Su, Y.; Brown, B.T.; Chan, X.; Yang, L.; Bobkova, E.V.; et al. Inhibition of Hematopoietic Protein Tyrosine Phosphatase Augments and Prolongs ERK1/2 and P38 Activation. ACS Chem. Biol. 2012, 7, 367–377. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Thumkeo, D.; Katsura, Y.; Nishimura, Y.; Kanchanawong, P.; Tohyama, K.; Ishizaki, T.; Kitajima, S.; Takahashi, C.; Hirata, T.; Watanabe, N.; et al. MDia1/3-Dependent Actin Polymerization Spatiotemporally Controls LAT Phosphorylation by Zap70 at the Immune Synapse. Sci. Adv. 2020, 6, eaay2432. [Google Scholar] [CrossRef][Green Version]
- Gomez, T.S.; Kumar, K.; Medeiros, R.B.; Shimizu, Y.; Leibson, P.J.; Billadeau, D.D. Formins Regulate the Actin-Related Protein 2/3 Complex-Independent Polarization of the Centrosome to the Immunological Synapse. Immunity 2007, 26, 177–190. [Google Scholar] [CrossRef][Green Version]
- Murugesan, S.; Hong, J.; Yi, J.; Li, D.; Beach, J.R.; Shao, L.; Meinhardt, J.; Madison, G.; Wu, X.; Betzig, E.; et al. Formin-Generated Actomyosin Arcs Propel T Cell Receptor Microcluster Movement at the Immune Synapse. J. Cell Biol. 2016, 215, 383–399. [Google Scholar] [CrossRef][Green Version]
- Vostakolaei, M.A.; Hatami-Baroogh, L.; Babaei, G.; Molavi, O.; Kordi, S.; Abdolalizadeh, J. Hsp70 in Cancer: A Double Agent in the Battle between Survival and Death. J. Cell Physiol. 2021, 236, 3420–3444. [Google Scholar] [CrossRef]
- Isogai, T.; van der Kammen, R.; Goerdayal, S.S.; Heck, A.J.; Altelaar, A.F.; Innocenti, M. Proteomic Analyses Uncover a New Function and Mode of Action for Mouse Homolog of Diaphanous 2 (MDia2). Mol. Cell Proteom. 2015, 14, 1064–1078. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cangkrama, M.; Wietecha, M.; Mathis, N.; Okumura, R.; Ferrarese, L.; Al-Nuaimi, D.; Antsiferova, M.; Dummer, R.; Innocenti, M.; Werner, S. A Paracrine Activin A-MDia2 Axis Promotes Squamous Carcinogenesis via Fibroblast Reprogramming. EMBO Mol. Med. 2020, 12, e11466. [Google Scholar] [CrossRef] [PubMed]
- Isogai, T.; van der Kammen, R.; Bleijerveld, O.B.; Goerdayal, S.S.; Argenzio, E.; Altelaar, A.F.; Innocenti, M. Quantitative Proteomics Illuminates a Functional Interaction between MDia2 and the Proteasome. J. Proteome Res. 2016, 15, 4624–4637. [Google Scholar] [CrossRef] [PubMed]
- Boysen, M.; Kityk, R.; Mayer, M.P. Hsp70- and Hsp90-Mediated Regulation of the Conformation of P53 DNA Binding Domain and P53 Cancer Variants. Mol. Cell 2019, 74, 831–843 e4. [Google Scholar] [CrossRef]
- Nishikawa, S.; Kaida, A.; Parrales, A.; Ranjan, A.; Alalem, M.; Ren, H.; Schoenen, F.J.; Johnson, D.K.; Iwakuma, T. DNAJA1- and Conformational Mutant P53-Dependent Inhibition of Cancer Cell Migration by a Novel Compound Identified through a Virtual Screen. Cell Death Discov. 2022, 8, 437. [Google Scholar] [CrossRef]
- Baell, J.B. Observations on Screening-Based Research and Some Concerning Trends in the Literature. Future Med. Chem. 2010, 2, 1529–1546. [Google Scholar] [CrossRef]
- Gauvin, T.J.; Fukui, J.; Peterson, J.R.; Higgs, H.N. Isoform-Selective Chemical Inhibition of MDia-Mediated Actin Assembly. Biochemistry 2009, 48, 9327–9329. [Google Scholar] [CrossRef][Green Version]

| Target Name (Organism) | AID | Source |
|---|---|---|
| Chain A, RIBONUCLEASE H (HIV-1) | 372 | Molecular Targets Development Program |
| Dusp6 (Rattus norvegicus) | 425 | Burnham Center for Chemical Genomics |
| PTPN7 (Homo sapiens) | 521 | Burnham Center for Chemical Genomics |
| HSPA1A (Homo sapiens) | 583 | Burnham Center for Chemical Genomics |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Innocenti, M. Investigating Mammalian Formins with SMIFH2 Fifteen Years in: Novel Targets and Unexpected Biology. Int. J. Mol. Sci. 2023, 24, 9058. https://doi.org/10.3390/ijms24109058
Innocenti M. Investigating Mammalian Formins with SMIFH2 Fifteen Years in: Novel Targets and Unexpected Biology. International Journal of Molecular Sciences. 2023; 24(10):9058. https://doi.org/10.3390/ijms24109058
Chicago/Turabian StyleInnocenti, Metello. 2023. "Investigating Mammalian Formins with SMIFH2 Fifteen Years in: Novel Targets and Unexpected Biology" International Journal of Molecular Sciences 24, no. 10: 9058. https://doi.org/10.3390/ijms24109058
APA StyleInnocenti, M. (2023). Investigating Mammalian Formins with SMIFH2 Fifteen Years in: Novel Targets and Unexpected Biology. International Journal of Molecular Sciences, 24(10), 9058. https://doi.org/10.3390/ijms24109058






