Marker-Assisted Selection in Breeding for Fruit Trait Improvement: A Review
Abstract
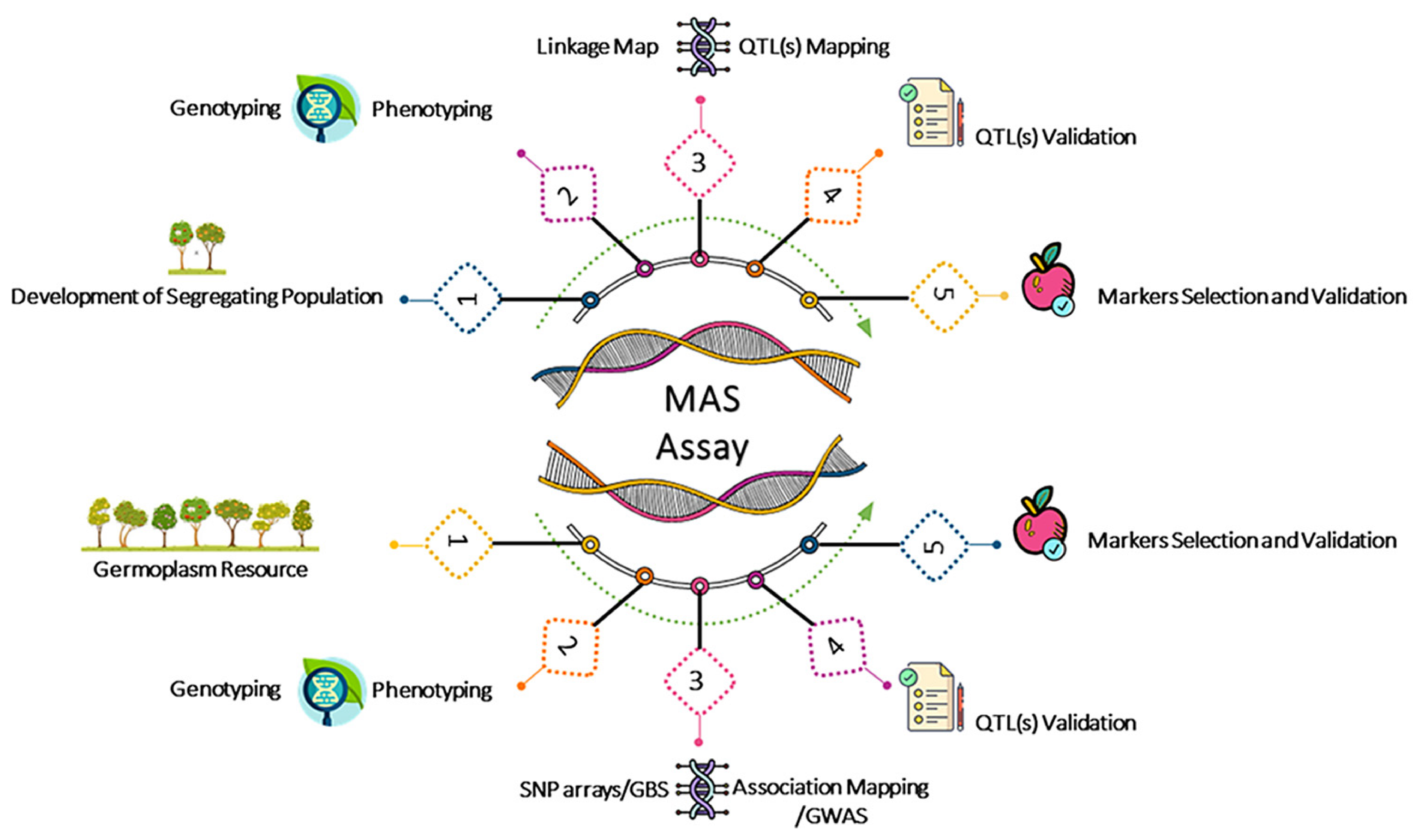
1. Introduction
2. Phenological Traits Related to Fruit Production
3. Skin Colour
4. Flesh Colour
5. Structural Qualities
5.1. Fruit Size and Weight
5.2. Seedlessness
5.3. Firmness, Crispness, Texture
6. Organoleptic Qualities
6.1. Soluble Soil Content (SSC) and Sugar Content
6.2. Pulp Acidity and pH
6.3. Dry Matter Content
6.4. Volatile Organic Compounds
7. Harvesting Date, Maturity Date, Ethylene, Ripening
8. Conclusions—Perspectives
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- González-Aguilar, G.; Robles-Sánchez, R.M.; Martínez-Téllez, M.A.; Olivas, G.I.; Alvarez-Parrilla, E.; De La Rosa, L.A. Bioactive Compounds in Fruits: Health Benefits and Effect of Storage Conditions. Stewart Postharvest Rev. 2008, 4, 1–10. [Google Scholar] [CrossRef]
- FAOSTAT. Available online: https://www.fao.org/faostat/en/#home (accessed on 20 April 2023).
- FAO STATISTICS. Statistical Yearbook 2021; Food and Argiculture Organization of the United Nations: Rome, Italy, 2021. [Google Scholar] [CrossRef]
- Moser, R.; Raffaelli, R.; Thilmany-McFadden, D. Consumer Preferences for Fruit and Vegetables with Credence-Based Attributes: A Review. Int. Food Agribus. Manag. Rev. 2011, 14, 121–142. [Google Scholar]
- Schreiner, M.; Korn, M.; Stenger, M.; Holzgreve, L.; Altmann, M. Current Understanding and Use of Quality Characteristics of Horticulture Products. Sci. Hortic. 2013, 163, 63–69. [Google Scholar] [CrossRef]
- Demattè, M.L.; Pojer, N.; Endrizzi, I.; Corollaro, M.L.; Betta, E.; Aprea, E.; Charles, M.; Biasioli, F.; Zampini, M.; Gasperi, F. Effects of the Sound of the Bite on Apple Perceived Crispness and Hardness. Food Qual. Prefer. 2014, 38, 58–64. [Google Scholar] [CrossRef]
- The European Parliament and the Council of the European Union. Regulation (EC) No 396/2005 of the European Parliament and of the Council of 23 February 2005 on Maximum Residue Levels of Pesticides in or on Food and Feed of Plant and Animal Origin and Amending Council Directive 91/414/EECText with EEA Relevance. Off. J. Eur. Union 2005, 50, 1–16. [Google Scholar]
- The European Parliament and the Council of the European Union. Regulation (EC) No 1107/2009 of the European Parliament and of the Council of 21 October 2009 Concerning the Placing of Plant Protection Products on the Market and Repealing Council Directives 79/117/EEC and 91/414/EEC. Available online: https://eur-lex.europa.eu/legal-content/en/TXT/?uri=celex%3A32005R0396 (accessed on 20 April 2023).
- Van Nocker, S.; Gardiner, S.E. Breeding Better Cultivars, Faster: Applications of New Technologies for the Rapid Deployment of Superior Horticultural Tree Crops. Hortic. Res. 2014, 1, 22. [Google Scholar] [CrossRef]
- Hackett, W.P. Horticultural Reviews. In Horticultural Reviews; Janick, J., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 1985; Volume 7, pp. 109–157. [Google Scholar]
- Visser, T. The Relation between Growth, Juvenile Period and Fruiting of Apple Seedlings and Its Use to Improve Breeding Efficiency. Euphytica 1970, 19, 293–302. [Google Scholar] [CrossRef]
- Aldwinckle, H.S. Flowering of Apple Seedlings 16–20 Months after Germination1. HortScience 1975, 10, 124–126. [Google Scholar] [CrossRef]
- Weigel, D.; Nilsson, O. A Developmental Switch Sufficient for Flower Initiation in Diverse Plants. Nature 1995, 377, 495–500. [Google Scholar] [CrossRef]
- Flachowsky, H.; Peil, A.; Sopanen, T.; Elo, A.; Hanke, V. Overexpression of BpMADS4 from Silver Birch (Betula pendula Roth.) Induces Early-Flowering in Apple (Malus × domestica Borkh.). Plant Breed. 2007, 126, 137–145. [Google Scholar] [CrossRef]
- Flachowsky, H.; Hanke, M.V.; Peil, A.; Strauss, S.H.; Fladung, M. A Review on Transgenic Approaches to Accelerate Breeding of Woody Plants. Plant Breed. 2009, 128, 217–226. [Google Scholar] [CrossRef]
- Singh, B.D.; Singh, A.K. Association Mapping. In Marker-Assisted Plant Breeding: Principles and Practices; Springer: New Delhi, India, 2015; pp. 217–256. [Google Scholar]
- Young, N.D. Constructing a Plant Genetic Linkage Map with DNA Markers. In DNA-Based Markers in Plants; Phillips, R.L., Vasil, I.K., Eds.; Springer: Dordrecht, The Netherlands, 1994; Volume 1, pp. 39–57. [Google Scholar]
- Kulwal, P.L. Trait Mapping Approaches through Linkage Mapping in Plants. In Plant Genetics and Molecular Biology; Varshney, R., Pandey, M., Chitikineni, A., Eds.; Springer Science and Business Media Deutschland GmbH: Berlin/Heidelberg, Germany, 2018; Volume 164, pp. 53–82. [Google Scholar]
- Michelmore, R.W.; Paran, I.; Kesseli, R.V. Identification of Markers Linked to Disease-Resistance Genes by Bulked Segregant Analysis: A Rapid Method to Detect Markers in Specific Genomic Regions by Using Segregating Populations. Proc. Natl. Acad. Sci. USA 1991, 88, 9828–9832. [Google Scholar] [CrossRef]
- Miles, C.; Wayne, M. Quantitative Trait Locus (QTL) Analysis | Learn Science at Scitable. Nat. Educ. 2008, 1, 208. [Google Scholar]
- Kumar, S.; Bink, M.C.A.M.; Volz, R.K.; Bus, V.G.M.; Chagné, D. Towards Genomic Selection in Apple (Malus × domestica Borkh.) Breeding Programmes: Prospects, Challenges and Strategies. Tree Genet. Genomes 2012, 8, 1–14. [Google Scholar] [CrossRef]
- Kumar, S.; Chagné, D.; Bink, M.C.A.M.; Volz, R.K.; Whitworth, C.; Carlisle, C. Genomic Selection for Fruit Quality Traits in Apple (Malus × domestica Borkh.). PLoS ONE 2012, 7, e36674. [Google Scholar] [CrossRef]
- Kumar, S.; Garrick, D.J.; Bink, M.C.A.M.; Whitworth, C.; Chagné, D.; Volz, R.K. Novel Genomic Approaches Unravel Genetic Architecture of Complex Traits in Apple. BMC Genom. 2013, 14, 393. [Google Scholar] [CrossRef]
- Gardiner, S.E.; Volz, R.K.; Chagné, D. Tools to Breed Better Cultivars Faster at Plant & Food Research. In Proceedings of the 1st International Rapid Cycle Crop Breeding Conference, Leesburg, VA, USA, 7–10 January 2014; pp. 7–9. [Google Scholar]
- Ru, S.; Main, D.; Evans, K.; Peace, C.; Ru, S.; Main, D.; Peace, C.; Evans, K. Current Applications, Challenges, and Perspectives of Marker-Assisted Seedling Selection in Rosaceae Tree Fruit Breeding. Tree Genet. Genomes 2015, 11, 8. [Google Scholar] [CrossRef]
- Wannemuehler, S.D.; Luby, J.J.; Yue, C.; Bedford, D.S.; Gallardo, R.K.; McCracken, V.A. A Cost–Benefit Analysis of DNA Informed Apple Breeding. HortScience 2019, 54, 1998–2004. [Google Scholar] [CrossRef]
- Luby, J.J.; Shaw, D.V. Does Marker-Assisted Selection Make Dollars and Sense in a Fruit Breeding Program? HortScience 2001, 36, 872–879. [Google Scholar] [CrossRef]
- Ru, S.; Hardner, C.; Evans, K.; Main, D.; Carter, P.A.; Harshman, J.; Sandefur, P.; Edge-Garza, D.; Peace, C. Empirical Evaluation of Multi-Trait DNA Testing in an Apple Seedling Population. Tree Genet. Genomes 2021, 17, 13. [Google Scholar] [CrossRef]
- Edge-Garza, D.A.; Luby, J.J.; Peace, C. Decision Support for Cost-Efficient and Logistically Feasible Marker-Assisted Seedling Selection in Fruit Breeding. Mol. Breed. 2015, 35, 223. [Google Scholar] [CrossRef]
- Wannemuehler, S.D.; Yue, C.; Shane, W.W.; Karina Gallardo, R.; McCracken, V. Estimated Implementation Costs of DNA-Informed Breeding in a Peach Breeding Program. Horttechnology 2020, 30, 356–364. [Google Scholar] [CrossRef]
- Campoy, J.A.; Ruiz, D.; Egea, J.; Jasper Rees, D.G.; Marc Celton, J.; Martínez-Gómez, P. Inheritance of Flowering Time in Apricot (Prunus Armeniaca L.) and Analysis of Linked Quantitative Trait Loci (QTLs) Using Simple Sequence Repeat (SSR) Markers. Plant Mol. Biol. Rep. 2011, 29, 404–410. [Google Scholar] [CrossRef]
- Urrestarazu, J.; Muranty, H.; Denancé, C.; Leforestier, D.; Ravon, E.; Guyader, A.; Guisnel, R.; Feugey, L.; Aubourg, S.; Celton, J.M.; et al. Genome-Wide Association Mapping of Flowering and Ripening Periods in Apple. Front. Plant Sci. 2017, 8, 1923. [Google Scholar] [CrossRef]
- da Silva Linge, C.; Cai, L.; Fu, W.; Clark, J.; Worthington, M.; Rawandoozi, Z.; Byrne, D.H.; Gasic, K. Multi-Locus Genome-Wide Association Studies Reveal Fruit Quality Hotspots in Peach Genome. Front. Plant Sci. 2021, 12, 644799. [Google Scholar] [CrossRef]
- Fan, S.; Bielenberg, D.G.; Zhebentyayeva, T.N.; Reighard, G.L.; Okie, W.R.; Holland, D.; Abbott, A.G. Mapping Quantitative Trait Loci Associated with Chilling Requirement, Heat Requirement and Bloom Date in Peach (Prunus Persica). New Phytol. 2010, 185, 917–930. [Google Scholar] [CrossRef]
- Romeu, J.F.; Monforte, A.J.; Sánchez, G.; Granell, A.; García-Brunton, J.; Badenes, M.L.; Ríos, G. Quantitative Trait Loci Affecting Reproductive Phenology in Peach. BMC Plant Biol. 2014, 14, 52. [Google Scholar] [CrossRef] [PubMed]
- Bielenberg, D.G.; Rauh, B.; Fan, S.; Gasic, K.; Abbott, A.G.; Reighard, G.L.; Okie, W.R.; Wells, C.E. Genotyping by Sequencing for SNP-Based Linkage Map Construction and QTL Analysis of Chilling Requirement and Bloom Date in Peach [Prunus Persica (L.) Batsch]. PLoS ONE 2015, 10, e0139406. [Google Scholar] [CrossRef]
- Rawandoozi, Z.J.; Hartmann, T.P.; Carpenedo, S.; Gasic, K.; da Silva Linge, C.; Cai, L.; Van de Weg, E.; Byrne, D.H. Mapping and Characterization QTLs for Phenological Traits in Seven Pedigree-Connected Peach Families. BMC Genom. 2021, 22, 187. [Google Scholar] [CrossRef] [PubMed]
- Aranzana, M.J.; Decroocq, V.; Dirlewanger, E.; Eduardo, I.; Gao, Z.S.; Gasic, K.; Iezzoni, A.; Jung, S.; Peace, C.; Prieto, H.; et al. Prunus Genetics and Applications after de Novo Genome Sequencing: Achievements and Prospects. Hortic. Res. 2019, 6, 58. [Google Scholar] [CrossRef]
- Hernández Mora, J.R.; Micheletti, D.; Bink, M.; Van de Weg, E.; Cantín, C.; Nazzicari, N.; Caprera, A.; Dettori, M.T.; Micali, S.; Banchi, E.; et al. Integrated QTL Detection for Key Breeding Traits in Multiple Peach Progenies. BMC Genom. 2017, 18, 404. [Google Scholar] [CrossRef] [PubMed]
- Castède, S.; Campoy, J.A.; García, J.Q.; Le Dantec, L.; Lafargue, M.; Barreneche, T.; Wenden, B.; Dirlewanger, E. Genetic Determinism of Phenological Traits Highly Affected by Climate Change in Prunus Avium: Flowering Date Dissected into Chilling and Heat Requirements. New Phytol. 2014, 202, 703–715. [Google Scholar] [CrossRef]
- Calle, A.; Cai, L.; Iezzoni, A.; Wünsch, A. Genetic Dissection of Bloom Time in Low Chilling Sweet Cherry (Prunus Avium L.) Using a Multi-Family QTL Approach. Front. Plant Sci. 2020, 10, 1647. [Google Scholar] [CrossRef]
- Kitamura, Y.; Takeuchi, T.; Yamane, H.; Tao, R. Simultaneous Down-Regulation of DORMANCY-ASSOCIATED MADS-Box6 and SOC1 during Dormancy Release in Japanese Apricot (Prunus mume) Flower Buds. J. Hortic. Sci. Biotechnol. 2016, 91, 476–482. [Google Scholar] [CrossRef]
- Castro, P.; Lewers, K.S. Identification of Quantitative Trait Loci (QTL) for Fruit-Quality Traits and Number of Weeks of Flowering in the Cultivated Strawberry. Mol. Breed. 2016, 36, 138. [Google Scholar] [CrossRef]
- Verma, S.; Zurn, J.D.; Salinas, N.; Mathey, M.M.; Denoyes, B.; Hancock, J.F.; Finn, C.E.; Bassil, N.V.; Whitaker, V.M. Clarifying Sub-Genomic Positions of QTLs for Flowering Habit and Fruit Quality in U.S. Strawberry (Fragaria × ananassa) Breeding Populations Using Pedigree-Based QTL Analysis. Hortic. Res. 2017, 4, 17062. [Google Scholar] [CrossRef] [PubMed]
- Del Pilar Moncada, M.; Tovar, E.; Carlos Montoya, J.; González, A.; Spindel, J.; McCouch, S. A Genetic Linkage Map of Coffee (Coffea arabica L.) and QTL for Yield, Plant Height, and Bean Size. Tree Genet. Genomes 2016, 12, 5. [Google Scholar] [CrossRef]
- Beltramo, C.; Valentini, N.; Portis, E.; Torello Marinoni, D.; Boccacci, P.; Sandoval Prando, A.M.; Roberto, B. Genetic Mapping and QTL Analysis in European Hazelnut (Corylus Avellana L.). Mol. Breed. 2016, 36, 27. [Google Scholar] [CrossRef]
- Gabay, G.; Dahan, Y.; Izhaki, Y.; Faigenboim, A.; Ben-Ari, G.; Elkind, Y.; Flaishman, M.A. High-Resolution Genetic Linkage Map of European Pear (Pyrus communis) and QTL Fine-Mapping of Vegetative Budbreak Time. BMC Plant Biol. 2018, 18, 175. [Google Scholar] [CrossRef]
- Dirlewanger, E.; Cosson, P.; Boudehri, K.; Renaud, C.; Capdeville, G.; Tauzin, Y.; Laigret, F.; Moing, A. Development of a Second-Generation Genetic Linkage Map for Peach [Prunus Persica (L.) Batsch] and Characterization of Morphological Traits Affecting Flower and Fruit. Tree Genet. Genomes 2006, 3, 1–13. [Google Scholar] [CrossRef]
- Marrano, A.; Sideli, G.M.; Leslie, C.A.; Cheng, H.; Neale, D.B. Deciphering of the Genetic Control of Phenology, Yield, and Pellicle Color in Persian Walnut (Juglans Regia L.). Front. Plant Sci. 2019, 10, 1140. [Google Scholar] [CrossRef] [PubMed]
- De Mori, G.; Testolin, R.; Cipriani, G. A Molecular Protocol for Early Sex Discrimination (ESD) in Actinidia Spp. J. Berry Res. 2022, 12, 249–266. [Google Scholar] [CrossRef]
- Vashistha, P.; Yadav, A.; Dwivedi, U.N.; Yadav, K. Genetics of Sex Chromosomes and Sex-Linked Molecular Markers in Papaya (Carica Papaya L.). Mol. Plant Breed. 2016, 7, 1–18. [Google Scholar] [CrossRef]
- Liao, L.; Liu, J.; Dai, Y.; Li, Q.; Xie, M.; Chen, Q.; Yin, H.; Qiu, G.; Liu, X. Development and Application of SCAR Markers for Sex Identification in the Dioecious Species Ginkgo biloba L. Euphytica 2009, 169, 49–55. [Google Scholar] [CrossRef]
- Akagi, T.; Kajita, K.; Kibe, T.; Morimura, H.; Tsujimoto, T.; Nishiyama, S.; Kawai, T.; Yamane, H.; Tao, R. Development of Molecular Markers Associated with Sexuality in Diospyros Lotus L. and Their Application in D. Kaki Thunb. J. Jpn. Soc. Hortic. Sci. 2014, 83, 214–221. [Google Scholar] [CrossRef]
- Atsumi, R.; Nishihara, R.; Tarora, K.; Urasaki, N.; Matsumura, H. Identification of Dominant Genetic Markers Relevant to Male Sex Determination in Mulberry (Morus Alba L.). Euphytica 2019, 215, 187. [Google Scholar] [CrossRef]
- Şahin, Z.N.; Sahin, E.C.; Aydin, Y.; Uncuoglu, A.A. Molecular Sexual Determinants in Pistacia Genus by KASP Assay. Mol. Biol. Rep. 2022, 49, 5473–5482. [Google Scholar] [CrossRef]
- Inoue, E.; Kasumi, M.; Sakuma, F.; Anzai, H.; Amano, K.; Hara, H. Identification of RAPD Marker Linked to Fruit Skin Color in Japanese Pear (Pyrus pyrifolia Nakai). Sci. Hortic. 2006, 107, 254–258. [Google Scholar] [CrossRef]
- Kayesh, E.; Lingfei, S.; Nicholas, K.K.; Sun, X.; Bilkish, N.; Zhang, Y.; Han, J.; Song, C.; Cheng, Z.M.; Fang, J. Fruit Skin Color and the Role of Anthocyanin. Acta Physiol. Plant 2013, 35, 2879–2890. [Google Scholar] [CrossRef]
- Lin-Wang, K.; Micheletti, D.; Palmer, J.; Volz, R.; Lozano, L.; Espley, R.; Hellens, R.P.; Chagnè, D.; Rowan, D.D.; Troggio, M.; et al. High Temperature Reduces Apple Fruit Colour via Modulation of the Anthocyanin Regulatory Complex. Plant Cell Environ. 2011, 34, 1176–1190. [Google Scholar] [CrossRef]
- Chang, Y.-C.; Lin, T.-C. Temperature Effects on Fruit Development and Quality Performance of Nagami Kumquat (Fortunella Margarita [Lour.] Swingle). Hortic. J. 2020, 89, 351–358. [Google Scholar] [CrossRef]
- Tan, Y.; Wen, B.; Xu, L.; Zong, X.; Sun, Y.; Wei, G.; Wei, H. High Temperature Inhibited the Accumulation of Anthocyanin by Promoting ABA Catabolism in Sweet Cherry Fruits. Front. Plant Sci. 2023, 14, 379. [Google Scholar] [CrossRef] [PubMed]
- Labadie, M.; Vallin, G.; Potier, A.; Petit, A.; Ring, L.; Hoffmann, T.; Gaston, A.; Munoz-Blanco, J.; Caballero, J.L.; Schwab, W.; et al. High-Resolution Quantitative Trait Locus Mapping and Whole Genome Sequencing Enable the Design of an Anthocyanidin Reductase-Specific Homoeo-Allelic Marker for Fruit Colour Improvement in Octoploid Strawberry (Fragaria × ananassa). Front. Plant Sci. 2022, 13, 869655. [Google Scholar] [CrossRef]
- Moriya, S.; Kunihisa, M.; Okada, K.; Shimizu, T.; Honda, C.; Yamamoto, T.; Muranty, H.; Denancé, C.; Katayose, Y.; Iwata, H.; et al. Allelic Composition of MdMYB1 Drives Red Skin Color Intensity in Apple (Malus 3 Domestica Borkh.) and Its Application to Breeding. Euphytica 2017, 213, 78. [Google Scholar] [CrossRef]
- Velasco, R.; Zharkikh, A.; Affourtit, J.; Dhingra, A.; Cestaro, A.; Kalyanaraman, A.; Fontana, P.; Bhatnagar, S.K.; Troggio, M.; Pruss, D.; et al. The Genome of the Domesticated Apple (Malus × domestica Borkh.). Nat. Genet. 2010, 42, 833–839. [Google Scholar] [CrossRef]
- El-Sharkawy, I.; Liang, D.; Xu, K. Transcriptome Analysis of an Apple (Malus × domestica) Yellow Fruit Somatic Mutation Identifies a Gene Network Module Highly Associated with Anthocyanin and Epigenetic Regulation. J. Exp. Bot. 2015, 66, 7359–7376. [Google Scholar] [CrossRef]
- Iglesias, I.; Echeverría, G.; Soria, Y. Differences in Fruit Colour Development, Anthocyanin Content, Fruit Quality and Consumer Acceptability of Eight “Gala” Apple Strains. Sci. Hortic. 2008, 119, 32–40. [Google Scholar] [CrossRef]
- Chagné, D.; Kirk, C.; How, N.; Whitworth, C.; Fontic, C.; Reig, G.; Sawyer, G.; Rouse, S.; Poles, L.; Gardiner, S.E.; et al. A Functional Genetic Marker for Apple Red Skin Coloration across Different Environments. Tree Genet. Genomes 2016, 12, 67. [Google Scholar] [CrossRef]
- Chagné, D.; Vanderzande, S.; Kirk, C.; Profitt, N.; Weskett, R.; Gardiner, S.E.; Peace, C.P.; Volz, R.K.; Bassil, N. V Validation of SNP Markers for Fruit Quality and Disease Resistance Loci in Apple (Malus × domestica Borkh.) Using the OpenArray® Platform. Hortic. Res. 2019, 6, 30. [Google Scholar] [CrossRef]
- Salazar, J.A.; Pacheco, I.; Shinya, P.; Zapata, P.; Silva, C.; Aradhya, M.; Velasco, D.; Ruiz, D.; Martínez-Gómez, P.; Infante, R. Genotyping by Sequencing for Snp-Based Linkage Analysis and Identification of QTLs Linked to Fruit Quality Traits in Japanese Plum (Prunus salicina Lindl.). Front. Plant Sci. 2017, 8, 476. [Google Scholar] [CrossRef]
- González, M.; Salazar, E.; Castillo, J.; Morales, P.; Mura-Jornet, I.; Maldonado, J.; Silva, H.; Carrasco, B. Genetic Structure Based on EST-SSR: A Putative Tool for Fruit Color Selection in Japanese Plum (Prunus Salicina L.) Breeding Programs. Mol. Breed. 2016, 36, 68. [Google Scholar] [CrossRef]
- Connors, C.H. Some Notes on the Inheritance of Unit Characters in the Peach. Proc. Am. Soc. Hortic. Sci. 1920, 16, 24–36. [Google Scholar]
- Falchi, R.; Vendramin, E.; Zanon, L.; Scalabrin, S.; Cipriani, G.; Verde, I.; Vizzotto, G.; Morgante, M. Three Distinct Mutational Mechanisms Acting on a Single Gene Underpin the Origin of Yellow Flesh in Peach. Plant J. 2013, 76, 175–187. [Google Scholar] [CrossRef]
- Cantín, C.M.; Crisosto, C.H.; Ogundiwin, E.A.; Gradziel, T.; Torrents, J.; Moreno, M.A.; Gogorcena, Y. Chilling Injury Susceptibility in an Intra-Specific Peach [Prunus persica (L.) Batsch] Progeny. Postharvest Biol. Technol. 2010, 58, 79–87. [Google Scholar] [CrossRef]
- Eduardo, I.; Pacheco, I.; Chietera, G.; Bassi, D.; Pozzi, C.; Vecchietti, A.; Rossini, L. QTL Analysis of Fruit Quality Traits in Two Peach Intraspecific Populations and Importance of Maturity Date Pleiotropic Effect. Tree Genet. Genomes 2011, 7, 323–335. [Google Scholar] [CrossRef]
- Frett, T.J.; Reighard, G.L.; Okie, W.R.; Gasic, K. Mapping Quantitative Trait Loci Associated with Blush in Peach [Prunus persica (L.) Batsch]. Tree Genet. Genomes 2014, 10, 367–381. [Google Scholar] [CrossRef]
- Beckman, T.G.; Rodriguez Alcazar, J.; Sherman, W.B.; Werner, D.J. Evidence for Qualitative Suppression of Red Skin Color in Peach. HortScience 2005, 40, 523–524. [Google Scholar] [CrossRef]
- Tuan, P.A.; Bai, S.; Yaegaki, H.; Tamura, T.; Hihara, S.; Moriguchi, T.; Oda, K. The Crucial Role of PpMYB10.1 in Anthocyanin Accumulation in Peach and Relationships between Its Allelic Type and Skin Color Phenotype. BMC Plant Biol. 2015, 15, 280. [Google Scholar] [CrossRef]
- Bretó, M.P.C.M.; Cantín, I.; Iglesias, P.; Arús, I.E. Mapping a Major Gene for Red Skin Color Suppression (Highlighter) in Peach. Euphytica 2017, 213, 14. [Google Scholar] [CrossRef]
- Costa dos Santos, F.H.; Cavalcanti, J.J.V.; Silva, F.P. Detection of Quantitative Trait Loci for Physical Traits of Cashew Apple. Crop Breed. Appl. Biotechnol. 2010, 10, 101–109. [Google Scholar] [CrossRef]
- Imai, A.; Nonaka, K.; Kuniga, T.; Yoshioka, T.; Hayashi, T. Genome-Wide Association Mapping of Fruit-Quality Traits Using Genotyping-by-Sequencing Approach in Citrus Landraces, Modern Cultivars, and Breeding Lines in Japan. Tree Genet. Genomes 2018, 14, 24. [Google Scholar] [CrossRef]
- Yu, Y.; Chen, C.; Gmitter, F.G. QTL Mapping of Mandarin (Citrus Reticulata) Fruit Characters Using High-Throughput SNP Markers. Tree Genet. Genomes 2016, 12, 77. [Google Scholar] [CrossRef]
- Yamamoto, T.; Terakami, S.; Takada, N.; Nishio, S.; Onoue, N.; Nishitani, C.; Kunihisa, M.; Inoue, E.; Iwata, H.; Hayashi, T.; et al. Identification of QTLs Controlling Harvest Time and Fruit Skin Color in Japanese Pear (Pyrus pyrifolia Nakai). Breed. Sci. 2014, 64, 351. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.S.; French, A.P. The Inheritance of Certain Fruit and Foliage Characteristics in the Peach. Mass. Agr. Expt. Sta. Bul. 1949, 452, 11–12. [Google Scholar]
- Brandi, F.; Bar, E.; Mourgues, F.; Horváth, G.; Turcsi, E.; Giuliano, G.; Liverani, A.; Tartarini, S.; Lewinsohn, E.; Rosati, C. Study of “Redhaven” Peach and Its White-Fleshed Mutant Suggests a Key Role of CCD4 Carotenoid Dioxygenase in Carotenoid and Norisoprenoid Volatile Metabolism. BMC Plant Biol. 2011, 11, 24. [Google Scholar] [CrossRef]
- Adami, M.; De Franceschi, P.; Brandi, F.; Liverani, A.; Giovannini, D.; Rosati, C.; Dondini, L.; Tartarini, S. Identifying a Carotenoid Cleavage Dioxygenase (Ccd4) Gene Controlling Yellow/White Fruit Flesh Color of Peach. Plant Mol. Biol. Rep. 2013, 31, 1166–1175. [Google Scholar] [CrossRef]
- Salazar, J.A.; Ruiz, D.; Egea, J.; Martínez-Gómez, P. Transmission of Fruit Quality Traits in Apricot (Prunus Armeniaca L.) and Analysis of Linked Quantitative Trait Loci (QTLs) Using Simple Sequence Repeat (SSR) Markers. Plant Mol. Biol. Rep. 2013, 31, 1506–1517. [Google Scholar] [CrossRef]
- Blas, A.L.; Ming, R.; Liu, Z.; Veatch, O.J.; Paull, R.E.; Moore, P.H.; Yu, Q. Cloning of the Papaya Chromoplast-Specific Lycopene b-Cyclase, CpCYC-b, Controlling Fruit Flesh Color Reveals Conserved Microsynteny and a Recombination Hot Spot. Plant Physiol. 2010, 152, 2013–2022. [Google Scholar] [CrossRef]
- Vázquez Calderón, M.; Zavala León, M.J.; Sánchez Teyer, L.F.; Mijangos Cortés, J.O.; Ortiz García, M.M.; Fuentes Ortiz, G.; Santamaría, J.M.; Vázquez Calderón, M.; Zavala León, M.J.; Sánchez Teyer, L.F.; et al. Validation of the Use of Molecular Markers of Sex and Color in Hybrids Derived from Crosses of Maradol x Creole Papaya. Rev. Mex. Cienc. Agric. 2016, 7, 767–780. [Google Scholar]
- Campos-Rivero, G.; Cazares-Sanchez, E.; Tamayo-Ordonez, M.C.; Tamayo-Ordonez, Y.J.; Padilla-Ramírez, J.S.; Quiroz-Moreno, A.; Sanchez-Teyer, L.F. Application of Sequence Specific Amplified Polymorphism (SSAP) and Simple Sequence Repeat (SSR) Markers for Variability and Molecular Assisted Selection (MAS) Studies of the Mexican Guava. Afr. J. Agric. Res. 2017, 12, 2372–2387. [Google Scholar] [CrossRef]
- Lila, M.A. Anthocyanins and Human Health: An In Vitro Investigative Approach. J. Biomed. Biotechnol. 2004, 2004, 306–313. [Google Scholar] [CrossRef]
- Dong, Y.; Wu, X.; Han, L.; Bian, J.; He, C.; El-Omar, E.; Gong, L.; Wang, M. The Potential Roles of Dietary Anthocyanins in Inhibiting Vascular Endothelial Cell Senescence and Preventing Cardiovascular Diseases. Nutrients 2022, 14, 2836. [Google Scholar] [CrossRef] [PubMed]
- Nistor, M.; Pop, R.; Daescu, A.; Pintea, A.; Socaciu, C.; Rugina, D. Anthocyanins as Key Phytochemicals Acting for the Prevention of Metabolic Diseases: An Overview. Molecules 2022, 27, 4254. [Google Scholar] [CrossRef]
- Werner, D.J.; Creller, M.A.; Chaparro, J.X. Inheritance of the Blood-Flesh Trait in Peach. HortScience 1998, 33, 1243–1246. [Google Scholar] [CrossRef]
- Gillen, A.M.; Bliss, F.A. Identification and Mapping of Markers Linked to the Mi Gene for Root-Knot Nematode Resistance in Peach. J. Am. Soc. Hortic. Sci. 2005, 130, 24–33. [Google Scholar] [CrossRef]
- Shen, Z.; Confolent, C.; Lambert, P.; Poëssel, J.-L.; Quilot-Turion, B.; Yu, M.; Ma, R.; Pascal, T. Characterization and Genetic Mapping of a New Blood-Flesh Trait Controlled by the Single Dominant Locus DBF in Peach. Tree Genet. Genomes 2013, 9, 1435–1446. [Google Scholar] [CrossRef]
- Donoso, J.M.; Picañol, R.; Serra, O.; Howad, W.; Alegre, S.; Arú, P.; Eduardo, I. Exploring Almond Genetic Variability Useful for Peach Improvement: Mapping Major Genes and QTLs in Two Interspecific Almond x Peach Populations. Mol. Breed. 2016, 36, 16. [Google Scholar] [CrossRef]
- Yamamoto, T.; Shimada, T.; Imai, T.; Yaegaki, H.; Haji, T.; Matsuta, N.; Yamaguchi, M.; Hayashi, T. Characterization of Morphological Traits Based on a Genetic Linkage Map in Peach. Breed. Sci. 2001, 51, 271–278. [Google Scholar] [CrossRef]
- Yamamoto, T.; Yamaguchi, M.; Hayashi, T. An Integrated Genetic Linkage Map of Peach by SSR, STS, AFLP and RAPD. J. Jpn. Soc. Hortic. Sci. 2005, 74, 204–213. [Google Scholar] [CrossRef]
- Guo, J.; Cao, K.; Deng, C.; Li, Y.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; Wu, J.; Guan, L.; et al. An Integrated Peach Genome Structural Variation Map Uncovers Genes Associated with Fruit Traits. Genome Biol. 2020, 21, 258. [Google Scholar] [CrossRef]
- Zaracho, N.; Reig, G.; Kalluri, N.; Arús, P.; Eduardo, I. Inheritance of Fruit Red-Flesh Patterns in Peach. Plants 2023, 12, 394. [Google Scholar] [CrossRef] [PubMed]
- Sooriyapathirana, S.S.; Khan, A.; Sebolt, A.M.; Wang, D.; Bushakra, J.M.; Lin-Wang, K.; Allan, A.C.; Gardiner, S.E.; Chagné, D.; Iezzoni, A.F. QTL Analysis and Candidate Gene Mapping for Skin and Flesh Color in Sweet Cherry Fruit (Prunus avium L.). Tree Genet. Genomes 2010, 6, 821–832. [Google Scholar] [CrossRef]
- Stegmeir, T.; Lichun, C.; Basundari, F.R.A.; Sebolt, A.M.; Iezzoni, A.F. A DNA Test for Fruit Flesh Color in Tetraploid Sour Cherry (Prunus cerasus L.). Mol. Breed. 2015, 35, 149. [Google Scholar] [CrossRef]
- Purugganan, M.D.; Fuller, D.Q. The Nature of Selection during Plant Domestication. Nature 2009, 457, 843–849. [Google Scholar] [CrossRef]
- Grandillo, S.; Ku, H.M.; Tanksley, S.D. Identifying the Loci Responsible for Natural Variation in Fruit Size and Shape in Tomato. Theor. Appl. Genet. 1999, 99, 978–987. [Google Scholar] [CrossRef]
- Guo, M.; Rupe, M.A.; Dieter, J.A.; Zou, J.; Spielbauer, D.; Duncan, K.E.; Howard, R.J.; Hou, Z.; Simmons, C.R. Cell Number Regulator1 Affects Plant and Organ Size in Maize: Implications for Crop Yield Enhancement and Heterosis. Plant Cell 2010, 22, 1057–1073. [Google Scholar] [CrossRef]
- De Franceschi, P.; Stegmeir, T.; Cabrera, A.; Van Der Knaap, E.; Rosyara, U.R.; Sebolt, A.M.; Dondini, L.; Dirlewanger, E.; Quero-Garcia, J.; Campoy, J.A.; et al. Cell Number Regulator Genes in Prunus Provide Candidate Genes for the Control of Fruit Size in Sweet and Sour Cherry. Mol. Breed. 2013, 32, 311–326. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, J.; Wang, Y.; Wang, H.; Wang, L.; Zhang, L.; Xiong, M.; He, W.; Yang, S.; Chen, Q.; et al. Development and Cross-Species Transferability of Novel Genomic-SSR Markers and Their Utility in Hybrid Identification and Trait Association Analysis in Chinese Cherry. Horticulturae 2022, 8, 222. [Google Scholar] [CrossRef]
- Ji, F.; Wei, W.; Liu, Y.; Wang, G.; Zhang, Q.; Xing, Y.; Zhang, S.; Liu, Z.; Cao, Q.; Qin, L. Construction of a SNP-Based High-Density Genetic Map Using Genotyping by Sequencing (GBS) and QTL Analysis of Nut Traits in Chinese Chestnut (Castanea mollissima Blume). Front. Plant Sci. 2018, 9, 816. [Google Scholar] [CrossRef]
- Khefifi, H.; Dumont, D.; Costantino, G.; Doligez, A.; Brito, A.C.; Bérard, A.; Morillon, R.; Ollitrault, P.; Luro, F. Mapping of QTLs for Citrus Quality Traits throughout the Fruit Maturation Process on Clementine (Citrus reticulata × C. sinensis) and Mandarin (C. reticulata Blanco) Genetic Maps. Tree Genet. Genomes 2022, 18, 40. [Google Scholar] [CrossRef]
- Razi, M.; Darvishzadeh, R.; Amiri, M.E.; Doulati-Banehd, H.; Martínez-Gómez, P. Molecular Characterization of a Diverse Iranian Table Grapevine Germplasm Using REMAP Markers: Population Structure, Linkage Disequilibrium and Association Mapping of Berry Yield and Quality Traits. Biologia 2019, 74, 173–185. [Google Scholar] [CrossRef]
- Doligez, A.; Bertrand, Y.; Farnos, M.; Grolier, M.; Romieu, C.; Esnault, F.; Dias, S.; Berger, G.; François, P.; Pons, T.; et al. New Stable QTLs for Berry Weight Do Not Colocalize with QTLs for Seed Traits in Cultivated Grapevine (Vitis vinifera L.). BMC Plant Biol. 2013, 13, 217. [Google Scholar] [CrossRef] [PubMed]
- Ban, Y.; Mitani, N.; Sato, A.; Kono, A.; Hayashi, T. Genetic Dissection of Quantitative Trait Loci for Berry Traits in Interspecific Hybrid Grape (Vitis labruscana × Vitis vinifera). Euphytica 2016, 211, 295–310. [Google Scholar] [CrossRef]
- Salazar, J.A.; Pacheco, I.; Zapata, P.; Shinya, P.; Ruiz, D.; Martínez-Gómez, P.; Infante, R. Identification of Loci Controlling Phenology, Fruit Quality and Post-Harvest Quantitative Parameters in Japanese Plum (Prunus salicina Lindl.). Postharvest Biol. Technol. 2020, 169, 111292. [Google Scholar] [CrossRef]
- Jue, D.; Liu, L.; Sang, X.; Shu, B.; Wang, J.; Wang, Y.; Zhang, C.; Shi, S. SNP-Based High-Density Genetic Map Construction and Candidate Gene Identification for Fruit Quality Traits of Dimocarpus Longan Lour. Sci. Hortic. 2021, 284, 110086. [Google Scholar] [CrossRef]
- Mathiazhagan, M.; Chidambara, B.; Hunashikatti, L.R.; Ravishankar, K.V. Genomic Approaches for Improvement of Tropical Fruits: Fruit Quality, Shelf Life and Nutrient Content. Genes 2021, 12, 1881. [Google Scholar] [CrossRef]
- da Silva Linge, C.; Bassi, D.; Bianco, L.; Pacheco, I.; Pirona, R.; Rossini, L. Genetic Dissection of Fruit Weight and Size in an F2 Peach (Prunus persica (L.) Batsch) Progeny. Mol. Breed. 2015, 35, 71. [Google Scholar] [CrossRef]
- Fresnedo-Ramírez, J.; Frett, T.J.; Sandefur, P.J.; Salgado-Rojas, A.; Clark, J.R.; Gasic, K.; Peace, C.P.; Anderson, N.; Hartmann, T.P.; Byrne, D.H.; et al. QTL Mapping and Breeding Value Estimation through Pedigree-Based Analysis of Fruit Size and Weight in Four Diverse Peach Breeding Programs. Tree Genet. Genomes 2016, 12, 25. [Google Scholar] [CrossRef]
- Zeballos, J.L.; Abidi, W.; Giménez, R.; Monforte, A.J.; Moreno, M.Á.; Gogorcena, Y. Mapping QTLs Associated with Fruit Quality Traits in Peach [Prunus persica (L.) Batsch] Using SNP Maps. Tree Genet. Genomes 2016, 12, 37. [Google Scholar] [CrossRef]
- Cao, K.; Li, Y.; Deng, C.H.; Gardiner, S.E.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; Wang, L. Comparative Population Genomics Identified Genomic Regions and Candidate Genes Associated with Fruit Domestication Traits in Peach. Plant Biotechnol. J. 2019, 17, 1954–1970. [Google Scholar] [CrossRef]
- Abdelghafar, A.; da Silva Linge, C.; Okie, W.R.; Gasic, K. Mapping QTLs for Phytochemical Compounds and Fruit Quality in Peach. Mol. Breed. 2020, 40, 32. [Google Scholar] [CrossRef]
- Shi, P.; Xu, Z.; Zhang, S.; Wang, X.; Ma, X.; Zheng, J.; Xing, L.; Zhang, D.; Ma, J.; Han, M.; et al. Construction of a High-Density SNP-Based Genetic Map and Identification of Fruit-Related QTLs and Candidate Genes in Peach [Prunus persica (L.) Batsch]. BMC Plant Biol. 2020, 20, 438. [Google Scholar] [CrossRef] [PubMed]
- Bernard, A.; Crabier, J.; Donkpegan, A.S.L.; Marrano, A.; Lheureux, F.; Dirlewanger, E. Genome-Wide Association Study Reveals Candidate Genes Involved in Fruit Trait Variation in Persian Walnut (Juglans regia L.). Front. Plant Sci. 2021, 11, 2163. [Google Scholar] [CrossRef]
- Striem, M.J.; Ben-Hayyim, G.; Spiegel-Roy, P. Identifying Molecular Genetic Markers Associated with Seedlessness in Grape. J. Am. Soc. Hortic. Sci. 1996, 121, 758–763. [Google Scholar] [CrossRef]
- Doligez, A.; Bouquet, A.; Danglot, Y.; Lahogue, F.; Riaz, S.; Meredith, C.P.; Edwards, K.J.; This, P. Genetic Mapping of Grapevine (Vitis Vinifera L.) Applied to the Detection of QTLs for Seedlessness and Berry Weight. Theor. Appl. Genet. 2002, 105, 780–795. [Google Scholar] [CrossRef]
- Cabezas, J.A.; Cervera, M.T.; Ruiz-García, L.; Carreño, J.; Martínez-Zapater, J.M. A Genetic Analysis of Seed and Berry Weight in Grapevine. Genome 2006, 49, 1572–1585. [Google Scholar] [CrossRef]
- Wang, Y. Analysis of Sequencing the RAPD Marker Linked to Seedless Genes in Grapes. Acta Univ. Agric. Boreali-Occident. 1997, 25, 1–5. [Google Scholar]
- Lahogue, F.; This, P.; Bouquet, A. Identification of a Codominant Scar Marker Linked to the Seedlessness Character in Grapevine. Theor. Appl. Genet. 1998, 97, 950–959. [Google Scholar] [CrossRef]
- Adam-Blondon, A.-F.; Lahogue-Esnault, F.; Bouquet, A.; Boursiquot, J.M.; This, P. Usefulness of Two SCAR Markers for Marker-Assisted Selection of Seedless Grapevine Cultivars. VITIS–J. Grapevine Res. 2001, 40, 147. [Google Scholar] [CrossRef]
- Mejía, N.; Hinrichsen, P. A New, Highly Assertive Scar Marker Potentially Useful to Assist Selection for Seedlessness in Table Grape Breeding. Acta Hortic. 2003, 603, 559–564. [Google Scholar] [CrossRef]
- Royo, C.; Torres-Pérez, R.; Mauri, N.; Diestro, N.; Cabezas, J.A.; Marchal, C.; Lacombe, T.; Ibáñez, J.; Tornel, M.; Carreño, J.; et al. The Major Origin of Seedless Grapes Is Associated with a Missense Mutation in the MADS-Box Gene VviAGL11. Plant Physiol. 2018, 177, 1234–1253. [Google Scholar] [CrossRef]
- Conner, P.J.; Gunawan, G.; Clark, J.R. Characterization of the P3-VvAGL11 Marker for Stenospermocarpic Seedlessness in Euvitis × muscadinia Grape Hybrid Progenies. J. Am. Soc. Hortic. Sci. 2018, 143, 167–172. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, S.; Jiao, C.; Li, Z.; Liu, C.; Wang, X. ping QTL-Seq Analysis of the Seed Size Trait in Grape Provides New Molecular Insights on Seedlessness. J. Integr. Agric. 2022, 21, 2910–2925. [Google Scholar] [CrossRef]
- Yamasaki, A.; Kitajima, A.; Ohara, N.; Tanaka, M.; Hasegawa, K. Histological Study of Expression of Seedlessness in Citrus Kinokuni ‘Mukaku Kishu’ and Its Progenies. J. Am. Soc. Hortic. Sci. 2007, 132, 869–875. [Google Scholar] [CrossRef]
- Gmitter, F.G.; Soneji, J.R.; Rao, M.N. Citrus Breeding. In Breeding Plantation Tree Crops: Temperate Species; Jain, S.M., Priyadarshan, P.M., Eds.; Springer: New York, NY, USA, 2009; pp. 105–134. ISBN 9780387712031. [Google Scholar]
- Chavez, D.J.; Chaparro, J.X. Identification of Markers Linked to Seedlessness in Citrus Kinokuni Hort. Ex Tanaka and Its Progeny Using Bulked Segregant Analysis. HortScience 2011, 46, 693–697. [Google Scholar] [CrossRef]
- Xiao, J.; Chen, L.; Xie, M.; Liu, H.; Ye, W. Identification of AFLP Fragments Linked to Seedlessness in Ponkan Mandarin (Citrus reticulata Blanco) and Conversion to SCAR Markers. Sci. Hortic. 2009, 121, 505–510. [Google Scholar] [CrossRef]
- Goto, S.; Yoshioka, T.; Ohta, S.; Kita, M.; Hamada, H.; Shimizu, T. QTL Mapping of Male Sterility and Transmission Pattern in Progeny of Satsuma Mandarin. PLoS ONE 2018, 13, e0200844. [Google Scholar] [CrossRef]
- Kanzaki, S.; Akagi, T.; Masuko, T.; Kimura, M.; Yamada, M.; Sato, A.; Mitani, N.; Ustunomiya, N.; Yonemori, K. SCAR Markers for Practical Application of Marker-Assisted Selection in Persimmon (Diospyros kaki Thunb.) Breeding. J. Jpn. Soc. Hortic. Sci. 2010, 79, 150–155. [Google Scholar] [CrossRef]
- Kono, A.; Kobayashi, S.; Onoue, N.; Sato, A. Characterization of a Highly Polymorphic Region Closely Linked to the AST Locus and Its Potential Use in Breeding of Hexaploid Persimmon (Diospyros kaki Thunb.). Mol. Breed. 2016, 36, 56. [Google Scholar] [CrossRef]
- Blasco, M.; Gil-Muñoz, F.; del Mar Naval, M.; Badenes, M.L. Molecular Assisted Selection for Pollination-Constant and Non-Astringent Type without Male Flowers in Spanish Germplasm for Persimmon Breeding. Agronomy 2020, 10, 1172. [Google Scholar] [CrossRef]
- Costa, F.; Peace, C.P.; Stella, S.; Serra, S.; Musacchi, S.; Bazzani, M.; Sansavini, S.; Van de Weg, W.E. QTL Dynamics for Fruit Firmness and Softening around an Ethylene-Dependent Polygalacturonase Gene in Apple (Malus×domestica Borkh.). J. Exp. Bot. 2010, 61, 3029–3039. [Google Scholar] [CrossRef] [PubMed]
- Longhi, S.; Cappellin, L.; Guerra, W.; Costa, F. Validation of a Functional Molecular Marker Suitable for Marker-Assisted Breeding for Fruit Texture in Apple (Malus 3 Domestica Borkh.). Mol. Breed. 2013, 32, 841–852. [Google Scholar] [CrossRef]
- Migicovsky, Z.; Yeats, T.H.; Watts, S.; Song, J.; Forney, C.F.; Burgher-MacLellan, K.; Somers, D.J.; Gong, Y.; Zhang, Z.; Vrebalov, J.; et al. Apple Ripening Is Controlled by a NAC Transcription Factor. Front. Genet. 2021, 12, 908. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, J.; Liu, W.; Liu, N.; Zhang, Y.; Xu, M.; Liu, S.; Ma, X.; Zhang, Y. Construction of a High-Density Genetic Map and Identification of Quantitative Trait Loci Linked to Fruit Quality Traits in Apricots Using Specific-Locus Amplified Fragment Sequencing. Front. Plant Sci. 2022, 13, 103. [Google Scholar] [CrossRef]
- Biabiany, S.; Araou, E.; Cormier, F.; Martin, G.; Carreel, F.; Hervouet, C.; Salmon, F.; Efile, J.C.; Lopez-Lauri, F.; D’Hont, A.; et al. Detection of Dynamic QTLs for Traits Related to Organoleptic Quality during Banana Ripening. Sci. Hortic. 2022, 293, 110690. [Google Scholar] [CrossRef]
- Carreño, I.; Cabezas, J.A.; Martínez-Mora, C.; Arroyo-García, R.; Cenis, J.L.; Martínez-Zapater, J.M.; Carreño, J.; Ruiz-García, L. Quantitative Genetic Analysis of Berry Firmness in Table Grape (Vitis vinifera L.). Tree Genet. Genomes 2015, 11, 818. [Google Scholar] [CrossRef]
- Correa, J.; Mamani, M.; Muñoz-Espinoza, C.; González-Agüero, M.; Defilippi, B.G.; Campos-Vargas, R.; Pinto, M.; Hinrichsen, P. New Stable QTLs for Berry Firmness in Table Grapes. Am. J. Enol. Vitic. 2016, 67, 212–217. [Google Scholar] [CrossRef]
- Jiang, J.; Fan, X.; Zhang, Y.; Tang, X.; Li, X.; Liu, C.; Zhang, Z. Construction of a High-Density Genetic Map and Mapping of Firmness in Grapes (Vitis vinifera L.) Based on Whole-Genome Resequencing. Int. J. Mol. Sci. 2020, 21, 797. [Google Scholar] [CrossRef] [PubMed]
- Peace, C.P.; Crisosto, C.H.; Gradziel, T.M. Endopolygalacturonase: A Candidate Gene for Freestone and Melting Flesh in Peach. Mol. Breed. 2005, 16, 21–31. [Google Scholar] [CrossRef]
- Eduardo, I.; Picañol, R.; Rojas, E.; Batlle, I.; Howad, W.; Aranzana, M.J.; Arús, P. Mapping of a Major Gene for the Slow Ripening Character in Peach: Co-Location with the Maturity Date Gene and Development of a Candidate Gene-Based Diagnostic Marker for Its Selection. Euphytica 2015, 205, 627–636. [Google Scholar] [CrossRef]
- Martínez-García, P.J.; Parfitt, D.E.; Ogundiwin, E.A.; Fass, J.; Chan, H.M.; Ahmad, R.; Lurie, S.; Dandekar, A.; Gradziel, T.M.; Crisosto, C.H. High Density SNP Mapping and QTL Analysis for Fruit Quality Characteristics in Peach (Prunus persica L.). Tree Genet. Genomes 2013, 9, 19–36. [Google Scholar] [CrossRef]
- Nuñez-Lillo, G.; Cifuentes-Esquivel, A.; Troggio, M.; Micheletti, D.; Infante, R.; Campos-Vargas, R.; Orellana, A.; Blanco-Herrera, F.; Meneses, C. Identification of Candidate Genes Associated with Mealiness and Maturity Date in Peach [Prunus persica (L.) Batsch] Using QTL Analysis and Deep Sequencing. Tree Genet. Genomes 2015, 11, 86. [Google Scholar] [CrossRef]
- Serra, O.; Giné-Bordonaba, J.; Eduardo, I.; Bonany, J.; Echeverria, G.; Larrigaudière, C.; Arús, P. Genetic Analysis of the Slow-Melting Flesh Character in Peach. Tree Genet. Genomes 2017, 13, 77. [Google Scholar] [CrossRef]
- Carrasco-Valenzuela, T.; Muñoz-Espinoza, C.; Riveros, A.; Pedreschi, R.; Arús, P.; Campos-Vargas, R.; Meneses, C. Expression QTL (EQTLs) Analyses Reveal Candidate Genes Associated With Fruit Flesh Softening Rate in Peach [Prunus persica (L.) Batsch]. Front. Plant Sci. 2019, 10, 1581. [Google Scholar] [CrossRef]
- Morgutti, S.; Negrini, N.; Nocito, F.F.; Ghiani, A.; Bassi, D.; Cocucci, M. Changes in Endopolygalacturonase Levels and Characterization of a Putative Endo-PG Gene during Fruit Softening in Peach Genotypes with Nonmelting and Melting Flesh Fruit Phenotypes. New Phytol. 2006, 171, 315–328. [Google Scholar] [CrossRef]
- Font i Forcada, C.; Oraguzie, N.; Igartua, E.; Moreno, M.A.; Gogorcena, Y. Population Structure and Marker-Trait Associations for Pomological Traits in Peach and Nectarine Cultivars. Tree Genet. Genomes 2013, 9, 331–349. [Google Scholar] [CrossRef]
- Cai, L.; Quero-García, J.; Barreneche, T.; Dirlewanger, E.; Saski, C.; Iezzoni, A. A Fruit Firmness QTL Identified on Linkage Group 4 in Sweet Cherry (Prunus avium L.) Is Associated with Domesticated and Bred Germplasm. Sci. Rep. 2019, 9, 5008. [Google Scholar] [CrossRef]
- Calle, A.; Balas, F.; Cai, L.; Iezzoni, A.; López-Corrales, M.; Serradilla, M.J.; Wünsch, A. Fruit Size and Firmness QTL Alleles of Breeding Interest Identified in a Sweet Cherry ‘Ambrunés’ × ‘Sweetheart’ Population. Mol. Breed. 2020, 40, 86. [Google Scholar] [CrossRef]
- Wei, K.; Ma, C.; Sun, K.; Liu, Q.; Zhao, N.; Sun, Y.; Tu, K.; Pan, L. Relationship between Optical Properties and Soluble Sugar Contents of Apple Flesh during Storage. Postharvest Biol. Technol. 2020, 159, 111021. [Google Scholar] [CrossRef]
- Ma, B.; Zhao, S.; Wu, B.; Wang, D.; Peng, Q.; Owiti, A.; Fang, T.; Liao, L.; Ogutu, C.; Korban, S.S.; et al. Construction of a High Density Linkage Map and Its Application in the Identification of QTLs for Soluble Sugar and Organic Acid Components in Apple. Tree Genet. Genomes 2016, 12, 1. [Google Scholar] [CrossRef]
- Westwood, M.N. Temperate-Zone Pomology; Timber Press: Portland, OR, USA, 1993. [Google Scholar]
- Etienne, C.; Rothan, C.; Moing, A.; Plomion, C.; Bodénès, C.; Svanella-Dumas, L.; Cosson, P.; Pronier, V.; Monet, R.; Dirlewanger, E. Candidate Genes and QTLs for Sugar and Organic Acid Content in Peach [Prunus persica (L.) Batsch]. Theor. Appl. Genet. 2002, 105, 145–159. [Google Scholar] [CrossRef]
- Zorrilla-Fontanesi, Y.; Cabeza, A.; Domínguez, P.; Medina, J.J.; Valpuesta, V.; Denoyes-Rothan, B.; Sánchez-Sevilla, J.F.; Amaya, I. Quantitative Trait Loci and Underlying Candidate Genes Controlling Agronomical and Fruit Quality Traits in Octoploid Strawberry (Fragaria × ananassa). Theor. Appl. Genet. 2011, 123, 755–778. [Google Scholar] [CrossRef]
- dos Santos, F.H.C.; Cavalcanti, J.J.V.; da Silva, F.P. QTL Detection for Physicochemical Characteristics of Cashew Apple. Crop Breed. Appl. Biotechnol. 2011, 11, 17–26. [Google Scholar] [CrossRef]
- Yang, S.; Fresnedo-Ramírez, J.; Sun, Q.; Manns, D.C.; Sacks, G.L.; Mansfield, A.K.; Luby, J.J.; Londo, J.P.; Reisch, B.I.; Cadle-Davidson, L.E.; et al. Next Generation Mapping of Enological Traits in an F2 Interspecific Grapevine Hybrid Family. PLoS ONE 2016, 11, e0149560. [Google Scholar] [CrossRef]
- Kenis, K.; Keulemans, J.; Davey, M.W. Identification and Stability of QTLs for Fruit Quality Traits in Apple. Tree Genet. Genomes 2008, 4, 647–661. [Google Scholar] [CrossRef]
- Guan, Y.; Peace, C.; Rudell, D.; Verma, S.; Evans, K. QTLs Detected for Individual Sugars and Soluble Solids Content in Apple. Mol. Breed. 2015, 35, 135. [Google Scholar] [CrossRef]
- Zhen, Q.; Fang, T.; Peng, Q.; Liao, L.; Zhao, L.; Owiti, A.; Han, Y. Developing Gene-Tagged Molecular Markers for Evaluation of Genetic Association of Apple SWEET Genes with Fruit Sugar Accumulation. Hortic. Res. 2018, 5, 14. [Google Scholar] [CrossRef]
- Klemens, P.A.W.; Patzke, K.; Deitmer, J.; Spinner, L.; Le Hir, R.; Bellini, C.; Bedu, M.; Chardon, F.; Krapp, A.; Ekkehard Neuhaus, H. Overexpression of the Vacuolar Sugar Carrier AtSWEET16 Modifies Germination, Growth, and Stress Tolerance in Arabidopsis. Plant Physiol. 2013, 163, 1338–1352. [Google Scholar] [CrossRef] [PubMed]
- Chardon, F.; Bedu, M.; Calenge, F.; Klemens, P.A.W.; Spinner, L.; Clement, G.; Chietera, G.; Léran, S.; Ferrand, M.; Lacombe, B.; et al. Leaf Fructose Content Is Controlled by the Vacuolar Transporter SWEET17 in Arabidopsis. Curr. Biol. 2013, 23, 697–702. [Google Scholar] [CrossRef]
- Kunihisa, M.; Moriya, S.; Abe, K.; Okada, K.; Haji, T.; Hayashi, T.; Kim, H.; Nishitani, C.; Terakami, S.; Yamamoto, T. Identification of QTLs for Fruit Quality Traits in Japanese Apples: QTLs for Early Ripening Are Tightly Related to Preharvest Fruit Drop. Breed. Sci. 2014, 64, 240. [Google Scholar] [CrossRef] [PubMed]
- Verde, I.; Abbott, A.G.; Scalabrin, S.; Jung, S.; Shu, S.; Marroni, F.; Zhebentyayeva, T.; Dettori, M.T.; Grimwood, J.; Cattonaro, F.; et al. The High-Quality Draft Genome of Peach (Prunus Persica) Identifies Unique Patterns of Genetic Diversity, Domestication and Genome Evolution. Nat. Genet. 2013, 45, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Illa, E.; Sargent, D.J.; Girona, E.L.; Bushakra, J.; Cestaro, A.; Crowhurst, R.; Pindo, M.; Cabrera, A.; Van Der Knaap, E.; Iezzoni, A.; et al. Comparative Analysis of Rosaceous Genomes and the Reconstruction of a Putative Ancestral Genome for the Family. BMC Evol. Biol. 2011, 11, 9. [Google Scholar] [CrossRef] [PubMed]
- Rawandoozi, Z.J.; Hartmann, T.P.; Carpenedo, S.; Gasic, K.; Da Silva Linge, C.; Cai, L.; Van De Weg, E.; Byrne, D.H. Identification and Characterization of QTLs for Fruit Quality Traits in Peach through a Multi-Family Approach. BMC Genom. 2020, 21, 522. [Google Scholar] [CrossRef]
- Zurn, J.D.; Driskill, M.; Jung, S.; Main, D.; Yin, M.H.; Clark, M.C.; Cheng, L.; Ashrafi, H.; Aryal, R.; Clark, J.R.; et al. A Rosaceae Family-Level Approach To Identify Loci Influencing Soluble Solids Content in Blackberry for DNA-Informed Breeding. G3 Genes|Genomes|Genet. 2020, 10, 3729–3740. [Google Scholar] [CrossRef]
- Lerceteau-Köhler, E.; Moing, A.; Guérin, G.; Renaud, C.; Petit, A.; Rothan, C.; Denoyes, B. Genetic Dissection of Fruit Quality Traits in the Octoploid Cultivated Strawberry Highlights the Role of Homoeo-QTL in Their Control. Theor. Appl. Genet. 2012, 124, 1059–1077. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, N.; Fang, L.C.; Liang, Z.C.; Li, S.H.; Wu, B.H. Construction of a High-Density Genetic Map and QTLs Mapping for Sugars and Acids in Grape Berries. BMC Plant Biol. 2015, 15, 28. [Google Scholar] [CrossRef]
- Viana, A.P.; de Resende, M.D.V.; Riaz, S.; Walker, M.A. Genome Selection in Fruit Breeding: Application to Table Grapes. Sci. Agric. 2016, 73, 142–149. [Google Scholar] [CrossRef]
- Bates, R.P.; Morris, J.R.; Crandall, P.G.; FAO. Principles and Practices of Small- and Medium-Scale Fruit Juice Processing; Food and Agriculture Organization of the United Nations: Rome, Italy, 2001; ISBN 9251046611. [Google Scholar]
- Quarta, R.; Dettori, M.T.; Sartori, A.; Verde, I. Genetic Linkage Map and QTL Analysis in Peach. Acta Hortic. 2000, 521, 233–241. [Google Scholar] [CrossRef]
- Quilot, B.; Wu, B.H.; Kervella, J.; Génard, M.; Foulongne, M.; Moreau, K. QTL Analysis of Quality Traits in an Advanced Backcross between Prunus Persica Cultivars and the Wild Relative Species P. Davidiana. Theor. Appl. Genet. 2004, 109, 884–897. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, D.; Lambert, P.; Audergon, J.M.; Dondini, L.; Tartarini, S.; Adami, M.; Gennari, F.; Cervellati, C.; De Franceschi, P.; Sansavini, S.; et al. Identification of QTLS for fruit quality traits in apricot. Acta Hortic. 2010, 862, 587–592. [Google Scholar] [CrossRef]
- Bugaud, C.; Maraval, I.; Daribo, M.O.; Leclerc, N.; Salmon, F. Optimal and Acceptable Levels of Sweetness, Sourness, Firmness, Mealiness and Banana Aroma in Dessert Banana (Musa sp.). Sci. Hortic. 2016, 211, 399–409. [Google Scholar] [CrossRef]
- Bugaud, C.; Cazevieille, P.; Daribo, M.O.; Telle, N.; Julianus, P.; Fils-Lycaon, B.; Mbéguié-A-Mbéguié, D. Rheological and Chemical Predictors of Texture and Taste in Dessert Banana (Musa spp.). Postharvest Biol. Technol. 2013, 84, 1–8. [Google Scholar] [CrossRef]
- Mengist, M.F.; Grace, M.H.; Mackey, T.; Munoz, B.; Pucker, B.; Bassil, N.; Luby, C.; Ferruzzi, M.; Lila, M.A.; Iorizzo, M. Dissecting the Genetic Basis of Bioactive Metabolites and Fruit Quality Traits in Blueberries (Vaccinium corymbosum L.). Front. Plant Sci. 2022, 13, 2925. [Google Scholar] [CrossRef]
- Fong, S.K.; Kawash, J.; Wang, Y.; Johnson-Cicalese, J.; Polashock, J.; Vorsa, N. A Low Citric Acid Trait in Cranberry: Genetics and Molecular Mapping of a Locus Impacting Fruit Acidity. Tree Genet. Genomes 2020, 16, 42. [Google Scholar] [CrossRef]
- Fong, S.K.; Kawash, J.; Wang, Y.; Johnson-Cicalese, J.; Polashock, J.; Vorsa, N. A Low Malic Acid Trait in Cranberry Fruit: Genetics, Molecular Mapping, and Interaction with a Citric Acid Locus. Tree Genet. Genomes 2021, 17, 4. [Google Scholar] [CrossRef]
- Song, S.; del Mar Hernández, M.; Provedo, I.; Menéndez, C.M. Segregation and Associations of Enological and Agronomic Traits in Graciano × Tempranillo Wine Grape Progeny (Vitis vinifera L.). Euphytica 2014, 195, 259–277. [Google Scholar] [CrossRef]
- Viana, A.P.; Riaz, S.; Walker, M.A. Genetic Dissection of Agronomic Traits within a Segregating Population of Breeding Table Grapes. Genet. Mol. Res. 2013, 12, 951–964. [Google Scholar] [CrossRef]
- Borsani, J.; Budde, C.O.; Porrini, L.; Lauxmann, M.A.; Lombardo, V.A.; Murray, R.; Andreo, C.S.; Drincovich, M.F.; Lara, M.V. Carbon Metabolism of Peach Fruit after Harvest: Changes in Enzymes Involved in Organic Acid and Sugar Level Modifications. J. Exp. Bot. 2009, 60, 1823–1837. [Google Scholar] [CrossRef] [PubMed]
- Boudehri, K.; Belka, M.A.; Cardinet, G.; Capdeville, G.; Renaud, C.; Tauzin, Y.; Dirlewanger, E.; Dirlewanger, A.M.; Troadec, C.; Jublot, D.; et al. Toward the Isolation of the d Gene Controlling the Acidity of Peach Fruit by Positional Cloning. Acta Hortic. 2009, 814, 507–510. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, L.; Zhu, G.; Cao, K.; Fang, W.; Chen, C.; Wang, X. DNA Marker-Assisted Evaluation of Fruit Acidity in Diverse Peach (Prunus persica) Germplasm. Euphytica 2016, 210, 413–426. [Google Scholar] [CrossRef]
- Eduardo, I.; López-Girona, E.; BatlIe, I.; Reig, G.; Iglesias, I.; Howad, W.; Arús, P.; Aranzana, M.J. Development of Diagnostic Markers for Selection of the Subacid Trait in Peach. Tree Genet. Genomes 2014, 10, 1695–1709. [Google Scholar] [CrossRef]
- Fresnedo-Ramírez, J.; Bink, M.C.A.M.; van de Weg, E.; Famula, T.R.; Crisosto, C.H.; Frett, T.J.; Gasic, K.; Peace, C.P.; Gradziel, T.M. QTL Mapping of Pomological Traits in Peach and Related Species Breeding Germplasm. Mol. Breed. 2015, 35, 166. [Google Scholar] [CrossRef]
- Serra, S.; Goke, A.; Diako, C.; Vixie, B.; Ross, C.; Musacchi, S. Consumer Perception of d’Anjou Pear Classified by Dry Matter at Harvest Using near-Infrared Spectroscopy. Int. J. Food Sci. Technol. 2019, 54, 2256–2265. [Google Scholar] [CrossRef]
- Palmer, J.W.; Harker, F.R.; Tustin, D.S.; Johnston, J. Fruit Dry Matter Concentration: A New Quality Metric for Apples. J. Sci. Food Agric. 2010, 90, 2586–2594. [Google Scholar] [CrossRef] [PubMed]
- Escribano, S.; Biasi, W.V.; Lerud, R.; Slaughter, D.C.; Mitcham, E.J. Non-Destructive Prediction of Soluble Solids and Dry Matter Content Using NIR Spectroscopy and Its Relationship with Sensory Quality in Sweet Cherries. Postharvest Biol. Technol. 2017, 128, 112–120. [Google Scholar] [CrossRef]
- Gibert, O.; Dufour, D.; Giraldo, A.; Sánchez, T.; Reynes, M.; Pain, J.P.; González, A.; Fernández, A.; Díaz, A. Differentiation between Cooking Bananas and Dessert Bananas. 1. Morphological and Compositional Characterization of Cultivated Colombian Musaceae (Musa sp.) in Relation to Consumer Preferences. J. Agric. Food Chem. 2009, 57, 7857–7869. [Google Scholar] [CrossRef]
- Kouassi, H.A.; Assemand, E.F.; Gibert, O.; Maraval, I.; Ricci, J.; Thiemele, D.E.F.; Bugaud, C. Textural and Physicochemical Predictors of Sensory Texture and Sweetness of Boiled Plantain. Int. J. Food Sci. Technol. 2021, 56, 1160–1170. [Google Scholar] [CrossRef]
- Burdon, J.; McLeod, D.; Lallu, N.; Gamble, J.; Petley, M.; Gunson, A. Consumer Evaluation of “Hayward” Kiwifruit of Different at-Harvest Dry Matter Contents. Postharvest Biol. Technol. 2004, 34, 245–255. [Google Scholar] [CrossRef]
- Jaeger, S.R.; Harker, R.; Triggs, C.M.; Gunson, A.; Campbell, R.L.; Jackman, R.; Requejo-Jackman, C. Determining Consumer Purchase Intentions: The Importance of Dry Matter, Size, and Price of Kiwifruit. J. Food Sci. 2011, 76, S177–S184. [Google Scholar] [CrossRef]
- Chagné, D.; Dayatilake, D.; Diack, R.; Oliver, M.; Ireland, H.; Watson, A.; Gardiner, S.E.; Johnston, J.W.; Schaffer, R.J.; Tustin, S. Genetic and Environmental Control of Fruit Maturation, Dry Matter and Firmness in Apple (Malus × Domestica Borkh.). Hortic. Res. 2014, 1, 14046. [Google Scholar] [CrossRef]
- Knudsen, J.T.; Eriksson, R.; Gershenzon, J.; Ståhl, B. Diversity and Distribution of Floral Scent. Bot. Rev. 2006, 72, 1. [Google Scholar] [CrossRef]
- Cappellin, L.; Farneti, B.; Di Guardo, M.; Busatto, N.; Khomenko, I.; Romano, A.; Velasco, R.; Costa, G.; Biasioli, F.; Costa, F. QTL Analysis Coupled with PTR-ToF-MS and Candidate Gene-Based Association Mapping Validate the Role of Md-AAT1 as a Major Gene in the Control of Flavor in Apple Fruit. Plant Mol. Biol. Rep. 2015, 33, 239–252. [Google Scholar] [CrossRef]
- Schaffer, R.J.; Friel, E.N.; Souleyre, E.J.F.; Bolitho, K.; Thodey, K.; Ledger, S.; Bowen, J.H.; Ma, J.H.; Nain, B.; Cohen, D.; et al. A Genomics Approach Reveals That Aroma Production in Apple Is Controlled by Ethylene Predominantly at the Final Step in Each Biosynthetic Pathway. Plant Physiol. 2007, 144, 1899–1912. [Google Scholar] [CrossRef] [PubMed]
- Larsen, B.; Migicovsky, Z.; Jeppesen, A.A.; Gardner, K.M.; Toldam-Andersen, T.B.; Myles, S.; Ørgaard, M.; Petersen, M.A.; Pedersen, C. Genome-Wide Association Studies in Apple Reveal Loci for Aroma Volatiles, Sugar Composition, and Harvest Date. Plant Genome 2019, 12, 180104. [Google Scholar] [CrossRef] [PubMed]
- Olbricht, K.; Grafe, C.; Weis, K.; Ulrich, D. Inheritance of Aroma Compounds in a Model Population of Fragaria · Ananassa Duch. Plant Breed. 2008, 127, 87–93. [Google Scholar] [CrossRef]
- Olbricht, K.; Ulrich, D.; Weiss, K.; Grafe, C. Variation in the Amounts of Selected Volatiles in a Model Population of Fragaria  Ananassa Duch. As Influenced by Harvest Year. J. Agric. Food Chem. 2011, 59, 944–952. [Google Scholar] [CrossRef]
- Chambers, A.H.; Pillet, J.; Plotto, A.; Bai, J.; Whitaker, V.M.; Folta, K.M. Identification of a Strawberry Flavor Gene Candidate Using an Integrated Genetic-Genomic-Analytical Chemistry Approach. BMC Genom. 2014, 15, 217. [Google Scholar] [CrossRef]
- Noh, Y.H.; Lee, S.; Whitaker, V.M.; Cearley, K.R.; Cha, J.S. A High-Throughput Marker-Assisted Selection System Combining Rapid DNA Extraction High-Resolution Melting and Simple Sequence Repeat Analysis: Strawberry As A Model for Fruit Crops. J. Berry Res. 2017, 7, 23–31. [Google Scholar] [CrossRef]
- Yu, Y.; Bai, J.; Chen, C.; Plotto, A.; Yu, Q.; Baldwin, E.A.; Gmitter, F.G. Identification of QTLs Controlling Aroma Volatiles Using a “Fortune” x “Murcott” (Citrus reticulata) Population. BMC Genom. 2017, 18, 646. [Google Scholar] [CrossRef]
- Eduardo, I.; Chietera, G.; Pirona, R.; Pacheco, I.; Troggio, M.; Banchi, E.; Bassi, D.; Rossini, L.; Vecchietti, A.; Pozzi, C.; et al. Genetic Dissection of Aroma Volatile Compounds from the Essential Oil of Peach Fruit: QTL Analysis and Identification of Candidate Genes Using Dense SNP Maps. Tree Genet. Genomes 2013, 9, 189–204. [Google Scholar] [CrossRef]
- Doligez, A.; Audiot, E.; Baumes, R.; This, P. QTLs for Muscat Flavor and Monoterpenic Odorant Content in Grapevine (Vitis vinifera L.). Mol. Breed. 2006, 18, 109–125. [Google Scholar] [CrossRef]
- Battilana, J.; Costantini, L.; Emanuelli, F.; Sevini, F.; Segala, C.; Moser, S.; Velasco, R.; Versini, G.; Grando, M.S. The 1-Deoxy-d-Xylulose 5-Phosphate Synthase Gene Co-Localizes with a Major QTL Affecting Monoterpene Content in Grapevine. Theor. Appl. Genet. 2009, 118, 653–669. [Google Scholar] [CrossRef] [PubMed]
- Duchêne, E.; Butterlin, G.; Claudel, P.; Dumas, V.; Jaegli, N.; Merdinoglu, D. A Grapevine (Vitis vinifera L.) Deoxy-D: -Xylulose Synthase Gene Colocates with a Major Quantitative Trait Loci for Terpenol Content. Theor. Appl. Genet. 2009, 118, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Lü, P.; Yu, S.; Zhu, N.; Chen, Y.R.; Zhou, B.; Pan, Y.; Tzeng, D.; Fabi, J.P.; Argyris, J.; Garcia-Mas, J.; et al. Genome Encode Analyses Reveal the Basis of Convergent Evolution of Fleshy Fruit Ripening. Nat. Plants 2018, 4, 784–791. [Google Scholar] [CrossRef] [PubMed]
- Migicovsky, Z.; Gardner, K.M.; Money, D.; Sawler, J.; Bloom, J.S.; Moffett, P.; Chao, C.T.; Schwaninger, H.; Fazio, G.; Zhong, G.; et al. Genome to Phenome Mapping in Apple Using Historical Data. Plant Genome 2016, 9, plantgenome2015-11. [Google Scholar] [CrossRef]
- McClure, K.A.; Gardner, K.M.; Douglas, G.M.; Song, J.; Forney, C.F.; DeLong, J.; Fan, L.; Du, L.; Toivonen, P.M.A.; Somers, D.J.; et al. A Genome-Wide Association Study of Apple Quality and Scab Resistance. Plant Genome 2018, 11, 170075. [Google Scholar] [CrossRef]
- Moyano, E.; Martínez-Rivas, F.J.; Blanco-Portales, R.; Molina-Hidalgo, F.J.; Ric-Varas, P.; Matas-Arroyo, A.J.; Caballero, J.L.; Muñoz-Blanco, J.; Rodríguez-Franco, A. Genome-Wide Analysis of the NAC Transcription Factor Family and Their Expression during the Development and Ripening of the Fragaria × Ananassa Fruits. PLoS ONE 2018, 13, e0196953. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Y.; Turečková, V.; Xue, G.P.; Fernie, A.R.; Mueller-Roeber, B.; Balazadeh, S. The NAC Transcription Factor SLNAP2 Regulates Leaf Senescence and Fruit Yield in Tomato. Plant Physiol. 2018, 177, 1286–1302. [Google Scholar] [CrossRef] [PubMed]
- Pirona, R.; Eduardo, I.; Pacheco, I.; Da Silva Linge, C.; Miculan, M.; Verde, I.; Tartarini, S.; Dondini, L.; Pea, G.; Bassi, D.; et al. Fine Mapping and Identification of a Candidate Gene for a Major Locus Controlling Maturity Date in Peach. BMC Plant Biol. 2013, 13, 166. [Google Scholar] [CrossRef]
- Shan, W.; Kuang, J.F.; Chen, L.; Xie, H.; Peng, H.H.; Xiao, Y.Y.; Li, X.P.; Chen, W.X.; He, Q.G.; Chen, J.Y.; et al. Molecular Characterization of Banana NAC Transcription Factors and Their Interactions with Ethylene Signalling Component EIL during Fruit Ripening. J. Exp. Bot. 2012, 63, 5171–5187. [Google Scholar] [CrossRef]
- Nishio, S.; Terakami, S.; Matsumoto, T.; Yamamoto, T.; Takada, N.; Kato, H.; Katayose, Y.; Saito, T. Identification of QTLs for Agronomic Traits in the Japanese Chestnut (Castanea Crenata Sieb. et Zucc.) Breeding. Hortic. J. 2018, 87, 43–54. [Google Scholar] [CrossRef]
- Harada, T.; Sunako, T.; Wakasa, Y.; Soejima, J.; Satoh, T.; Niizeki, M. An Allele of the 1-Aminocyclopropane-1-Carboxylate Synthase Gene (Md-ACS1) Accounts for the Low Level of Ethylene Production in Climacteric Fruits of Some Apple Cultivars. Theor. Appl. Genet. 2000, 101, 742–746. [Google Scholar] [CrossRef]
- Bai, S.; Wang, A.; Igarashi, M.; Kon, T.; Fukasawa-Akada, T.; Li, T.; Harada, T.; Hatsuyama, Y. Distribution of MdACS3 Null Alleles in Apple (Malus × Domestica Borkh.) and Its Relevance to the Fruit Ripening Characters. Breed. Sci. 2012, 62, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Verde, I.; Bassil, N.; Scalabrin, S.; Gilmore, B.; Lawley, C.T.; Gasic, K.; Micheletti, D.; Rosyara, U.R.; Cattonaro, F.; Vendramin, E.; et al. Development and Evaluation of a 9k Snp Array for Peach by Internationally Coordinated Snp Detection and Validation in Breeding Germplasm. PLoS ONE 2012, 7, e35668. [Google Scholar] [CrossRef]
- Dirlewanger, E.; Quero-García, J.; Le Dantec, L.; Lambert, P.; Ruiz, D.; Dondini, L.; Illa, E.; Quilot-Turion, B.; Audergon, J.M.; Tartarini, S.; et al. Comparison of the Genetic Determinism of Two Key Phenological Traits, Flowering and Maturity Dates, in Three Prunus Species: Peach, Apricot and Sweet Cherry. Heredity 2012, 109, 280–292. [Google Scholar] [CrossRef]
- Sánchez-Pérez, R.; Howad, W.; Dicenta, F.; Arús, P.; Martínez-Gómez, P. Mapping Major Genes and Quantitative Trait Loci Controlling Agronomic Traits in Almond. Plant Breed. 2007, 126, 310–318. [Google Scholar] [CrossRef]
- Meneses, C.; Ulloa-Zepeda, L.; Cifuentes-Esquivel, A.; Infante, R.; Cantin, C.M.; Batlle, I.; Arús, P.; Eduardo, I. A Codominant Diagnostic Marker for the Slow Ripening Trait in Peach. Mol. Breed. 2016, 36, 77. [Google Scholar] [CrossRef]
- Danilevskaya, O.N.; Meng, X.; Selinger, D.A.; Deschamps, S.; Hermon, P.; Vansant, G.; Gupta, R.; Ananiev, E.V.; Muszynski, M.G. Involvement of the MADS-Box Gene ZMM4 in Floral Induction and Inflorescence Development in Maize. Plant Physiol. 2008, 147, 2054–2069. [Google Scholar] [CrossRef]
- Graham, J.; Hackett, C.A.; Smith, K.; Woodhead, M.; Hein, I.; McCallum, S. Mapping QTLs for Developmental Traits in Raspberry from Bud Break to Ripe Fruit. Theor. Appl. Genet. 2009, 118, 1143–1155. [Google Scholar] [CrossRef]


| Character | Phenotype | Species | Marker | Type |
|---|---|---|---|---|
| Phenological traits related to fruit production | Flowering time | Apricot | UDAp-423r, AMPA-105 | SSR |
| Pollen sterility | Peach | Ps | Gene | |
| Sex determination | Ginco | GBA, GBB | SCAR | |
| Sex determination | Kaki | DlSx-AF4S | SCAR | |
| Sex determination | Kiwifruit | SyGI, FrBy, Ank | Gene | |
| Sex determination | Mulberry | MBS markers | RAD-seq | |
| Sex determination | Papaya | SCART1, SCART12, SCARW11 | SCAR | |
| Sex determination | Pistacio | SNP-PIS-167992, P-ATL-91951–565 | KASP | |
| Skin colour | Red skin colour | Apple | Mdo.chr9.4 | SSR |
| Red skin colour | Apple | ss475879531 | SNP | |
| Red skin colour | Peach | PpMYB10.1 | Indel polymorphism | |
| Red skin colour | Strawberry | 18bp-del-5’UTR-ANR | Indel polymorphism | |
| Flesh colour | Dark purple-red/yellow colour | Cherry | LG3_13.146 | SSR |
| Red/yellow colour | Papaya | CPFC1 | Indel polymorphism | |
| White/yellow colour | Peach | ccd4-INS1-f/ ccd4-E2-r | Indel polymorphism | |
| Fruit size and weight | Increase fruit size | Sweet cherry | PavCNR12, PavCNR20 | SSR |
| Increase weight | Grapevine | VVIB19 | SSR | |
| Increase weight | Japanese plum | UDAp456 | SSR | |
| Seedlessness | No seed | Citrus | OPAI11-0.8, OPAJ19-1.0, OPM06r-0.85, OPAJ04r-0.6 | RAPD |
| No seed | Citrus | AFLP-2, AFLP-5 | SCAR | |
| No seed | Grapevine | UBC-269500 | RAPD | |
| No seed | Grapevine | SCC8 | SCAR | |
| No seed | Grapevine | SCF27 | SCAR | |
| No seed | Grapevine | VMC7F2 | SSR | |
| No seed | Grapevine | VviAGL11 | Gene | |
| No seed | Grapevine | chrG8:11240814 | SNP | |
| Firmness, crispness, texture | Firmness loss | Apple | Md-PG1 | SNP |
| Firmness loss | Apple | NAC18.1 | Gene | |
| High-texture performance | Apple | Md-PG1SSR | SSR | |
| Firmness loss | Grapevine | VMC2E7 | SSR | |
| Firmness loss | Sweet cherry | qtnFF 4.1, qtnFF 4.2 | SNP | |
| Pulp acidity and pH | Acidity | Apple | AHFBAZU, AHHS7CA, AH89247, AHBAIAO | SNP |
| pH and malic acid | Apricot | PaCITA7, UDAp-471 | SSR | |
| Acidity | Cranberry | scf258d SSR | SSR | |
| Malic acid | Cranberry | MA_476 | KASP | |
| Titratable acidity | Grapevine | VVIM63, VVS1, VVIC51 | SSR | |
| Titratable acidity (acid/non-acid) | Peach | D_allele_SNP | SNP | |
| Titratable acidity (acid/non-acid) | Peach | CPPCT040 | SSR | |
| VOCs | γ-Decalactone flavor | Strawberry | FaFAD1 | SSR |
| Harvesting date, maturity date, ethylene, ripening | Harvesting date | Apple | chr3:31409362 | SNP |
| Slow ripening | Peach | del_Prupe.4G186800 | Indel polymorphism | |
| Slow ripening | Raspberry | P12M121-194 | AFLP |
| Species | Trait | Mapping Strategy | Linkage Group | Candidate Gene(s) | References |
|---|---|---|---|---|---|
| Sweet cherry | Size | BP | 2 | FW2.2/CNR | [105] |
| Chinese cherry | Size | AM | 8 | Auxin response, cell differentiation, pectin biosynthesis | [106] |
| Chinese chestnut | Nut weight, width, thickness, height | BP | 1 | [107] | |
| Citrus | Size, weight, diameter | BP, AM | 2, 4, 5, 7, 8 | [79,108] | |
| Coffee | Yield, bean size | BP | 2, 4, 6 | [45] | |
| Grape | Weight | AM, BP | 1, 8,11, 17, 18 | Aux/IAA9, DELLA protein | [109,110,111] |
| Japanese plum | Weight | BP | 2 | [112] | |
| Logan | Weight | BP | 1, 4, 10, 14 | FW2.2/CNR, P450, EXP4 | [113] |
| Peach | Weight, diameter | BP, AM | 1, 2, 3, 4, 5, 6, 7 | [33,115,116,117,118,119,120] | |
| Guava | Weight, size, yield | [114] | |||
| Papaya | Weight, size | [114] | |||
| Banana | Weight | 3 | [114] | ||
| Walnut | Weight, size | AM | 1, 3, 6, 8, 12, 5, 11, 15, 7, 9, 14 | Beta-galactosidase, RBK1, BEL1-like | [121] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Mori, G.; Cipriani, G. Marker-Assisted Selection in Breeding for Fruit Trait Improvement: A Review. Int. J. Mol. Sci. 2023, 24, 8984. https://doi.org/10.3390/ijms24108984
De Mori G, Cipriani G. Marker-Assisted Selection in Breeding for Fruit Trait Improvement: A Review. International Journal of Molecular Sciences. 2023; 24(10):8984. https://doi.org/10.3390/ijms24108984
Chicago/Turabian StyleDe Mori, Gloria, and Guido Cipriani. 2023. "Marker-Assisted Selection in Breeding for Fruit Trait Improvement: A Review" International Journal of Molecular Sciences 24, no. 10: 8984. https://doi.org/10.3390/ijms24108984
APA StyleDe Mori, G., & Cipriani, G. (2023). Marker-Assisted Selection in Breeding for Fruit Trait Improvement: A Review. International Journal of Molecular Sciences, 24(10), 8984. https://doi.org/10.3390/ijms24108984






