Hypoxia Is Associated with Increased Immune Infiltrates and Both Anti-Tumour and Immune Suppressive Signalling in Muscle-Invasive Bladder Cancer
Abstract
1. Introduction
2. Results
2.1. ChIP-Seq Identified HIF Binding Sites with High Specificity and Low Background
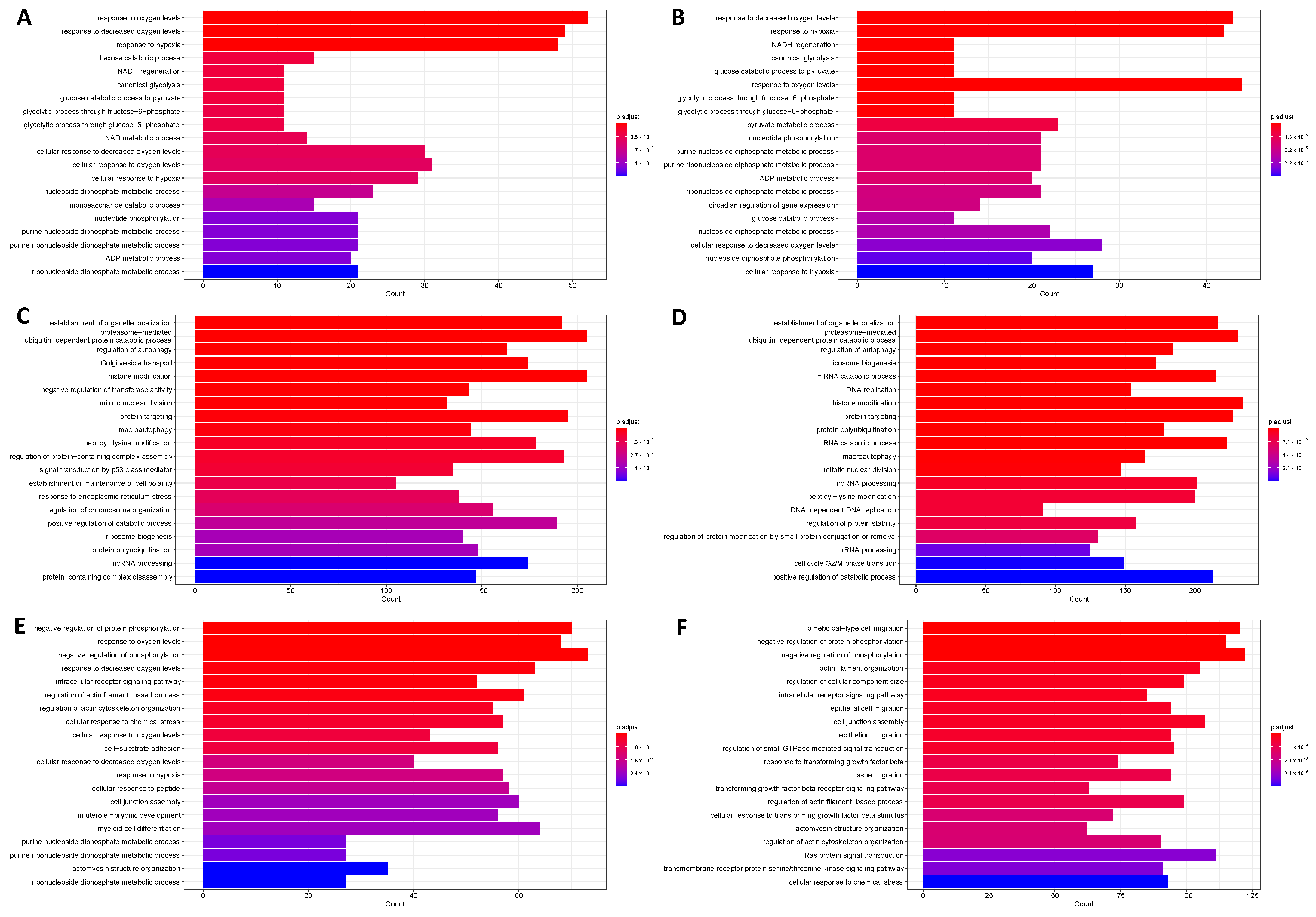
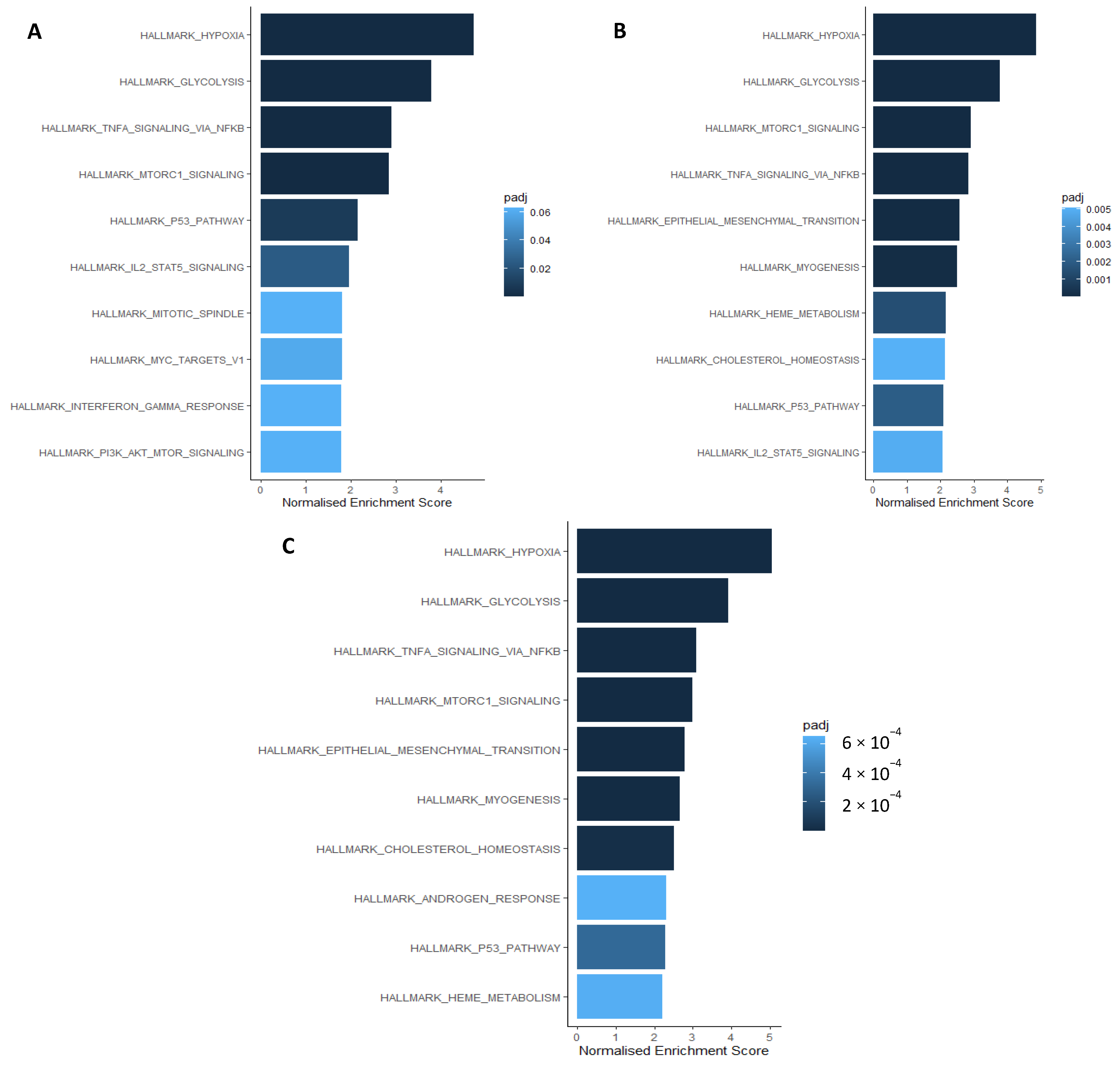
2.2. HIF1 and HIF2 Are Associated with Distinct Biological Processes
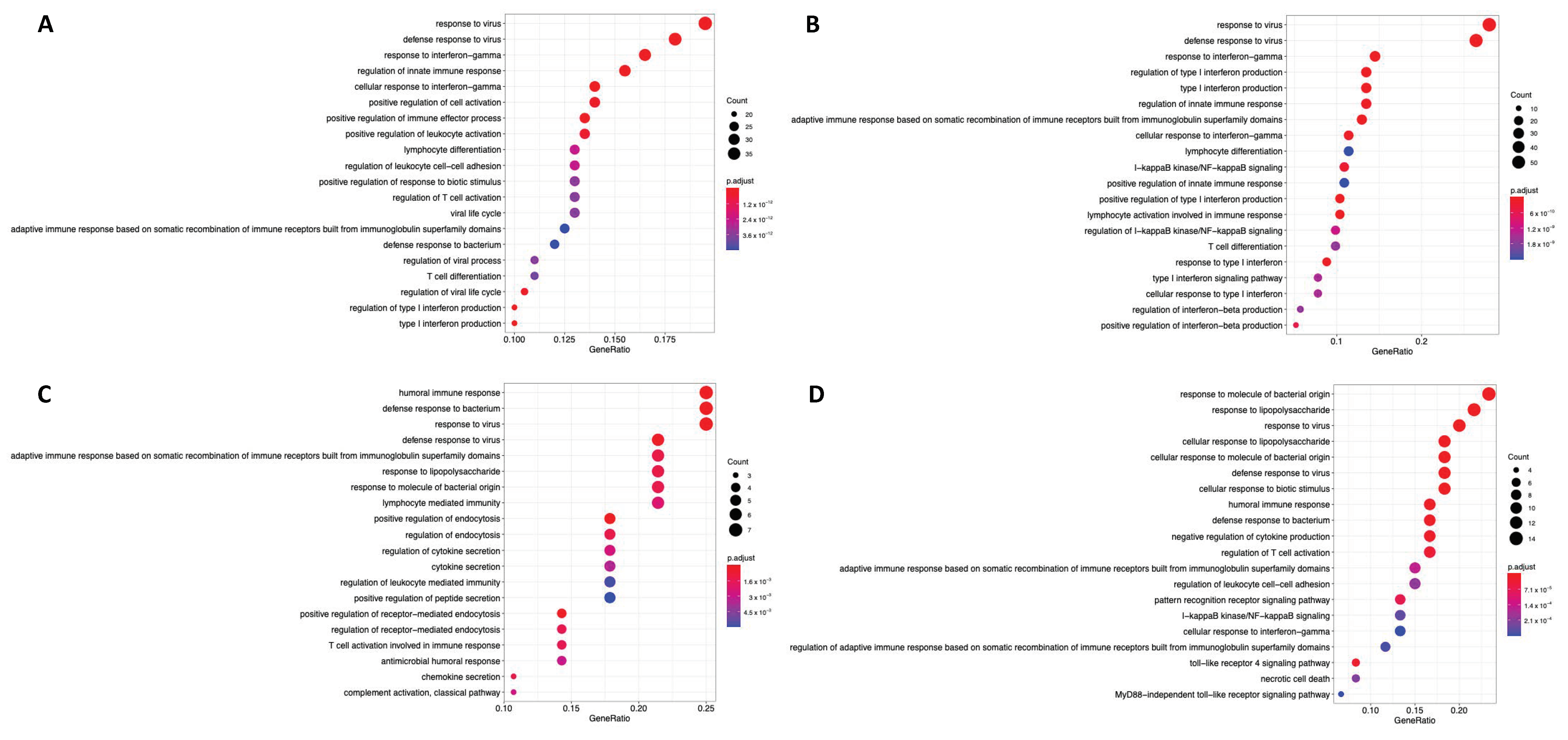
2.3. HIF1 and HIF2 Are Associated with Unique Immune-Related Processes
2.4. Hypoxia Associates with Myeloid, Neutrophil, and CD4+ T Cell Signalling Processes
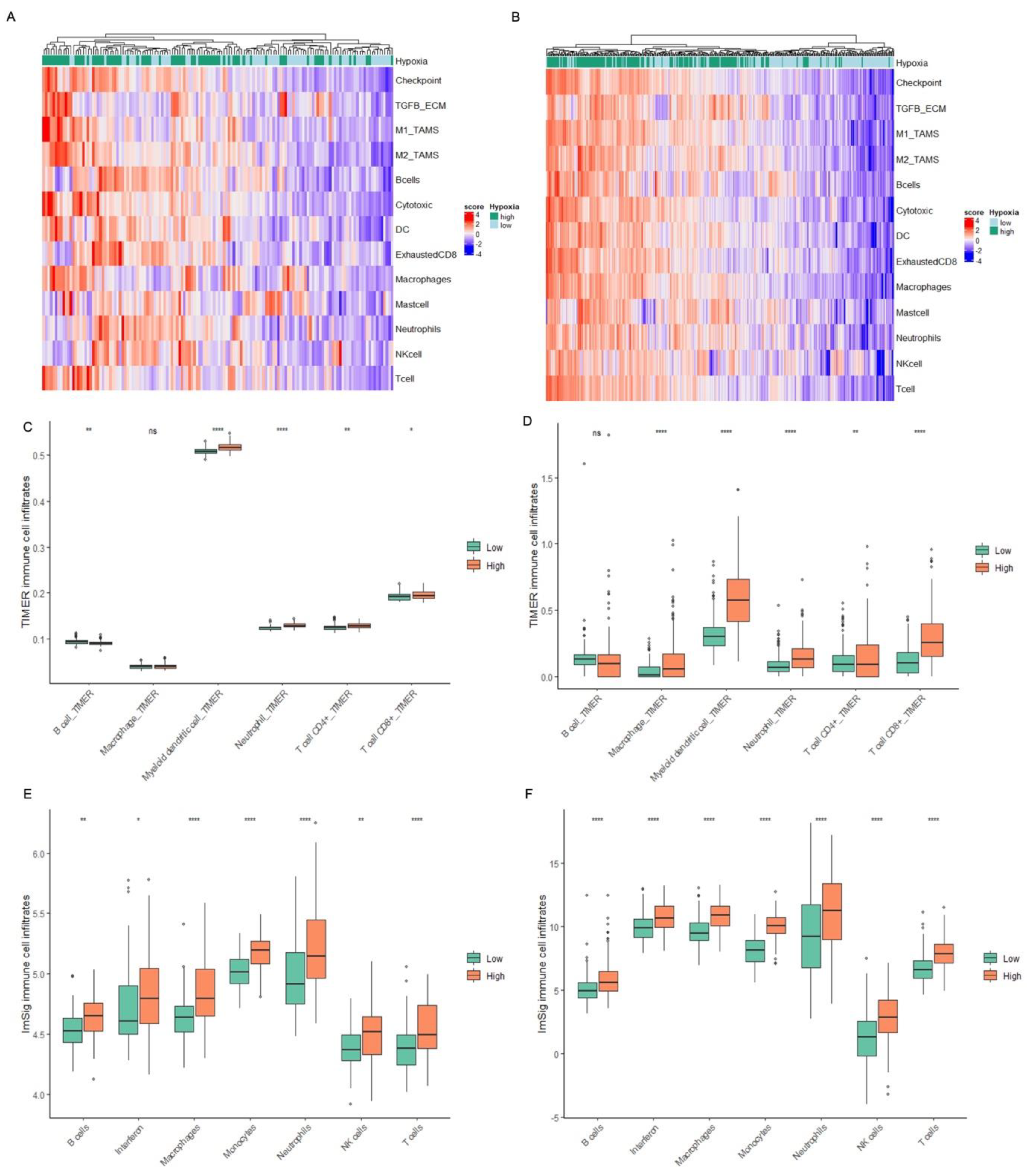
2.5. Hypoxia Associates with an Inflamed TME in MIBC Patient Tumours
3. Discussion
4. Materials and Methods
4.1. Cohorts
4.2. ChIP-Seq Data Generation
4.3. Microarray Data Generation
4.4. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- National Institute for Health and Care Excellence Bladder Cancer: Diagnosis and Management. Available online: https://www.nice.org.uk/guidance/ng2/chapter/1-Recommendations#treating-muscle-invasive-bladder-cancer-2 (accessed on 8 March 2022).
- Kachikwu, E.L.; Iwamoto, K.S.; Liao, Y.-P.; DeMarco, J.J.; Agazaryan, N.; Economou, J.S.; McBride, W.H.; Schaue, D. Radiation Enhances Regulatory T Cell Representation. Int. J. Radiat. Oncol. 2011, 81, 1128–1135. [Google Scholar] [CrossRef] [PubMed]
- Barker, H.E.; Paget, J.T.E.; Khan, A.A.; Harrington, K.J. The Tumour Microenvironment after Radiotherapy: Mechanisms of Resistance and Recurrence. Nat. Rev. Cancer 2015, 15, 409–425. [Google Scholar] [CrossRef] [PubMed]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, Inflammation, and Cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef] [PubMed]
- Höckel, M.; Schlenger, K.; Mitze, M.; Schäffer, U.; Vaupel, P. Hypoxia and Radiation Response in Human Tumors. Semin. Radiat. Oncol. 1996, 6, 3–9. [Google Scholar] [CrossRef]
- Theodoropoulos, V.E.; Lazaris, A.C.; Sofras, F.; Gerzelis, I.; Tsoukala, V.; Ghikonti, I.; Manikas, K.; Kastriotis, I. Hypoxia-Inducible Factor 1α Expression Correlates with Angiogenesis and Unfavorable Prognosis in Bladder Cancer. Eur. Urol. 2004, 46, 200–208. [Google Scholar] [CrossRef]
- Hunter, B.A.; Eustace, A.; Irlam, J.J.; Valentine, H.R.; Denley, H.; Oguejiofor, K.K.; Swindell, R.; Hoskin, P.J.; Choudhury, A.; West, C.M. Expression of Hypoxia-Inducible Factor-1α Predicts Benefit from Hypoxia Modification in Invasive Bladder Cancer. Br. J. Cancer 2014, 111, 437. [Google Scholar] [CrossRef]
- Vaupel, P.; Harrison, L. Tumor Hypoxia: Causative Factors, Compensatory Mechanisms, and Cellular Response. Oncologist 2004, 9 (Suppl. S5), 4–9. [Google Scholar] [CrossRef]
- Manoochehri Khoshinani, H.; Afshar, S.; Najafi, R. Hypoxia: A Double-Edged Sword in Cancer Therapy. Cancer Investig. 2016, 34, 536–545. [Google Scholar] [CrossRef]
- Elia, A.R.; Cappello, P.; Puppo, M.; Fraone, T.; Vanni, C.; Eva, A.; Musso, T.; Novelli, F.; Varesio, L.; Giovarelli, M. Human Dendritic Cells Differentiated in Hypoxia Down-Modulate Antigen Uptake and Change Their Chemokine Expression Profile. J. Leukoc. Biol. 2008, 84, 1472–1482. [Google Scholar] [CrossRef]
- Corzo, C.A.; Condamine, T.; Lu, L.; Cotter, M.J.; Youn, J.-I.; Cheng, P.; Cho, H.-I.; Celis, E.; Quiceno, D.G.; Padhya, T.; et al. HIF1α Regulates Function and Differentiation of Myeloid-Derived Suppressor Cells in the Tumor Microenvironment. J. Exp. Med. 2010, 207, 2439–2453. [Google Scholar] [CrossRef]
- Walmsley, S.R.; Print, C.; Farahi, N.; Peyssonnaux, C.; Johnson, R.S.; Cramer, T.; Sobolewski, A.; Condliffe, A.M.; Cowburn, A.S.; Johnson, N.; et al. Hypoxia-Induced Neutrophil Survival Is Mediated by HIF-1α-Dependent NF-κB Activity. J. Exp. Med. 2005, 201, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Cramer, T.; Yamanishi, Y.; Clausen, B.E.; Förster, I.; Pawlinski, R.; Mackman, N.; Haase, V.H.; Jaenisch, R.; Corr, M.; Nizet, V.; et al. HIF1α Is Essential for Myeloid Cell-Mediated Inflammation. Cell 2003, 112, 645. [Google Scholar] [CrossRef] [PubMed]
- Talks, K.L.; Turley, H.; Gatter, K.C.; Maxwell, P.H.; Pugh, C.W.; Ratcliffe, P.J.; Harris, A.L. The Expression and Distribution of the Hypoxia-Inducible Factors HIF-1α and HIF-2α in Normal Human Tissues, Cancers, and Tumor-Associated Macrophages. Am. J. Pathol. 2000, 157, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Doedens, A.L.; Stockmann, C.; Rubinstein, M.P.; Liao, D.; Zhang, N.; DeNardo, D.G.; Coussens, L.M.; Karin, M.; Goldrath, A.W.; Johnson, R.S. Macrophage Expression of HIF1α Suppresses T Cell Function and Promotes Tumor Progression. Cancer Res. 2010, 70, 7465. [Google Scholar] [CrossRef]
- Murdoch, C.; Giannoudis, A.; Lewis, C.E.; Weich, H.A.; Mantovani, A.; Marmé, D. Mechanisms Regulating the Recruitment of Macrophages into Hypoxic Areas of Tumors and Other Ischemic Tissues. Blood 2004, 104, 2224–2234. [Google Scholar] [CrossRef]
- Ohta, A.; Gorelik, E.; Prasad, S.J.; Ronchese, F.; Lukashev, D.; Wong, M.K.K.; Huang, X.; Caldwell, S.; Liu, K.; Smith, P.; et al. A2A Adenosine Receptor Protects Tumors from Antitumor T Cells. Proc. Natl. Acad. Sci. USA 2006, 103, 13132. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, Y.; Yang, M.; Zhang, Y.; Xie, Q.; Li, Z.; Dong, Z.; Yang, Y.; Deng, B.; Feng, A.; et al. Hypoxia Induces T-Cell Apoptosis by Inhibiting Chemokine C Receptor 7 Expression: The Role of Adenosine Receptor A2. Cell. Mol. Immunol. 2009, 7, 77–82. [Google Scholar] [CrossRef]
- Hatfield, S.M.; Kjaergaard, J.; Lukashev, D.; Schreiber, T.H.; Belikoff, B.; Abbott, R.; Sethumadhavan, S.; Philbrook, P.; Ko, K.; Cannici, R.; et al. Immunological Mechanisms of the Antitumor Effects of Supplemental Oxygenation. Sci. Transl. Med. 2015, 7, 277ra30. [Google Scholar] [CrossRef]
- Jayaprakash, P.; Ai, M.; Liu, A.; Budhani, P.; Bartkowiak, T.; Sheng, J.; Ager, C.; Nicholas, C.; Jaiswal, A.R.; Sun, Y.; et al. Targeted Hypoxia Reduction Restores T Cell Infiltration and Sensitizes Prostate Cancer to Immunotherapy. J. Clin. Investig. 2018, 128, 5137–5149. [Google Scholar] [CrossRef]
- Chai, C.Y.; Chen, W.T.; Hung, W.C.; Kang, W.Y.; Huang, Y.C.; Su, Y.C.; Yang, C.H. Hypoxia-Inducible Factor-1α Expression Correlates with Focal Macrophage Infiltration, Angiogenesis and Unfavourable Prognosis in Urothelial Carcinoma. J. Clin. Pathol. 2008, 61, 658–664. [Google Scholar] [CrossRef]
- Palazon, A.; Tyrakis, P.A.; Macias, D.; Veliça, P.; Rundqvist, H.; Fitzpatrick, S.; Vojnovic, N.; Phan, A.T.; Loman, N.; Hedenfalk, I.; et al. An HIF1α/VEGF-A Axis in Cytotoxic T Cells Regulates Tumor Progression. Cancer Cell 2017, 32, 669–683.e5. [Google Scholar] [CrossRef] [PubMed]
- Doedens, A.L.; Phan, A.T.; Stradner, M.H.; Fujimoto, J.K.; Nguyen, J.V.; Yang, E.; Johnson, R.S.; Goldrath, A.W. Hypoxia-Inducible Factors Enhance the Effector Responses of CD8+ T Cells to Persistent Antigen. Nat. Immunol. 2013, 14, 1173–1182. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Li, L.; Li, M.; Wang, X. HIF1A Expression Correlates with Increased Tumor Immune and Stromal Signatures and Aggressive Phenotypes in Human Cancers. Cell. Oncol. 2020, 43, 877–888. [Google Scholar] [CrossRef]
- Smythies, J.A.; Sun, M.; Masson, N.; Salama, R.; Simpson, P.D.; Murray, E.; Neumann, V.; Cockman, M.E.; Choudhry, H.; Ratcliffe, P.J.; et al. Inherent DNA -binding Specificities of the HIF -1α and HIF -2α Transcription Factors in Chromatin. EMBO Rep. 2019, 20, e46401. [Google Scholar] [CrossRef] [PubMed]
- Schödel, J.; Oikonomopoulos, S.; Ragoussis, J.; Pugh, C.W.; Ratcliffe, P.J.; Mole, D.R. High-Resolution Genome-Wide Mapping of HIF-Binding Sites by ChIP-Seq. Blood 2011, 117, e207–e217. [Google Scholar] [CrossRef] [PubMed]
- Mole, D.R.; Blancher, C.; Copley, R.R.; Pollard, P.J.; Gleadle, J.M.; Ragousis, J.; Ratcliffe, P.J. Genome-Wide Association of Hypoxia-Inducible Factor (HIF)-1α and HIF2α DNA Binding with Expression Profiling of Hypoxia-Inducible Transcripts. J. Biol. Chem. 2009, 284, 16767–16775. [Google Scholar] [CrossRef]
- Hill, R.P.; Bristow, R.G.; Fyles, A.; Koritzinsky, M.; Milosevic, M.; Wouters, B.G. Hypoxia and Predicting Radiation Response. Semin. Radiat. Oncol. 2015, 25, 260–272. [Google Scholar] [CrossRef]
- Barsoum, I.B.; Smallwood, C.A.; Siemens, D.R.; Graham, C.H. A Mechanism of Hypoxia-Mediated Escape from Adaptive Immunity in Cancer Cells. Cancer Res. 2014, 74, 665–674. [Google Scholar] [CrossRef]
- Noman, M.Z.; Desantis, G.; Janji, B.; Hasmim, M.; Karray, S.; Dessen, P.; Bronte, V.; Chouaib, S. PD-L1 Is a Novel Direct Target of HIF1α, and Its Blockade under Hypoxia Enhanced: MDSC-Mediated T Cell Activation. J. Exp. Med. 2014, 211, 781–790. [Google Scholar] [CrossRef]
- Fabrizio, F.P.; Trombetta, D.; Rossi, A.; Sparaneo, A.; Castellana, S.; Muscarella, L.A. Gene Code CD274/PD-L1: From Molecular Basis toward Cancer Immunotherapy. Ther. Adv. Med. Oncol. 2018, 10, 1758835918815598. [Google Scholar] [CrossRef]
- Smith, V.; Mukherjee, D.; Lunj, S.; Choudhury, A.; Hoskin, P.; West, C.; Illidge, T. The Effect of Hypoxia on PD-L1 Expression in Bladder Cancer. BMC Cancer 2021, 21, 1271. [Google Scholar] [CrossRef]
- Bruns, I.B.; Beltman, J.B. Quantifying the Contribution of Transcription Factor Activity, Mutations and MicroRNAs to CD274 Expression in Cancer Patients. Sci. Rep. 2022, 12, 4374. [Google Scholar] [CrossRef] [PubMed]
- Ruf, M.; Moch, H.; Schraml, P. PD-L1 Expression Is Regulated by Hypoxia Inducible Factor in Clear Cell Renal Cell Carcinoma. Int. J. Cancer 2016, 139, 396–403. [Google Scholar] [CrossRef]
- Jorgovanovic, D.; Song, M.; Wang, L.; Zhang, Y. Roles of IFN-γ in Tumor Progression and Regression: A Review. Biomark. Res. 2020, 8, 49. [Google Scholar] [CrossRef]
- Aggarwal, B.B. Signalling Pathways of the TNF Superfamily: A Double-Edged Sword. Nat. Rev. Immunol. 2003, 3, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Montfort, A.; Colacios, C.; Levade, T.; Andrieu-Abadie, N.; Meyer, N.; Ségui, B. The TNF Paradox in Cancer Progression and Immunotherapy. Front. Immunol. 2019, 10, 1818. [Google Scholar] [CrossRef] [PubMed]
- Parameswaran, N.; Patial, S. Tumor Necrosis Factor-α Signaling in Macrophages. Crit. Rev. Eukaryot. Gene Expr. 2010, 20, 87. [Google Scholar] [CrossRef]
- Mahmud, S.A.; Manlove, L.S.; Farrar, M.A. Interleukin-2 and STAT5 in Regulatory T Cell Development and Function. JAK-STAT 2013, 2, e23154. [Google Scholar] [CrossRef]
- Jones, D.M.; Read, K.A.; Oestreich, K.J. Dynamic Roles for IL-2–STAT5 Signaling in Effector and Regulatory CD4+ T Cell Populations. J. Immunol. 2020, 205, 1721–1730. [Google Scholar] [CrossRef]
- Yang, L.; Taylor, J.; Eustace, A.; Irlam, J.J.; Denley, H.; Hoskin, P.J.; Alsner, J.; Buffa, F.M.; Harris, A.L.; Choudhury, A.; et al. A Gene Signature for Selecting Benefit from Hypoxia Modification of Radiotherapy for High-Risk Bladder Cancer Patients. Clin. Cancer Res. 2017, 23, 4761–4768. [Google Scholar] [CrossRef]
- Nirmal, A.J.; Regan, T.; Shih, B.B.-J.; Hume, D.A.; Sims, A.H.; Freeman, T.C. ImSig: A Resource for the Identification and Quantification of Immune Signatures in Blood and Tissue Transcriptomics Data. bioRxiv 2016, bioRxiv:077487. [Google Scholar] [CrossRef]
- Li, B.; Severson, E.; Pignon, J.C.; Zhao, H.; Li, T.; Novak, J.; Jiang, P.; Shen, H.; Aster, J.C.; Rodig, S.; et al. Comprehensive Analyses of Tumor Immunity: Implications for Cancer Immunotherapy. Genome Biol. 2016, 17, 174. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Ren, L.; Chen, Y.; Wang, H.; Wu, H.; Cheng, S.; Li, G.; Yu, S. Identification of a Hypoxia-Related Signature for Predicting Prognosis and the Immune Microenvironment in Bladder Cancer. Front. Mol. Biosci. 2021, 8, 380. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, X.; Bai, Y.; Hu, H.; Yang, Y.; Wang, J.; Tang, Y.; Ma, H.; Feng, D.; Li, D.; et al. Development and Validation of a Hypoxia-Related Signature for Predicting Survival Outcomes in Patients With Bladder Cancer. Front. Genet. 2021, 12, 670384. [Google Scholar] [CrossRef]
- Liu, Z.; Tang, Q.; Qi, T.; Othmane, B.; Yang, Z.; Chen, J.; Hu, J.; Zu, X. A Robust Hypoxia Risk Score Predicts the Clinical Outcomes and Tumor Microenvironment Immune Characters in Bladder Cancer. Front. Immunol. 2021, 12, 725223. [Google Scholar] [CrossRef]
- Hoskin, P.J.; Rojas, A.M.; Bentzen, S.M.; Saunders, M.I. Radiotherapy with Concurrent Carbogen and Nicotinamide in Bladder Carcinoma. J. Clin. Oncol. 2010, 28, 4912–4918. [Google Scholar] [CrossRef]
- Song, Y.P.; Mistry, H.; Irlam, J.; Valentine, H.; Yang, L.; Lane, B.; West, C.; Choudhury, A.; Hoskin, P.J. Long-Term Outcomes of Radical Radiation Therapy with Hypoxia Modification with Biomarker Discovery for Stratification: 10-Year Update of the BCON (Bladder Carbogen Nicotinamide) Phase 3 Randomized Trial (ISRCTN45938399). Int. J. Radiat. Oncol. Biol. Phys. 2021, 110, 1407–1415. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data [Online]. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 11 January 2022).
- Krueger, F. TrimGalore 2021. Available online: https://github.com/FelixKrueger/TrimGalore (accessed on 11 January 2022).
| Oxygen Concentration | HIF Subunit | Percent of Immune Genes Bound |
|---|---|---|
| 1% | HIF1α | 11.79 |
| HIF1β | 2.20 | |
| HIF2α | 4.77 | |
| 0.1% | HIF1α | 13.43 |
| HIF1β | 1.76 | |
| HIF2α | 7.54 |
| Term | ID | Ont | n | DE | P.DE |
|---|---|---|---|---|---|
| mitigation of host immune response by virus | GO:0030683 | BP | 2 | 2 | 0.04 |
| positive regulation of tolerance induction dependent upon immune response | GO:0002654 | BP | 2 | 2 | 0.04 |
| positive regulation of immune response to tumour cell | GO:0002839 | BP | 13 | 6 | 0.03 |
| positive regulation of myeloid leukocyte cytokine production | GO:0061081 | BP | 19 | 8 | 0.02 |
| Neutrophil-mediated immunity | GO:0002446 | BP | 501 | 127 | <0.001 |
| neutrophil activation involved in immune response | GO:0002283 | BP | 490 | 121 | 0.003 |
| myeloid-leukocyte-mediated immunity | GO:0002444 | BP | 555 | 136 | 0.003 |
| myeloid cell activation involved in immune response | GO:0002275 | BP | 549 | 127 | 0.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith, V.; Lee, D.; Reardon, M.; Shabbir, R.; Sahoo, S.; Hoskin, P.; Choudhury, A.; Illidge, T.; West, C.M.L. Hypoxia Is Associated with Increased Immune Infiltrates and Both Anti-Tumour and Immune Suppressive Signalling in Muscle-Invasive Bladder Cancer. Int. J. Mol. Sci. 2023, 24, 8956. https://doi.org/10.3390/ijms24108956
Smith V, Lee D, Reardon M, Shabbir R, Sahoo S, Hoskin P, Choudhury A, Illidge T, West CML. Hypoxia Is Associated with Increased Immune Infiltrates and Both Anti-Tumour and Immune Suppressive Signalling in Muscle-Invasive Bladder Cancer. International Journal of Molecular Sciences. 2023; 24(10):8956. https://doi.org/10.3390/ijms24108956
Chicago/Turabian StyleSmith, Vicky, Dave Lee, Mark Reardon, Rekaya Shabbir, Sudhakar Sahoo, Peter Hoskin, Ananya Choudhury, Timothy Illidge, and Catharine M. L. West. 2023. "Hypoxia Is Associated with Increased Immune Infiltrates and Both Anti-Tumour and Immune Suppressive Signalling in Muscle-Invasive Bladder Cancer" International Journal of Molecular Sciences 24, no. 10: 8956. https://doi.org/10.3390/ijms24108956
APA StyleSmith, V., Lee, D., Reardon, M., Shabbir, R., Sahoo, S., Hoskin, P., Choudhury, A., Illidge, T., & West, C. M. L. (2023). Hypoxia Is Associated with Increased Immune Infiltrates and Both Anti-Tumour and Immune Suppressive Signalling in Muscle-Invasive Bladder Cancer. International Journal of Molecular Sciences, 24(10), 8956. https://doi.org/10.3390/ijms24108956






