MncR: Late Integration Machine Learning Model for Classification of ncRNA Classes Using Sequence and Structural Encoding
Abstract
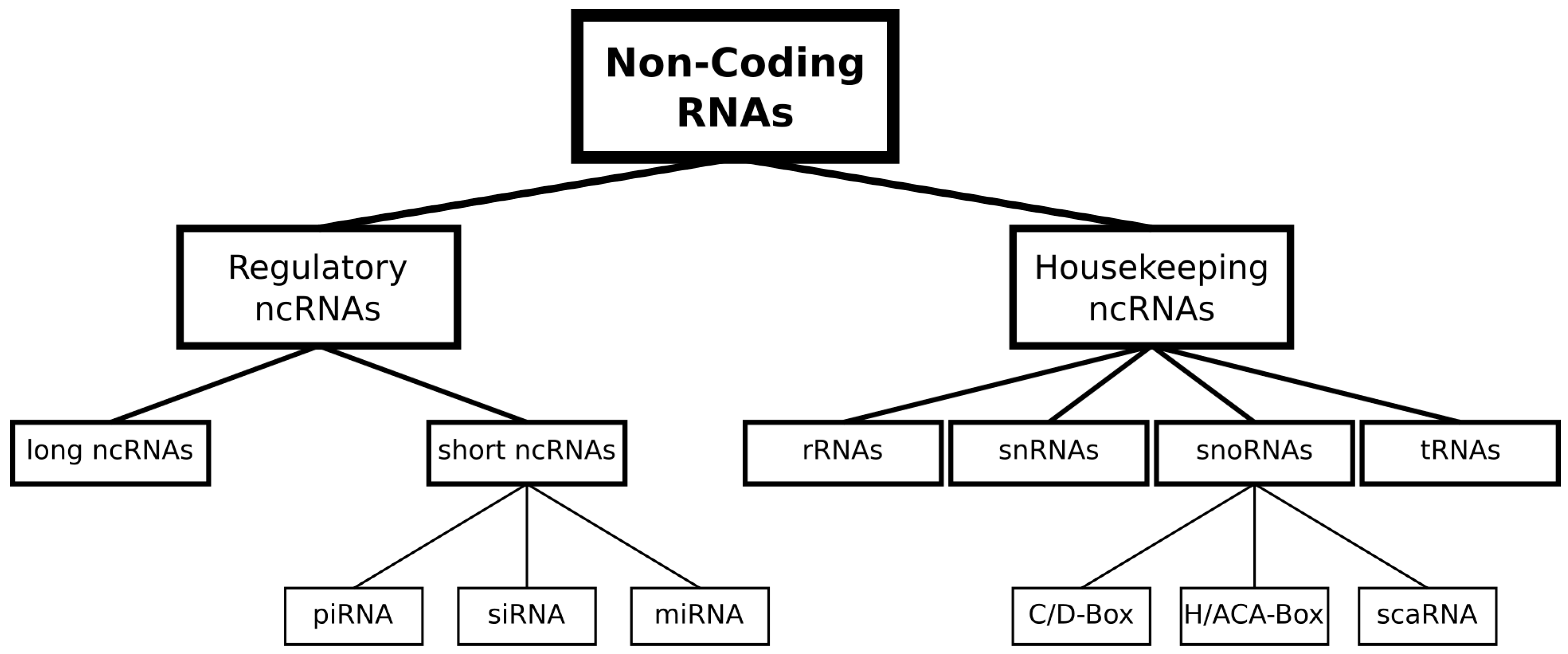
1. Introduction
2. Results
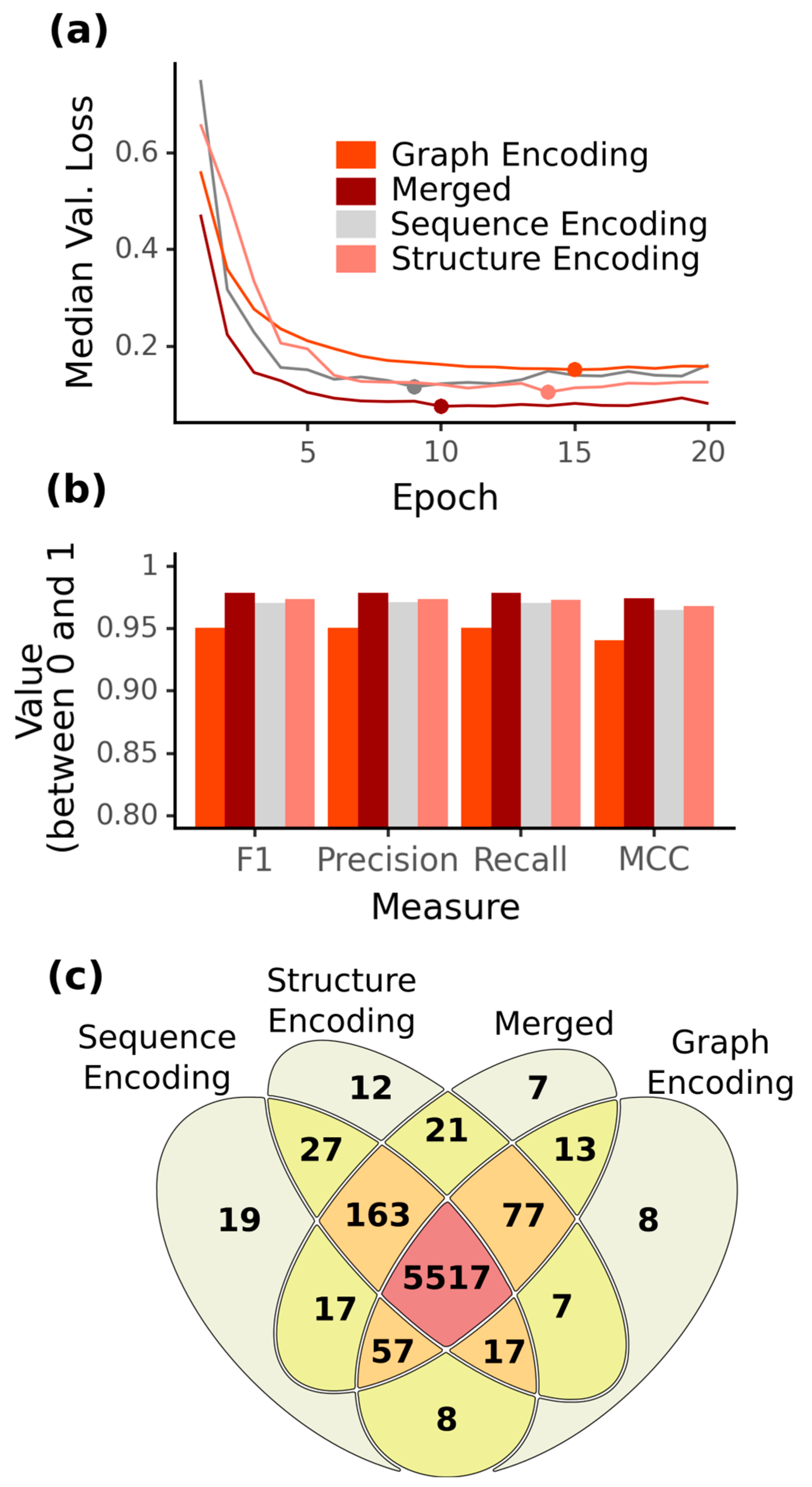
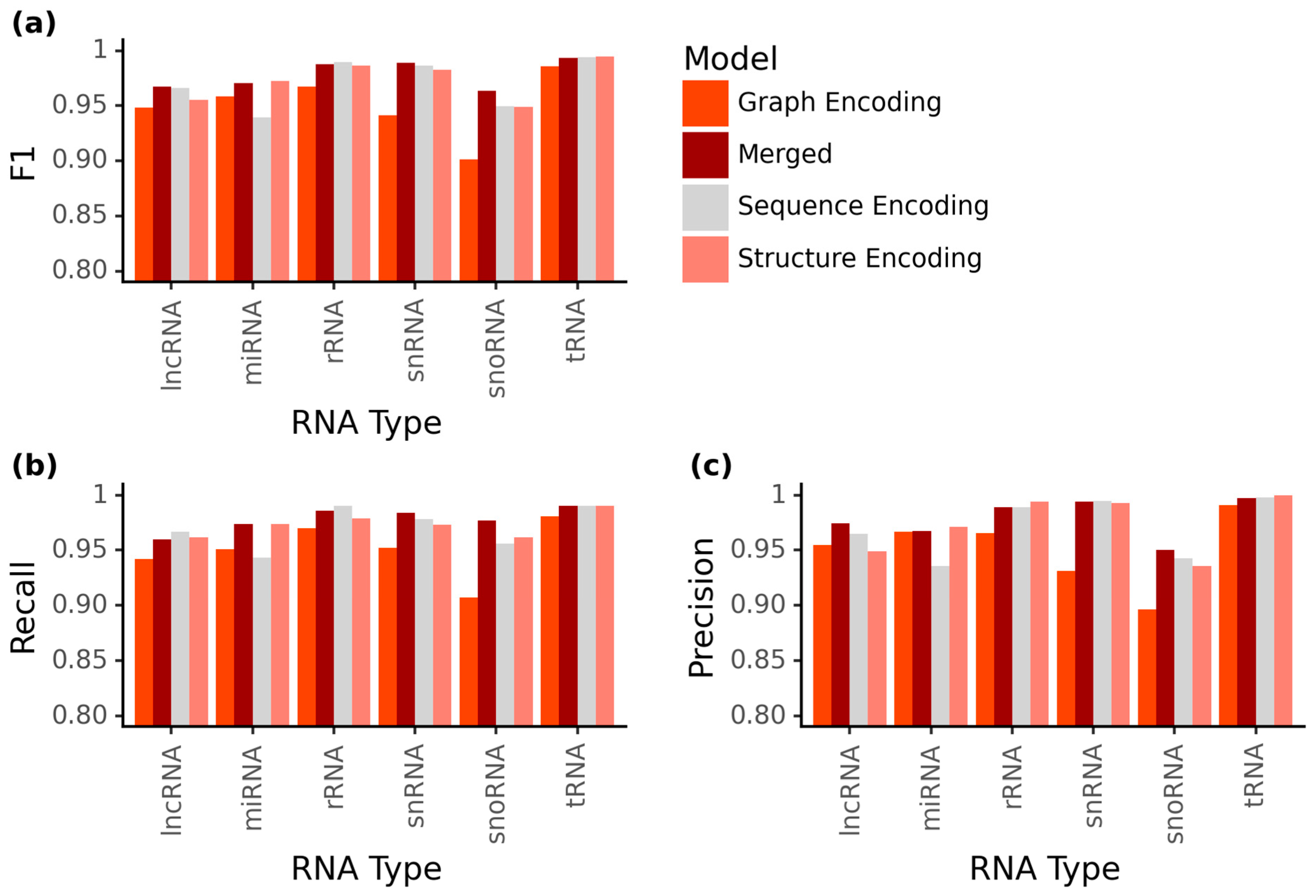
2.1. Increased Accuracy for ncRNA Prediction Combining Graph Vector Features and Primary Sequences
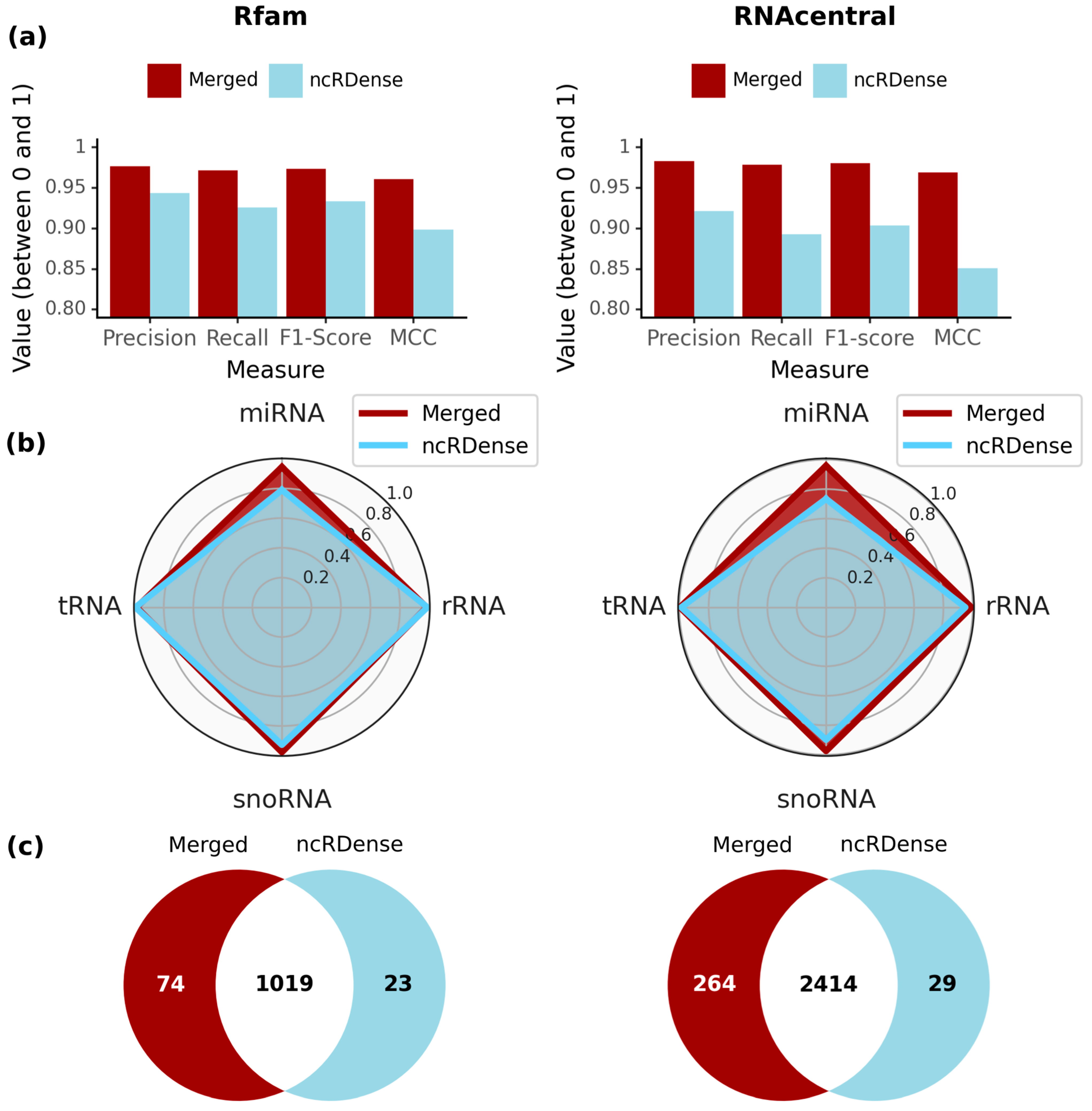
2.2. MncR Model Outperforms Benchmarked ncRNA Classification Tool Based on Two Different Test Sets
3. Discussion
4. Materials and Methods
4.1. Creation of Training, Validation and Test Datasets
4.2. ML Models for ncRNA Classification
4.3. Statistical Analysis
4.4. Benchmarking against Other ML ncRNA Classifiers
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Nucleotide | ||||||
|---|---|---|---|---|---|---|
| Structure | Struct. Encoding | A | C | G | T | Other |
| 5′-end | F | Q | U | D | L | N |
| Stem | S | W | I | F | Y | N |
| Inner Loop | I | E | O | G | X | N |
| Multi Loop | M | R | P | H | C | N |
| Hairpin Loop | H | T | A | J | V | N |
| 3′-end | T | Z | S | K | B | N |
References
- Qu, Z.; Adelson, D.L. Evolutionary conservation and functional roles of ncRNA. Front. Gene. 2012, 3, 205. [Google Scholar] [CrossRef] [PubMed]
- Salviano-Silva, A.; Lobo-Alves, S.; Almeida, R.; Malheiros, D.; Petzl-Erler, M. Besides Pathology: Long Non-Coding RNA in Cell and Tissue Homeostasis. ncRNA 2018, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Washietl, S.; Will, S.; Hendrix, D.A.; Goff, L.A.; Rinn, J.L.; Berger, B.; Kellis, M. Computational analysis of noncoding RNAs: Computational analysis of noncoding RNAs. WIREs RNA 2012, 3, 759–778. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.-D. Non-coding RNA: A new frontier in regulatory biology. Natl. Sci. Rev. 2014, 1, 190–204. [Google Scholar] [CrossRef]
- Wilson, D.N.; Doudna Cate, J.H. The Structure and Function of the Eukaryotic Ribosome. Cold Spring Harb. Perspect. Biol. 2012, 4, a011536. [Google Scholar] [CrossRef]
- Phizicky, E.M.; Hopper, A.K. tRNA biology charges to the front. Genes Dev. 2010, 24, 1832–1860. [Google Scholar] [CrossRef]
- McMahon, M.; Contreras, A.; Ruggero, D. Small RNAs with big implications: New insights into H/ACA snoRNA function and their role in human disease: H/ACA snoRNAs: Small RNAs with big implications. WIREs RNA 2015, 6, 173–189. [Google Scholar] [CrossRef]
- Bhartiya, D.; Scaria, V. Genomic variations in non-coding RNAs: Structure, function and regulation. Genomics 2016, 107, 59–68. [Google Scholar] [CrossRef]
- Machyna, M.; Heyn, P.; Neugebauer, K.M. Cajal bodies: Where form meets function: Cajal bodies. WIREs RNA 2013, 4, 17–34. [Google Scholar] [CrossRef]
- Morais, P.; Adachi, H.; Yu, Y.-T. Spliceosomal snRNA Epitranscriptomics. Front. Genet. 2021, 12, 652129. [Google Scholar] [CrossRef]
- Taft, R.J.; Pang, K.C.; Mercer, T.R.; Dinger, M.; Mattick, J.S. Non-coding RNAs: Regulators of disease: Non-coding RNAs: Regulators of disease. J. Pathol. 2010, 220, 126–139. [Google Scholar] [CrossRef]
- Wang, Z.; Wei, G.-H.; Liu, D.-P.; Liang, C.-C. Unravelling the world of cis-regulatory elements. Med. Bio. Eng. Comput. 2007, 45, 709–718. [Google Scholar] [CrossRef]
- Ong, C.-T.; Corces, V.G. Enhancer function: New insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 2011, 12, 283–293. [Google Scholar] [CrossRef]
- Cech, T.R.; Steitz, J.A. The Noncoding RNA Revolution—Trashing Old Rules to Forge New Ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef]
- Sana, J.; Faltejskova, P.; Svoboda, M.; Slaby, O. Novel classes of non-coding RNAs and cancer. J. Transl. Med. 2012, 10, 103. [Google Scholar] [CrossRef]
- Ayers, D.; Scerri, C. Non-coding RNA influences in dementia. Non-Coding RNA Res. 2018, 3, 188–194. [Google Scholar] [CrossRef]
- Chi, T.; Lin, J.; Wang, M.; Zhao, Y.; Liao, Z.; Wei, P. Non-Coding RNA as Biomarkers for Type 2 Diabetes Development and Clinical Management. Front. Endocrinol. 2021, 12, 630032. [Google Scholar] [CrossRef]
- Shi, Y.; Liu, Z.; Lin, Q.; Luo, Q.; Cen, Y.; Li, J.; Fang, X.; Gong, C. MiRNAs and Cancer: Key Link in Diagnosis and Therapy. Genes 2021, 12, 1289. [Google Scholar] [CrossRef]
- Zhang, X.; Xie, K.; Zhou, H.; Wu, Y.; Li, C.; Liu, Y.; Liu, Z.; Xu, Q.; Liu, S.; Xiao, D.; et al. Role of non-coding RNAs and RNA modifiers in cancer therapy resistance. Mol. Cancer 2020, 19, 47. [Google Scholar] [CrossRef]
- Bryant, R.J.; Pawlowski, T.; Catto, J.W.F.; Marsden, G.; Vessella, R.L.; Rhees, B.; Kuslich, C.; Visakorpi, T.; Hamdy, F.C. Changes in circulating microRNA levels associated with prostate cancer. Br. J. Cancer 2012, 106, 768–774. [Google Scholar] [CrossRef]
- Kumar, M.S.; Erkeland, S.J.; Pester, R.E.; Chen, C.Y.; Ebert, M.S.; Sharp, P.A.; Jacks, T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc. Natl. Acad. Sci. USA 2008, 105, 3903–3908. [Google Scholar] [CrossRef] [PubMed]
- Ishida, M.; Selaru, F.M. miRNA-Based Therapeutic Strategies. Curr. Pathobiol. Rep. 2013, 1, 63–70. [Google Scholar] [CrossRef]
- Melo, S.A.; Kalluri, R. Molecular Pathways: MicroRNAs as Cancer Therapeutics. Clin. Cancer Res. 2012, 18, 4234–4239. [Google Scholar] [CrossRef] [PubMed]
- The Athanasius F. Bompfünewerer Consortium; Backofen, R.; Bernhart, S.H.; Flamm, C.; Fried, C.; Fritzsch, G.; Hackermüller, J.; Hertel, J.; Hofacker, I.L.; Missal, K.; et al. RNAs everywhere: Genome-wide annotation of structured RNAs. J. Exp. Zool. 2007, 308B, 1–25. [Google Scholar] [CrossRef]
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, Biology and Functioning. In Non-Coding RNAs in Colorectal Cancer; Slaby, O., Calin, G.A., Eds.; Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Switzerland, 2016; Volume 937, pp. 3–17. ISBN 978-3-319-42057-8. [Google Scholar]
- Galasso, M.; Elena Sana, M.; Volinia, S. Non-coding RNAs: A key to future personalized molecular therapy? Genome Med. 2010, 2, 12. [Google Scholar] [CrossRef]
- Reis-Filho, J.S. Next-generation sequencing. Breast. Cancer Res. 2009, 11, S12. [Google Scholar] [CrossRef]
- Kang, Y.-J.; Yang, D.-C.; Kong, L.; Hou, M.; Meng, Y.-Q.; Wei, L.; Gao, G. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef]
- Li, M.; Liang, C. LncDC: A machine learning-based tool for long non-coding RNA detection from RNA-Seq data. Sci. Rep. 2022, 12, 19083. [Google Scholar] [CrossRef]
- Anuntakarun, S.; Lertampaiporn, S.; Laomettachit, T.; Wattanapornprom, W.; Ruengjitchatchawalya, M. mSRFR: A machine learning model using microalgal signature features for ncRNA classification. BioData Min. 2022, 15, 8. [Google Scholar] [CrossRef]
- Nithin, C.; Mukherjee, S.; Basak, J.; Bahadur, R.P. NcodR: A multi-class support vector machine classification to distinguish non-coding RNAs in Viridiplantae. Quant. Plant Bio. 2022, 3, e23. [Google Scholar] [CrossRef]
- Childs, L.; Nikoloski, Z.; May, P.; Walther, D. Identification and classification of ncRNA molecules using graph properties. Nucleic Acids Res. 2009, 37, e66. [Google Scholar] [CrossRef]
- Panwar, B.; Arora, A.; Raghava, G.P. Prediction and classification of ncRNAs using structural information. BMC Genom. 2014, 15, 127. [Google Scholar] [CrossRef]
- Fiannaca, A.; La Rosa, M.; La Paglia, L.; Rizzo, R.; Urso, A. nRC: Non-coding RNA Classifier based on structural features. BioData Min. 2017, 10, 27. [Google Scholar] [CrossRef]
- Wang, L.; Zheng, S.; Zhang, H.; Qiu, Z.; Zhong, X.; Liuliu, H.; Liu, Y. ncRFP: A Novel end-to-end Method for Non-Coding RNAs Family Prediction Based on Deep Learning. IEEE/ACM Trans. Comput. Biol. Bioinf. 2021, 18, 784–789. [Google Scholar] [CrossRef]
- Chantsalnyam, T.; Lim, D.Y.; Tayara, H.; Chong, K.T. ncRDeep: Non-coding RNA classification with convolutional neural network. Comput. Biol. Chem. 2020, 88, 107364. [Google Scholar] [CrossRef]
- Chantsalnyam, T.; Siraj, A.; Tayara, H.; Chong, K.T. ncRDense: A novel computational approach for classification of non-coding RNA family by deep learning. Genomics 2021, 113, 3030–3038. [Google Scholar] [CrossRef]
- Jha, M.; Gupta, R.; Saxena, R. Fast and precise prediction of non-coding RNAs (ncRNAs) using sequence alignment and k-mer counting. Int. J. Inf. Tecnol. 2023, 15, 577–585. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Bishop, C.M. Neural networks and their applications. Rev. Sci. Instrum. 1994, 65, 1803–1832. [Google Scholar] [CrossRef]
- Noviello, T.M.R.; Ceccarelli, F.; Ceccarelli, M.; Cerulo, L. Deep learning predicts short non-coding RNA functions from only raw sequence data. PloS Comput. Biol. 2020, 16, e1008415. [Google Scholar] [CrossRef] [PubMed]
- Hofacker, I.L. RNA Secondary Structure Analysis Using the ViennaRNA Package. CP Bioinform. 2003, 4, 12.2.1–12.2.12. [Google Scholar] [CrossRef] [PubMed]
- Borgelt, C.; Meinl, T.; Berthold, M. MoSS: A program for molecular substructure mining. In Proceedings of the 1st International Workshop on Open Source Data Mining: Frequent Pattern Mining Implementations, Chicago IL, USA, 21 August 2005; pp. 6–15. [Google Scholar]
- Kalvari, I.; Argasinska, J.; Quinones-Olvera, N.; Nawrocki, E.P.; Rivas, E.; Eddy, S.R.; Bateman, A.; Finn, R.D.; Petrov, A.I. Rfam 13.0: Shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 2018, 46, D335–D342. [Google Scholar] [CrossRef] [PubMed]
- Adams, L. Pri-miRNA processing: Structure is key. Nat. Rev. Genet. 2017, 18, 145. [Google Scholar] [CrossRef]
- Wu, T.; Du, Y. LncRNAs: From Basic Research to Medical Application. Int. J. Biol. Sci. 2017, 13, 295–307. [Google Scholar] [CrossRef]
- The RNAcentral Consortium; Sweeney, B.A.; Petrov, A.I.; Burkov, B.; Finn, R.D.; Bateman, A.; Szymanski, M.; Karlowski, W.M.; Gorodkin, J.; Seemann, S.E.; et al. RNAcentral: A hub of information for non-coding RNA sequences. Nucleic Acids Res. 2019, 47, D1250–D1251. [Google Scholar] [CrossRef]
- Kalvari, I.; Nawrocki, E.P.; Ontiveros-Palacios, N.; Argasinska, J.; Lamkiewicz, K.; Marz, M.; Griffiths-Jones, S.; Toffano-Nioche, C.; Gautheret, D.; Weinberg, Z.; et al. Rfam 14: Expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 2021, 49, D192–D200. [Google Scholar] [CrossRef]
- Chen, Y.; Qi, F.; Gao, F.; Cao, H.; Xu, D.; Salehi-Ashtiani, K.; Kapranov, P. Hovlinc is a recently evolved class of ribozyme found in human lncRNA. Nat. Chem. Biol. 2021, 17, 601–607. [Google Scholar] [CrossRef]
- Tuschl, T. RNA Interference and Small Interfering RNAs. ChemBioChem 2001, 2, 239–245. [Google Scholar] [CrossRef]
- Calcino, A.D.; Fernandez-Valverde, S.L.; Taft, R.J.; Degnan, B.M. Diverse RNA interference strategies in early-branching metazoans. BMC Evol. Biol. 2018, 18, 160. [Google Scholar] [CrossRef]
- Cunningham, F.; Allen, J.E.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Bennett, R.; et al. Ensembl 2022. Nucleic Acids Res. 2022, 50, D988–D995. [Google Scholar] [CrossRef]
- Harrison, P.W.; Ahamed, A.; Aslam, R.; Alako, B.T.F.; Burgin, J.; Buso, N.; Courtot, M.; Fan, J.; Gupta, D.; Haseeb, M.; et al. The European Nucleotide Archive in 2020. Nucleic Acids Res. 2021, 49, D82–D85. [Google Scholar] [CrossRef]
- Hestness, J.; Narang, S.; Ardalani, N.; Diamos, G.; Jun, H.; Kianinejad, H.; Patwary, M.M.A.; Yang, Y.; Zhou, Y. Deep Learning Scaling is Predictable, Empirically. arXiv 2017, arXiv:1712.00409. [Google Scholar] [CrossRef]
- Wang, S.; Liu, W.; Wu, J.; Cao, L.; Meng, Q.; Kennedy, P.J. Training deep neural networks on imbalanced data sets. In Proceedings of the 2016 International Joint Conference on Neural Networks (IJCNN), Vancouver, BC, Canada, 24–29 July 2016; pp. 4368–4374. [Google Scholar]
- Maticzka, D.; Lange, S.J.; Costa, F.; Backofen, R. GraphProt: Modeling binding preferences of RNA-binding proteins. Genome Biol. 2014, 15, R17. [Google Scholar] [CrossRef]
- Budach, S.; Marsico, A. pysster: Classification of biological sequences by learning sequence and structure motifs with convolutional neural networks. Bioinformatics 2018, 34, 3035–3037. [Google Scholar] [CrossRef]
- Picard, M.; Scott-Boyer, M.-P.; Bodein, A.; Périn, O.; Droit, A. Integration strategies of multi-omics data for machine learning analysis. Comput. Struct. Biotechnol. J. 2021, 19, 3735–3746. [Google Scholar] [CrossRef]
- Bachellerie, J.-P.; Cavaillé, J.; Hüttenhofer, A. The expanding snoRNA world. Biochimie 2002, 84, 775–790. [Google Scholar] [CrossRef]
- Steffen, P.; Voß, B.; Rehmsmeier, M.; Reeder, J.; Giegerich, R. RNAshapes: An integrated RNA analysis package based on abstract shapes. Bioinformatics 2006, 22, 500–503. [Google Scholar] [CrossRef] [PubMed]
- Seetin, M.G.; Mathews, D.H. RNA Structure Prediction: An Overview of Methods. In Bacterial Regulatory RNA; Keiler, K.C., Ed.; Humana Press: Totowa, NJ, USA, 2012; pp. 99–122. ISBN 978-1-61779-948-8. [Google Scholar]
- Ameres, S.L.; Zamore, P.D. Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell. Biol. 2013, 14, 475–488. [Google Scholar] [CrossRef]
- Fu, X.; Zhu, W.; Cai, L.; Liao, B.; Peng, L.; Chen, Y.; Yang, J. Improved Pre-miRNAs Identification Through Mutual Information of Pre-miRNA Sequences and Structures. Front. Genet. 2019, 10, 119. [Google Scholar] [CrossRef]
- Wilusz, J.E.; Freier, S.M.; Spector, D.L. 3′ End Processing of a Long Nuclear-Retained Noncoding RNA Yields a tRNA-like Cytoplasmic RNA. Cell 2008, 135, 919–932. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Cullen, B.R. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 2007, 13, 313–316. [Google Scholar] [CrossRef] [PubMed]
- Marz, M.; Stadler, P.F. Comparative analysis of eukaryotic U3 snoRNA. RNA Biol. 2009, 6, 503–507. [Google Scholar] [CrossRef] [PubMed]
- Anderson-Lee, J.; Fisker, E.; Kosaraju, V.; Wu, M.; Kong, J.; Lee, J.; Lee, M.; Zada, M.; Treuille, A.; Das, R. Principles for Predicting RNA Secondary Structure Design Difficulty. J. Mol. Biol. 2016, 428, 748–757. [Google Scholar] [CrossRef]
- Johnson, J.M.; Khoshgoftaar, T.M. Survey on deep learning with class imbalance. J. Big Data 2019, 6, 27. [Google Scholar] [CrossRef]
- Brant, E.J.; Budak, H. Plant Small Non-coding RNAs and Their Roles in Biotic Stresses. Front. Plant Sci. 2018, 9, 1038. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Navarin, N.; Costa, F. An efficient graph kernel method for non-coding RNA functional prediction. Bioinformatics 2017, 33, 2642–2650. [Google Scholar] [CrossRef]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neubock, R.; Hofacker, I.L. The Vienna RNA Websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef]
- TensorFlow Developers (2021). TensorFlow, v2.4.3; Google Brain Team: Mountain View, CA, USA, 2022. [CrossRef]
- Chollet, F. Keras. GitHub. 2015. Available online: https://github.com/fchollet/keras (accessed on 1 February 2022).
- Akiyama, M.; Sakakibara, Y. Informative RNA base embedding for RNA structural alignment and clustering by deep representation learning. NAR Genom. Bioinform. 2022, 4, lqac012. [Google Scholar] [CrossRef]
- Yi, D.; Ji, S.; Bu, S. An Enhanced Optimization Scheme Based on Gradient Descent Methods for Machine Learning. Symmetry 2019, 11, 942. [Google Scholar] [CrossRef]
- Matthews, B.W. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Et. Biophys. Acta (BBA)-Protein Struct. 1975, 405, 442–451. [Google Scholar] [CrossRef]
- Gold, V. (Ed.) The IUPAC Compendium of Chemical Terminology: The Gold Book, 4th ed.; International Union of Pure and Applied Chemistry (IUPAC): Research Triangle Park, NC, USA, 2019. [Google Scholar]

| ncRNA Class | Min. One Model Wrong | MncR Wrong | All Models Wrong | Only MncR Wrong |
|---|---|---|---|---|
| lncRNA | 87 | 40 | 12 | 6 |
| miRNA | 107 | 26 | 4 | 3 |
| rRNA | 50 | 14 | 1 | 2 |
| snRNA | 76 | 16 | 2 | 2 |
| snoRNA | 136 | 23 | 6 | 3 |
| tRNA | 28 | 10 | 4 | 1 |
| Sum | 484 | 129 | 29 | 17 |
| ncRNA Class | Subtype | Count |
|---|---|---|
| lncRNA | unspecified | 10,000 |
| miRNA | Mature miRNA | 5000 |
| Precursor miRNA | 5000 | |
| rRNA | 23S | 1435 |
| 25S | 1409 * | |
| 28S | 1435 | |
| 5.8S | 1420 * | |
| 5S | 1435 | |
| Small Subunit (SSU) | 1435 | |
| Mitochondrial (mt) | 1435 | |
| snRNA | unspecified | 10,000 |
| snoRNA | C/D-Box | 4044 |
| H/ACA-Box | 4044 | |
| scaRNA | 1913 * | |
| tRNA | unspecified | 10,000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dunkel, H.; Wehrmann, H.; Jensen, L.R.; Kuss, A.W.; Simm, S. MncR: Late Integration Machine Learning Model for Classification of ncRNA Classes Using Sequence and Structural Encoding. Int. J. Mol. Sci. 2023, 24, 8884. https://doi.org/10.3390/ijms24108884
Dunkel H, Wehrmann H, Jensen LR, Kuss AW, Simm S. MncR: Late Integration Machine Learning Model for Classification of ncRNA Classes Using Sequence and Structural Encoding. International Journal of Molecular Sciences. 2023; 24(10):8884. https://doi.org/10.3390/ijms24108884
Chicago/Turabian StyleDunkel, Heiko, Henning Wehrmann, Lars R. Jensen, Andreas W. Kuss, and Stefan Simm. 2023. "MncR: Late Integration Machine Learning Model for Classification of ncRNA Classes Using Sequence and Structural Encoding" International Journal of Molecular Sciences 24, no. 10: 8884. https://doi.org/10.3390/ijms24108884
APA StyleDunkel, H., Wehrmann, H., Jensen, L. R., Kuss, A. W., & Simm, S. (2023). MncR: Late Integration Machine Learning Model for Classification of ncRNA Classes Using Sequence and Structural Encoding. International Journal of Molecular Sciences, 24(10), 8884. https://doi.org/10.3390/ijms24108884






