Optimized De Novo Eriodictyol Biosynthesis in Streptomyces albidoflavus Using an Expansion of the Golden Standard Toolkit for Its Use in Actinomycetes
Abstract
1. Introduction
2. Results
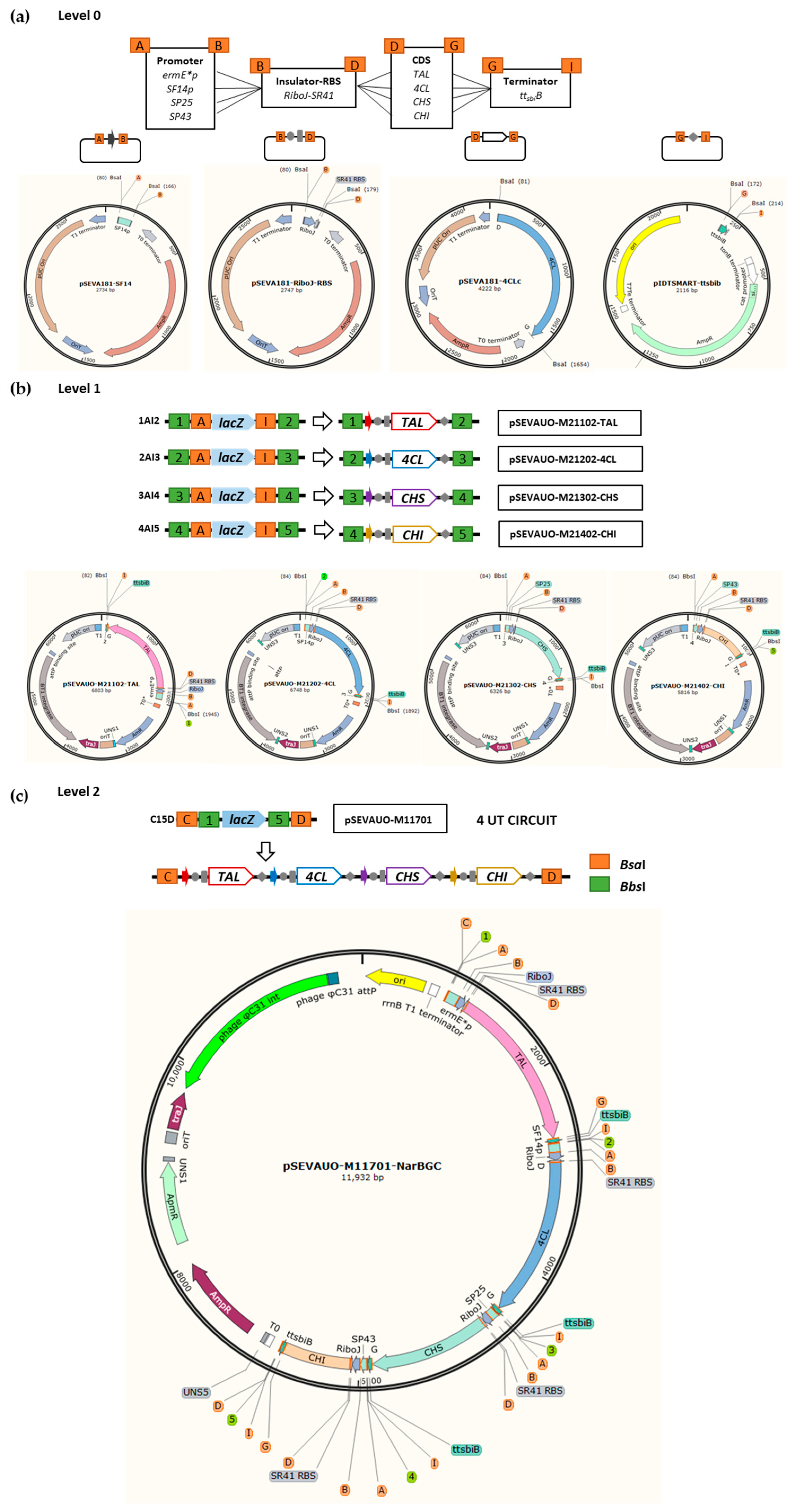
2.1. SEVA-Based Plasmid Library Design and Construction
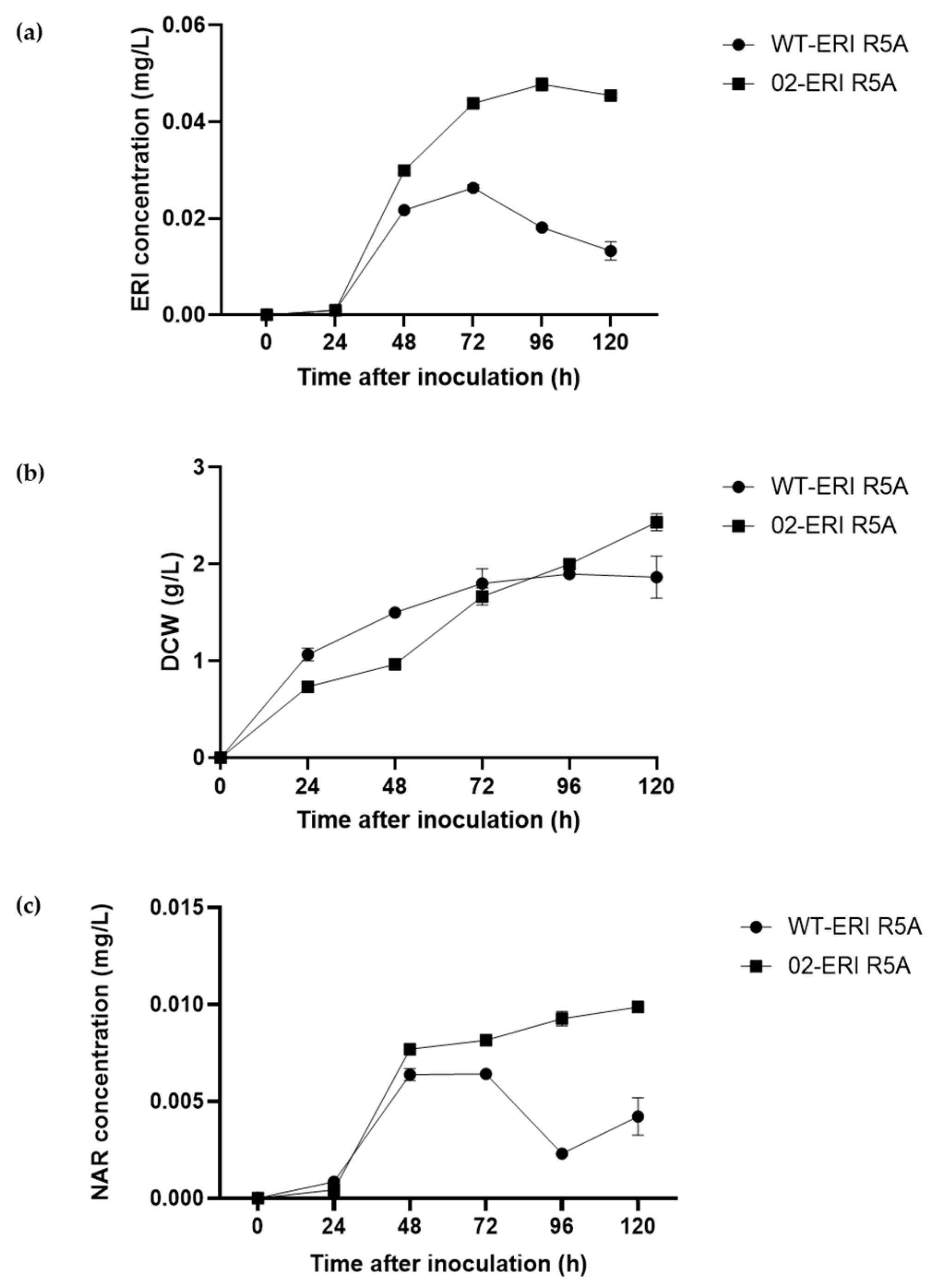
2.2. Eriodictyol Heterologous Biosynthesis in S. albidoflavus WT
2.3. Comparison of Conjugation and Genome Editing Efficiency between Wild-Type Cas9 and Cas9 Nickase
2.4. Generation of the S. albidoflavus UO-FLAV-002 Edited Bacterial Factory
2.5. Eriodictyol Heterologous Biosynthesis in S. albidoflavus UO-FLAV-002
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmids and Culture Conditions
4.2. Reagents and Biochemicals
4.3. Synthetic DNA and Enzymes
4.4. Construction of Plasmids
4.5. Strain Generation
4.6. Flavonoid Extraction and HPLC–HRESIMS Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaushal, N.; Singh, M.; Singh Sangwan, R. Flavonoids: Food Associations, Therapeutic Mechanisms, Metabolism and Nanoformulations. Food Res. Int. 2022, 157, 111442. [Google Scholar] [CrossRef]
- Roy, A.; Khan, A.; Ahmad, I.; Alghamdi, S.; Rajab, B.S.; Babalghith, A.O.; Alshahrani, M.Y.; Islam, S.; Islam, M.R. Flavonoids a Bioactive Compound from Medicinal Plants and Its Therapeutic Applications. BioMed Res. Int. 2022, 2022, 5445291. [Google Scholar] [CrossRef]
- Al-Khayri, J.M.; Sahana, G.R.; Nagella, P.; Joseph, B.V.; Alessa, F.M.; Al-Mssallem, M.Q. Flavonoids as Potential Anti-Inflammatory Molecules: A Review. Molecules 2022, 27, 2901. [Google Scholar] [CrossRef]
- Buranasudja, V.; Muangnoi, C.; Sanookpan, K.; Halim, H.; Sritularak, B.; Rojsitthisak, P. Eriodictyol Attenuates H2O2-Induced Oxidative Damage in Human Dermal Fibroblasts through Enhanced Capacity of Antioxidant Machinery. Nutrients 2022, 14, 2553. [Google Scholar] [CrossRef]
- Debnath, S.; Sarkar, A.; Das Mukherjee, D.; Ray, S.; Mahata, B.; Mahata, T.; Parida, P.K.; Das, T.; Mukhopadhyay, R.; Ghosh, Z.; et al. Eriodictyol Mediated Selective Targeting of the TNFR1/FADD/TRADD Axis in Cancer Cells Induce Apoptosis and Inhibit Tumor Progression and Metastasis. Transl. Oncol. 2022, 21, 101433. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Fu, H.; Li, C.; Deng, Z.; Chang, H. Eriodictyol Inhibits Breast Carcinogenesis by Targeting Circ_0007503 and Repressing PI3K/Akt Pathway. Phytomedicine 2022, 102, 154159. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Sim, W.; Han, S.-I.; Byeon, J.-H.; Jin, S.-B.; Morshidi, N.A.A.B.; Hong, Y.-Y.; Jung, Y.; Kim, J.H. Eriodictyol Attenuates Cholangiocarcinoma Malignancy by Regulating HMOX1 Expression: An In Vitro Study. Anticancer Res. 2022, 42, 3789–3798. [Google Scholar] [CrossRef]
- Zhao, L.; Qin, X.; Lin, T.; Xie, F.; Yao, L.; Li, Y.; Xiong, B.; Xu, Z.; Ye, Y.; Chen, H.; et al. Multi-Target Mechanisms against Coronaviruses of Constituents from Chinese Dagang Tea Revealed by Experimental and Docking Studies. J. Ethnopharmacol. 2022, 297, 115528. [Google Scholar] [CrossRef]
- Guo, P.; Zeng, M.; Wang, S.; Cao, B.; Liu, M.; Zhang, Y.; Jia, J.; Zhang, Q.; Zhang, B.; Wang, R.; et al. Eriodictyol and Homoeriodictyol Improve Memory Impairment in Aβ25-35-Induced Mice by Inhibiting the NLRP3 Inflammasome. Molecules 2022, 27, 2488. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, W.-J.; Zheng, X.-R.; Liu, Q.-L.; Du, Q.; Lai, Y.-J.; Liu, S.-Q. Eriodictyol Ameliorates Cognitive Dysfunction in APP/PS1 Mice by Inhibiting Ferroptosis via Vitamin D Receptor-Mediated Nrf2 Activation. Mol. Med. 2022, 28, 11. [Google Scholar] [CrossRef]
- Maquera-Huacho, P.M.; Spolidorio, D.P.; Manthey, J.A.; Grenier, D. Eriodictyol Suppresses Porphyromonas Gingivalis-Induced Reactive Oxygen Species Production by Gingival Keratinocytes and the Inflammatory Response of Macrophages. Front. Oral Health 2022, 3, 847914. [Google Scholar] [CrossRef]
- Xie, Y.; Ji, R.; Han, M. Eriodictyol Protects H9c2 Cardiomyocytes against the Injury Induced by Hypoxia/Reoxygenation by Improving the Dysfunction of Mitochondria. Exp. Ther. Med. 2019, 17, 551–557. [Google Scholar] [CrossRef]
- Hu, Q.; Qu, C.; Xiao, X.; Zhang, W.; Jiang, Y.; Wu, Z.; Song, D.; Peng, X.; Ma, X.; Zhao, Y. Flavonoids on Diabetic Nephropathy: Advances and Therapeutic Opportunities. Chin. Med. 2021, 16, 74. [Google Scholar] [CrossRef]
- Kwon, E.-Y.; Choi, M.-S. Dietary Eriodictyol Alleviates Adiposity, Hepatic Steatosis, Insulin Resistance, and Inflammation in Diet-Induced Obese Mice. Int. J. Mol. Sci. 2019, 20, 1227. [Google Scholar] [CrossRef]
- Yang, T.; Li, X.; Yu, J.; Deng, X.; Shen, P.-X.; Jiang, Y.-B.; Zhu, L.; Wang, Z.-Z.; Zhang, Y. Eriodictyol Suppresses Th17 Differentiation and the Pathogenesis of Experimental Autoimmune Encephalomyelitis. Food Funct. 2020, 11, 6875–6888. [Google Scholar] [CrossRef]
- Nisar, M.F.; Liu, T.; Wang, M.; Chen, S.; Chang, L.; Karisma, V.W.; Weixu; Diao, Q.; Xue, M.; Tang, X.; et al. Eriodictyol Protects Skin Cells from UVA Irradiation-Induced Photodamage by Inhibition of the MAPK Signaling Pathway. J. Photochem. Photobiol. B Biol. 2022, 226, 112350. [Google Scholar] [CrossRef]
- Liszt, K.I.; Hans, J.; Ley, J.P.; Köck, E.; Somoza, V. Characterization of Bitter Compounds via Modulation of Proton Secretion in Human Gastric Parietal Cells in Culture. J. Agric. Food Chem. 2018, 66, 2295–2300. [Google Scholar] [CrossRef] [PubMed]
- Beltrán, L.R.; Sterneder, S.; Hasural, A.; Paetz, S.; Hans, J.; Ley, J.P.; Somoza, V. Reducing the Bitter Taste of Pharmaceuticals Using Cell-Based Identification of Bitter-Masking Compounds. Pharmaceuticals 2022, 15, 317. [Google Scholar] [CrossRef] [PubMed]
- Islam, A.; Islam, M.S.; Rahman, M.K.; Uddin, M.N.; Akanda, M.R. The Pharmacological and Biological Roles of Eriodictyol. Arch. Pharmacal Res. 2020, 43, 582–592. [Google Scholar] [CrossRef]
- Deng, Z.; Hassan, S.; Rafiq, M.; Li, H.; He, Y.; Cai, Y.; Kang, X.; Liu, Z.; Yan, T. Pharmacological Activity of Eriodictyol: The Major Natural Polyphenolic Flavanone. Evid.-Based Complement. Altern. Med. 2020, 2020, 6681352. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Wu, J.; Du, G.; Zhou, J.; Chen, J. Efficient Synthesis of Eriodictyol from L-Tyrosine in Escherichia Coli. Appl. Environ. Microbiol. 2014, 80, 3072–3080. [Google Scholar] [CrossRef] [PubMed]
- Thuan, N.H.; Tatipamula, V.B.; Viet, T.T.; Tien, N.Q.D.; Loc, N.H. Bioproduction of Eriodictyol by Escherichia Coli Engineered Co-Culture. World J. Microbiol. Biotechnol. 2022, 38, 112. [Google Scholar] [CrossRef]
- Yan, Y.; Kohli, A.; Koffas, M.A.G. Biosynthesis of Natural Flavanones in Saccharomyces Cerevisiae. Appl. Environ. Microbiol. 2005, 71, 5610–5613. [Google Scholar] [CrossRef]
- Gao, S.; Xu, X.; Zeng, W.; Xu, S.; Lyv, Y.; Feng, Y.; Kai, G.; Zhou, J.; Chen, J. Efficient Biosynthesis of (2S)-Eriodictyol from (2S)-Naringenin in Saccharomyces cerevisiae through a Combination of Promoter Adjustment and Directed Evolution. ACS Synth. Biol. 2020, 9, 3288–3297. [Google Scholar] [CrossRef]
- Wu, X.; Liu, J.; Liu, D.; Yuwen, M.; Koffas, M.A.G.; Zha, J. Biosynthesis of Eriodictyol from Tyrosine by Corynebacterium Glutamicum. Microb. Cell Factories 2022, 21, 86. [Google Scholar] [CrossRef]
- Marín, L.; Gutiérrez-Del-Río, I.; Yagüe, P.; Manteca, Á.; Villar, C.J.; Lombó, F. De Novo Biosynthesis of Apigenin, Luteolin, and Eriodictyol in the Actinomycete Streptomyces albus and Production Improvement by Feeding and Spore Conditioning. Front. Microbiol. 2017, 8, 921. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Marsafari, M.; Koffas, M.; Zhou, J.; Xu, P. Optimizing Oleaginous Yeast Cell Factories for Flavonoids and Hydroxylated Flavonoids Biosynthesis. ACS Synth. Biol. 2019, 8, 2514–2523. [Google Scholar] [CrossRef]
- Fowler, Z.L.; Gikandi, W.W.; Koffas, M.A.G. Increased Malonyl Coenzyme A Biosynthesis by Tuning the Escherichia Coli Metabolic Network and Its Application to Flavanone Production. Appl. Environ. Microbiol. 2009, 75, 5831–5839. [Google Scholar] [CrossRef] [PubMed]
- Leonard, E.; Yan, Y.; Fowler, Z.L.; Li, Z.; Lim, C.-G.; Lim, K.-H.; Koffas, M.A.G. Strain Improvement of Recombinant Escherichia Coli for Efficient Production of Plant Flavonoids. Mol. Pharm. 2008, 5, 257–265. [Google Scholar] [CrossRef]
- Lombó, F.; Pfeifer, B.; Leaf, T.; Ou, S.; Kim, Y.S.; Cane, D.E.; Licari, P.; Khosla, C. Enhancing the atom economy of polyketide biosynthetic processes through metabolic engineering. Biotechnol. Prog. 2001, 17, 612–617. [Google Scholar] [CrossRef]
- Álvarez-Álvarez, R.; Botas, A.; Albillos, S.M.; Rumbero, A.; Martín, J.F.; Liras, P. Molecular Genetics of Naringenin Biosynthesis, a Typical Plant Secondary Metabolite Produced by Streptomyces clavuligerus. Microb. Cell Factories 2015, 14, 178. [Google Scholar] [CrossRef]
- Martín, J.F.; Liras, P. Comparative Molecular Mechanisms of Biosynthesis of Naringenin and Related Chalcones in Actinobacteria and Plants: Relevance for the Obtention of Potent Bioactive Metabolites. Antibiotics 2022, 11, 82. [Google Scholar] [CrossRef]
- Marín, L.; Gutiérrez-del-Río, I.; Entrialgo-Cadierno, R.; Claudio; Villar, J.; Lombó, F. De Novo Biosynthesis of Myricetin, Kaempferol and Quercetin in Streptomyces Albus and Streptomyces coelicolor. PLoS ONE 2018, 13, e0207278. [Google Scholar] [CrossRef]
- Wang, W.; Li, S.; Li, Z.; Zhang, J.; Fan, K.; Tan, G.; Ai, G.; Lam, S.M.; Shui, G.; Yang, Z.; et al. Harnessing the Intracellular Triacylglycerols for Titer Improvement of Polyketides in Streptomyces. Nat. Biotechnol. 2020, 38, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Ryu, Y.-G.; Butler, M.J.; Chater, K.F.; Lee, K.J. Engineering of Primary Carbohydrate Metabolism for Increased Production of Actinorhodin in Streptomyces coelicolor. Appl. Environ. Microbiol. 2006, 72, 7132–7139. [Google Scholar] [CrossRef]
- Gomez-Escribano, J.P.; Bibb, M.J. Engineering Streptomyces coelicolor for Heterologous Expression of Secondary Metabolite Gene Clusters. Microb. Biotechnol. 2011, 4, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Myronovskyi, M.; Rosenkränzer, B.; Nadmid, S.; Pujic, P.; Normand, P.; Luzhetskyy, A. Generation of a Cluster-Free Streptomyces Albus Chassis Strains for Improved Heterologous Expression of Secondary Metabolite Clusters. Metab. Eng. 2018, 49, 316–324. [Google Scholar] [CrossRef]
- Blázquez, B.; Torres-Bacete, J.; Leon, D.S.; Kniewel, R.; Martinez, I.; Sordon, S.; Wilczak, A.; Salgado, S.; Huszcza, E.; Popłoński, J.; et al. Golden Standard: A Complete Standard, Portable, and Interoperative MoClo Tool for Model and Non-Model Bacterial Hosts. bioRxiv 2022. [CrossRef]
- García-Gutiérrez, C.; Aparicio, T.; Torres-Sánchez, L.; Martínez-García, E.; de Lorenzo, V.; Villar, C.J.; Lombó, F. Multifunctional SEVA Shuttle Vectors for Actinomycetes and Gram-Negative Bacteria. Microbiologyopen 2020, 9, 1135–1149. [Google Scholar] [CrossRef] [PubMed]
- Torella, J.P.; Lienert, F.; Boehm, C.R.; Chen, J.-H.; Way, J.C.; Silver, P.A. Unique Nucleotide Sequence-Guided Assembly of Repetitive DNA Parts for Synthetic Biology Applications. Nat. Protoc. 2014, 9, 2075–2089. [Google Scholar] [CrossRef] [PubMed]
- Alberti, F.; Corre, C. Editing Streptomycete Genomes in the CRISPR/Cas9 Age. Nat. Prod. Rep. 2019, 36, 1237–1248. [Google Scholar] [CrossRef]
- Weber, E.; Engler, C.; Gruetzner, R.; Werner, S.; Marillonnet, S. A Modular Cloning System for Standardized Assembly of Multigene Constructs. PLoS ONE 2011, 6, e16765. [Google Scholar] [CrossRef] [PubMed]
- Maas, R.-M.; GUtz, J.; Wohlleben, W.; Muth, G. The Conjugative Plasmid PSG5 from Streptomyces Ghanaensis DSM 2932 Differs in Its Transfer Functions from Other Streptomyces Rolling-Circle-Type Plasmids. Microbiology 1998, 144, 2809–2817. [Google Scholar] [CrossRef][Green Version]
- Cobb, R.E.; Wang, Y.; Zhao, H. High-Efficiency Multiplex Genome Editing of Streptomyces Species Using an Engineered CRISPR/Cas System. ACS Synth. Biol. 2015, 4, 723–728. [Google Scholar] [CrossRef]
- Labes, G.; Bibb, M.; Wohlleben, W. Isolation and Characterization of a Strong Promoter Element from the Streptomyces Ghanaensis Phage 119 Using the Gentamicin Resistance Gene (AacC1) of Tn1696 as Reporter. Microbiology 1997, 143, 1503–1512. [Google Scholar] [CrossRef] [PubMed]
- Bibb, M.J.; White, J.; Ward, J.M.; Janssen, G.R. The MRNA for the 23S RRNA Methylase Encoded by the ErmE Gene of Saccharopolyspora Erythraea Is Translated in the Absence of a Conventional Ribosome-binding Site. Mol. Microbiol. 1994, 14, 533–545. [Google Scholar] [CrossRef]
- Bai, C.; Zhang, Y.; Zhao, X.; Hu, Y.; Xiang, S.; Miao, J.; Lou, C.; Zhang, L. Exploiting a Precise Design of Universal Synthetic Modular Regulatory Elements to Unlock the Microbial Natural Products in Streptomyces. Proc. Natl. Acad. Sci. USA 2015, 112, 12181–12186. [Google Scholar] [CrossRef] [PubMed]
- Martínez-García, E.; Goñi-Moreno, A.; Bartley, B.; McLaughlin, J.; Sánchez-Sampedro, L.; Pascual del Pozo, H.; Prieto Hernández, C.; Marletta, A.S.; De Lucrezia, D.; Sánchez-Fernández, G.; et al. SEVA 3.0: An Update of the Standard European Vector Architecture for Enabling Portability of Genetic Constructs among Diverse Bacterial Hosts. Nucleic Acids Res. 2019, 48, D1164–D1170. [Google Scholar] [CrossRef]
- Thorpe, H.M.; Smith, M.C.M. In Vitro Site-Specific Integration of Bacteriophage DNA Catalyzed by a Recombinase of the-Resolvase/Invertase Family. Proc. Natl. Acad. Sci. USA 1998, 95, 5505–5510. [Google Scholar] [CrossRef] [PubMed]
- Gregory, M.A.; Till, R.; Smith, M.C.M. Integration Site for Streptomyces Phage ΦBT1 and Development of Site-Specific Integrating Vectors. J. Bacteriol. 2003, 185, 5320–5323. [Google Scholar] [CrossRef] [PubMed]
- Boccard, F.; Smokvina, T.; Pernodet, J.L.; Friedmann, A.; Guérineau, M. The Integrated Conjugative Plasmid PSAM2 of Streptomyces Ambofaciens Is Related to Temperate Bacteriophages. EMBO J. 1989, 8, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Li, Y.; Shi, Z.; Hemme, C.L.; Li, Y.; Zhu, Y.; Van Nostrand, J.D.; He, Z.; Zhou, J. Efficient Genome Editing in Clostridium Cellulolyticum via CRISPR-Cas9 Nickase. Appl. Environ. Microbiol. 2015, 81, 4423–4431. [Google Scholar] [CrossRef] [PubMed]
- Olano, C.; García, I.; González, A.; Rodriguez, M.; Rozas, D.; Rubio, J.; Sánchez-Hidalgo, M.; Braña, A.F.; Méndez, C.; Salas, J.A. Activation and Identification of Five Clusters for Secondary Metabolites in Streptomyces Albus J1074. Microb. Biotechnol. 2014, 7, 242–256. [Google Scholar] [CrossRef]
- Zaburannyi, N.; Rabyk, M.; Ostash, B.; Fedorenko, V.; Luzhetskyy, A. Insights into Naturally Minimised Streptomyces Albidoflavus J1074 Genome. BMC Genom. 2014, 15, 97. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.T.; Medema, M.H.; Takano, E.; Breitling, R. Comparative Genome-Scale Metabolic Modeling of Actinomycetes: The Topology of Essential Core Metabolism. FEBS Lett. 2011, 585, 2389–2394. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Medema, M.H.; Kazempour, D.; Fischbach, M.A.; Breitling, R.; Takano, E.; Weber, T. AntiSMASH 2.—A Versatile Platform for Genome Mining of Secondary Metabolite Producers. Nucleic Acids Res. 2013, 41, W204–W212. [Google Scholar] [CrossRef]
- Gil, J.A.; Campelo-Diez, A.B. Candicidin Biosynthesis in Streptomyces Griseus. Appl. Microbiol. Biotechnol. 2003, 60, 633–642. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, X.; Kim, S.J.; Zhang, W. Antimycin-Type Depsipeptides: Discovery, Biosynthesis, Chemical Synthesis, and Bioactivities. Nat. Prod. Rep. 2016, 33, 1146–1165. [Google Scholar] [CrossRef]
- Wentzel, A.; Bruheim, P.; Øverby, A.; Jakobsen, Ø.M.; Sletta, H.; Omara, W.A.M.; Hodgson, D.A.; Ellingsen, T.E. Optimized Submerged Batch Fermentation Strategy for Systems Scale Studies of Metabolic Switching in Streptomyces Coelicolor A3(2). BMC Syst. Biol. 2012, 6, 59. [Google Scholar] [CrossRef]
- Kurth, D.G.; Gago, G.M.; de la Iglesia, A.; Bazet Lyonnet, B.; Lin, T.-W.; Morbidoni, H.R.; Tsai, S.-C.; Gramajo, H. ACCase 6 Is the Essential Acetyl-CoA Carboxylase Involved in Fatty Acid and Mycolic Acid Biosynthesis in Mycobacteria. Microbiology 2009, 155, 2664–2675. [Google Scholar] [CrossRef]
- Gago, G.; Diacovich, L.; Arabolaza, A.; Tsai, S.-C.; Gramajo, H. Fatty Acid Biosynthesis in Actinomycetes. FEMS Microbiol. Rev. 2011, 35, 475–497. [Google Scholar] [CrossRef]
- Combes, P.; Till, R.; Bee, S.; Smith, M.C.M. The Streptomyces Genome Contains Multiple Pseudo-AttB Sites for the ΦC31-Encoded Site-Specific Recombination System. J. Bacteriol. 2002, 184, 5746–5752. [Google Scholar] [CrossRef] [PubMed]
- Bilyk, B.; Luzhetskyy, A. Unusual Site-Specific DNA Integration into the Highly Active Pseudo-AttB of the Streptomyces Albus J1074 Genome. Appl. Microbiol. Biotechnol. 2014, 98, 5095–5104. [Google Scholar] [CrossRef] [PubMed]
- Bilyk, B.; Horbal, L.; Luzhetskyy, A. Chromosomal Position Effect Influences the Heterologous Expression of Genes and Biosynthetic Gene Clusters in Streptomyces Albus J1074. Microb. Cell Factories 2017, 16, 5. [Google Scholar] [CrossRef] [PubMed]
- Kieser, T.; Chater, K.; Bibb, M.; Buttner, M.; Hopwood, D. Practical Streptomyces Genetics; John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- Fernández, E.; Weissbach, U.; Reillo, C.S.; Braña, A.F.; Méndez, C.; Rohr, J.; Salas, J.A. Identification of Two Genes from Streptomyces Argillaceus Encoding Glycosyltransferases Involved in Transfer of a Disaccharide during Biosynthesis of the Antitumor Drug Mithramycin. J. Bacteriol. 1998, 180, 4929–4937. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; ISBN 0879695773. [Google Scholar]
- Martínez-García, E.; Fraile, S.; Algar, E.; Aparicio, T.; Velázquez, E.; Calles, B.; Tas, H.; Blázquez, B.; Martín, B.; Prieto, C.; et al. SEVA 4.0: An Update of the Standard European Vector Architecture Database for Advanced Analysis and Programming of Bacterial Phenotypes. Nucleic Acids Res. 2023, 51, D1558–D1567. [Google Scholar] [CrossRef]
- Myronovskyi, M.; Luzhetskyy, A. Native and Engineered Promoters in Natural Product Discovery. Nat. Prod. Rep. 2016, 33, 1006–1019. [Google Scholar] [CrossRef]
- Lou, C.; Stanton, B.; Chen, Y.-J.; Munsky, B.; Voigt, C.A. Ribozyme-Based Insulator Parts Buffer Synthetic Circuits from Genetic Context. Nat. Biotechnol. 2012, 30, 1137–1142. [Google Scholar] [CrossRef]
- Horbal, L.; Siegl, T.; Luzhetskyy, A. A Set of Synthetic Versatile Genetic Control Elements for the Efficient Expression of Genes in Actinobacteria. Sci. Rep. 2018, 8, 491. [Google Scholar] [CrossRef]
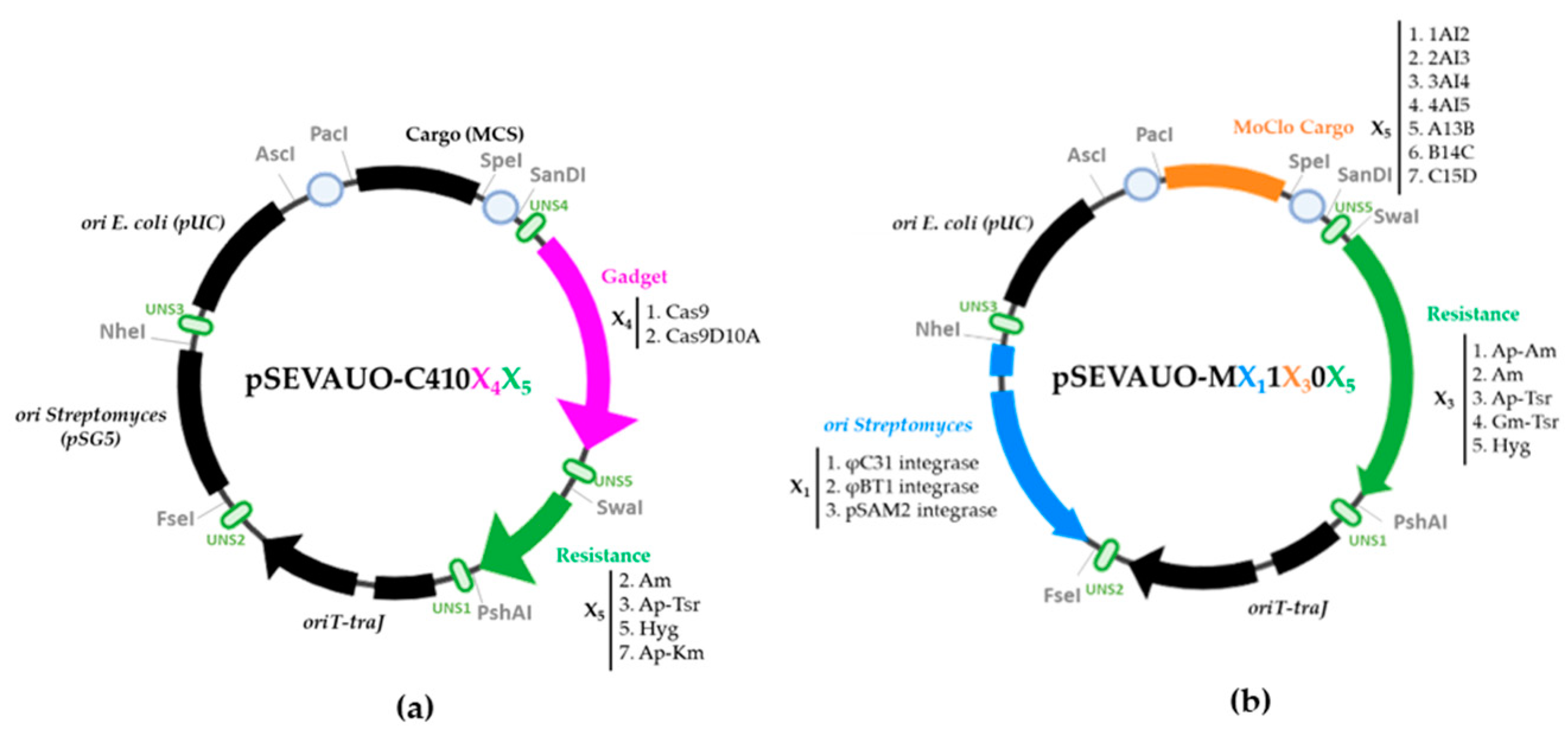
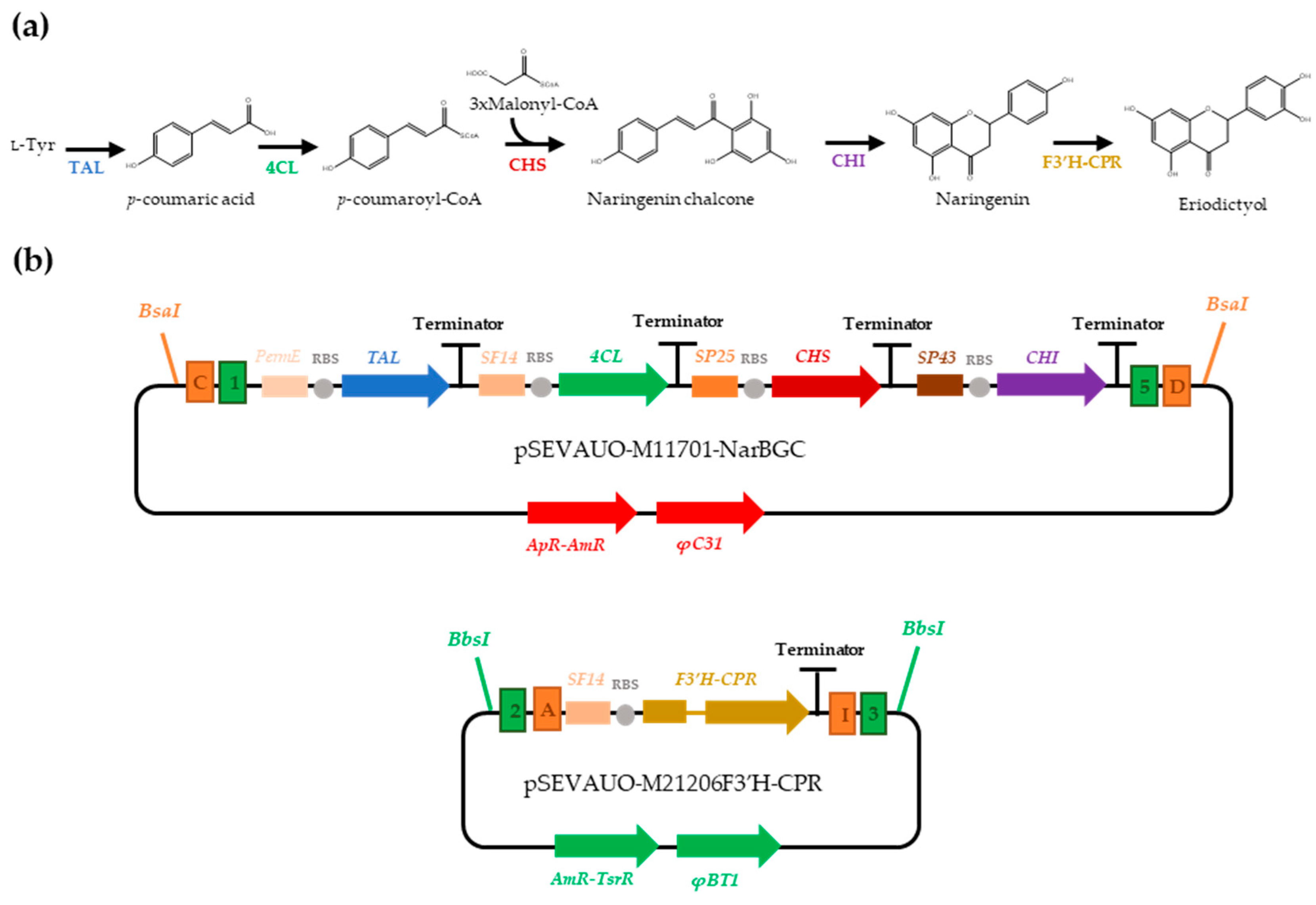
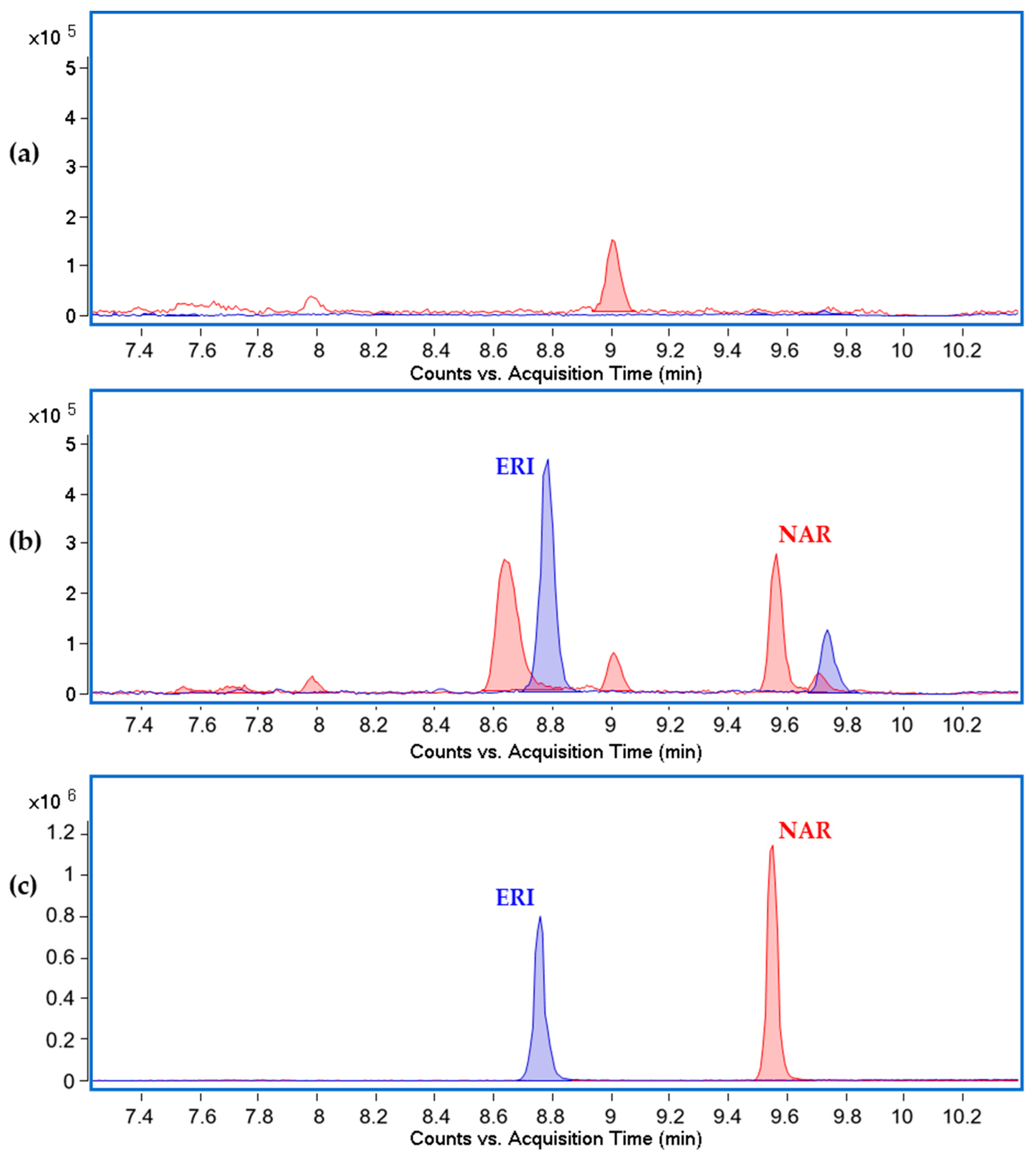
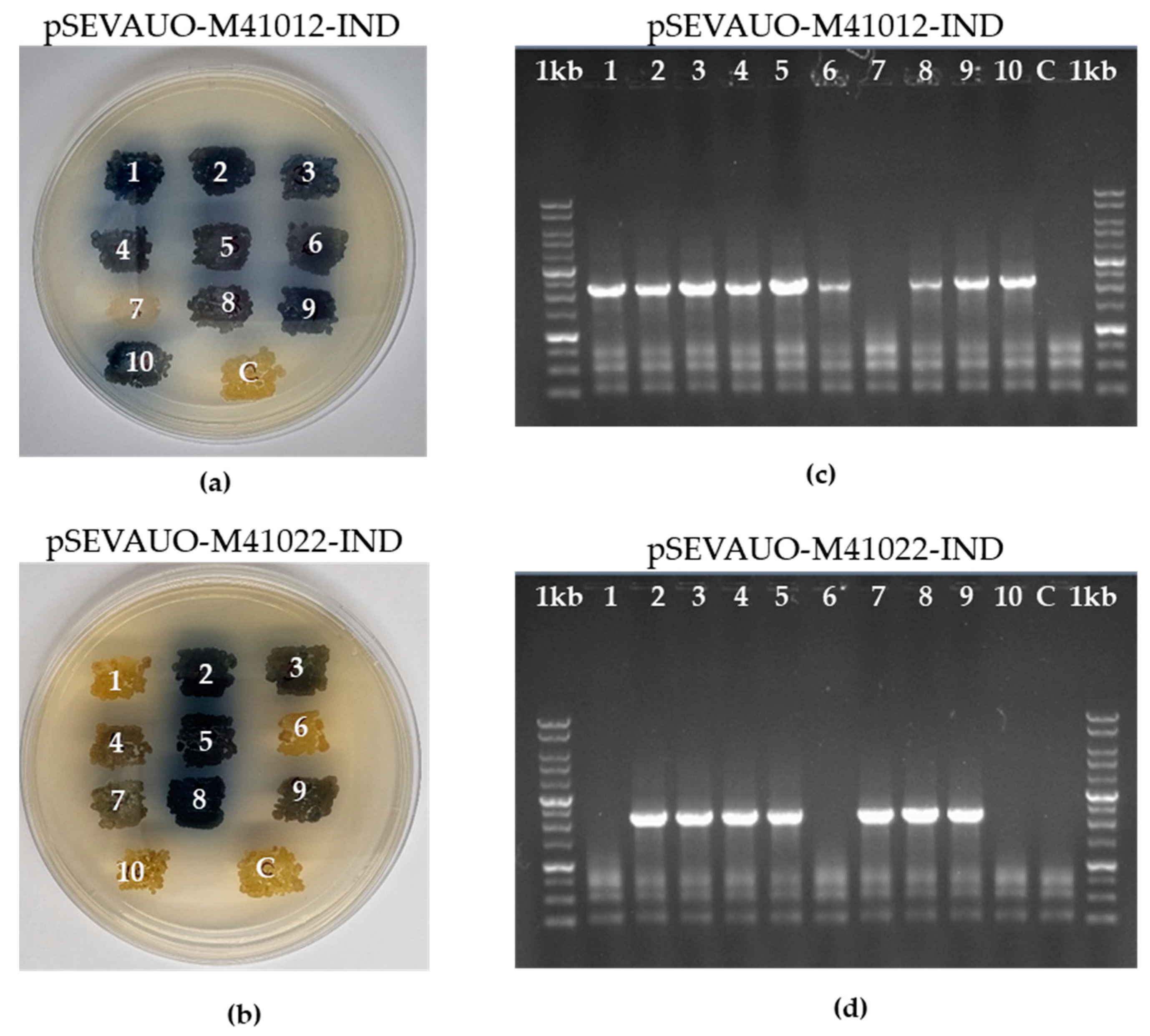
| Name | Origin Streptomyces | Origin E. coli | Cargo | Gadget | Resistance | Accession Number |
|---|---|---|---|---|---|---|
| pSEVAUO-C41012 | pSG5 | pUC | MCS | Cas9 | Am | OQ696801 |
| pSEVAUO-C41013 | pSG5 | pUC | MCS | Cas9 | Ap-Tsr | OQ696802 |
| pSEVAUO-C41017 | pSG5 | pUC | MCS | Cas9 | Ap-Km | OQ696803 |
| pSEVAUO-C41015 | pSG5 | pUC | MCS | Cas9 | Hyg | OQ696804 |
| pSEVAUO-C41022 | pSG5 | pUC | MCS | Cas9D10A | Am | OQ696805 |
| pSEVAUO-C41023 | pSG5 | pUC | MCS | Cas9D10A | Ap-Tsr | OQ696806 |
| pSEVAUO-C41027 | pSG5 | pUC | MCS | Cas9D10A | Ap-Km | OQ696807 |
| pSEVAUO-C41025 | pSG5 | pUC | MCS | Cas9D10A | Hyg | OQ696808 |
| pSEVAUO-M21102 | φBT1 | pUC | 1AI2 | - | Am | OQ696809 |
| pSEVAUO-M21202 | φBT1 | pUC | 2AI3 | - | Am | OQ696810 |
| pSEVAUO-M21302 | φBT1 | pUC | 3AI4 | - | Am | OQ696811 |
| pSEVAUO-M21402 | φBT1 | pUC | 4AI5 | - | Am | OQ696812 |
| pSEVAUO-M21503 | φBT1 | pUC | A13B | - | Ap-Tsr | OQ696813 |
| pSEVAUO-M21603 | φBT1 | pUC | B14C | - | Ap-Tsr | OQ696814 |
| pSEVAUO-M21703 | φBT1 | pUC | C15D | - | Ap-Tsr | OQ696815 |
| pSEVAUO-M21104 | φBT1 | pUC | 1AI2 | - | Gm-Tsr | OQ696816 |
| pSEVAUO-M21204 | φBT1 | pUC | 2AI3 | - | Gm-Tsr | OQ696817 |
| pSEVAUO-M21304 | φBT1 | pUC | 3AI4 | - | Gm-Tsr | OQ696818 |
| pSEVAUO-M21404 | φBT1 | pUC | 4AI5 | - | Gm-Tsr | OQ696819 |
| pSEVAUO-M21504 | φBT1 | pUC | A13B | - | Gm-Tsr | OQ696820 |
| pSEVAUO-M21604 | φBT1 | pUC | B14C | - | Gm-Tsr | OQ696821 |
| pSEVAUO-M21704 | φBT1 | pUC | C15D | - | Gm-Tsr | OQ696822 |
| pSEVAUO-M11101 | φC31 | pUC | 1AI2 | - | Ap-Am | OQ696823 |
| pSEVAUO-M11201 | φC31 | pUC | 2AI3 | - | Ap-Am | OQ696824 |
| pSEVAUO-M11301 | φC31 | pUC | 3AI4 | - | Ap-Am | OQ696825 |
| pSEVAUO-M11401 | φC31 | pUC | 4AI5 | - | Ap-Am | OQ696826 |
| pSEVAUO-M11501 | φC31 | pUC | A13B | - | Ap-Am | OQ696827 |
| pSEVAUO-M11601 | φC31 | pUC | B14C | - | Ap-Am | OQ696828 |
| pSEVAUO-M11701 | φC31 | pUC | C15D | - | Ap-Am | OQ696829 |
| pSEVAUO-M31105 | pSAM2 | pUC | 1AI2 | - | Hyg | OQ696830 |
| pSEVAUO-M31205 | pSAM2 | pUC | 2AI3 | - | Hyg | OQ696831 |
| pSEVAUO-M31305 | pSAM2 | pUC | 3AI4 | - | Hyg | OQ696832 |
| pSEVAUO-M31405 | pSAM2 | pUC | 4AI5 | - | Hyg | OQ696833 |
| pSEVAUO-M31505 | pSAM2 | pUC | A13B | - | Hyg | OQ696834 |
| pSEVAUO-M31605 | pSAM2 | pUC | B14C | - | Hyg | OQ696835 |
| pSEVAUO-M31705 | pSAM2 | pUC | C15D | - | Hyg | OQ696836 |
| pSEVAUO-C41012-Ind | pSG5 | pUC | PermE*-Ind DNA RT | Cas9-Ind Prot | Am | This work |
| pSEVAUO-C41022-Ind | pSG5 | pUC | PermE*-Ind DNA RT | Cas9D10A-Ind Prot | Am | This work |
| pSEVAUO-C41012-pSEB4 | pSG5 | pUC | pSEB4::UNS8 DNA RT | Cas9-pSEB4 Prot | Am | This work |
| pSEVAUO-C41012-C1-matBC | pSG5 | pUC | C1::PermE*-matBC DNA RT | Cas9-C1 Prot1-Prot2 | Am | OQ659402 |
| pSEVAUO-M21102-TAL | φBT1 | pUC | PermE*-TAL | - | Am | This work |
| pSEVAUO-M21202-4CL | φBT1 | pUC | SF14-4CL | - | Am | This work |
| pSEVAUO-M21302-CHS | φBT1 | pUC | SP25-CHS | - | Am | This work |
| pSEVAUO-M21402-CHI | φBT1 | pUC | SP43-CHI-I5 | - | Am | This work |
| pSEVAUO-M11701-NarBGC | φC31 | pUC | NAR BGC | - | Ap-Am | This work |
| pSEVAUO-M21206F3′H-CPR | φBT1 | pUC | SF14-F3′H-CPR | - | Ap-Tsr | OQ674225 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Magadán-Corpas, P.; Ye, S.; Pérez-Valero, Á.; McAlpine, P.L.; Valdés-Chiara, P.; Torres-Bacete, J.; Nogales, J.; Villar, C.J.; Lombó, F. Optimized De Novo Eriodictyol Biosynthesis in Streptomyces albidoflavus Using an Expansion of the Golden Standard Toolkit for Its Use in Actinomycetes. Int. J. Mol. Sci. 2023, 24, 8879. https://doi.org/10.3390/ijms24108879
Magadán-Corpas P, Ye S, Pérez-Valero Á, McAlpine PL, Valdés-Chiara P, Torres-Bacete J, Nogales J, Villar CJ, Lombó F. Optimized De Novo Eriodictyol Biosynthesis in Streptomyces albidoflavus Using an Expansion of the Golden Standard Toolkit for Its Use in Actinomycetes. International Journal of Molecular Sciences. 2023; 24(10):8879. https://doi.org/10.3390/ijms24108879
Chicago/Turabian StyleMagadán-Corpas, Patricia, Suhui Ye, Álvaro Pérez-Valero, Patrick L. McAlpine, Paula Valdés-Chiara, Jesús Torres-Bacete, Juan Nogales, Claudio J. Villar, and Felipe Lombó. 2023. "Optimized De Novo Eriodictyol Biosynthesis in Streptomyces albidoflavus Using an Expansion of the Golden Standard Toolkit for Its Use in Actinomycetes" International Journal of Molecular Sciences 24, no. 10: 8879. https://doi.org/10.3390/ijms24108879
APA StyleMagadán-Corpas, P., Ye, S., Pérez-Valero, Á., McAlpine, P. L., Valdés-Chiara, P., Torres-Bacete, J., Nogales, J., Villar, C. J., & Lombó, F. (2023). Optimized De Novo Eriodictyol Biosynthesis in Streptomyces albidoflavus Using an Expansion of the Golden Standard Toolkit for Its Use in Actinomycetes. International Journal of Molecular Sciences, 24(10), 8879. https://doi.org/10.3390/ijms24108879






