Genome-Wide Association Study Identifies Resistance Loci for Bacterial Blight in a Collection of Asian Temperate Japonica Rice Germplasm
Abstract
1. Introduction
2. Results
2.1. Genotypes and Population Structures of Rice Accessions
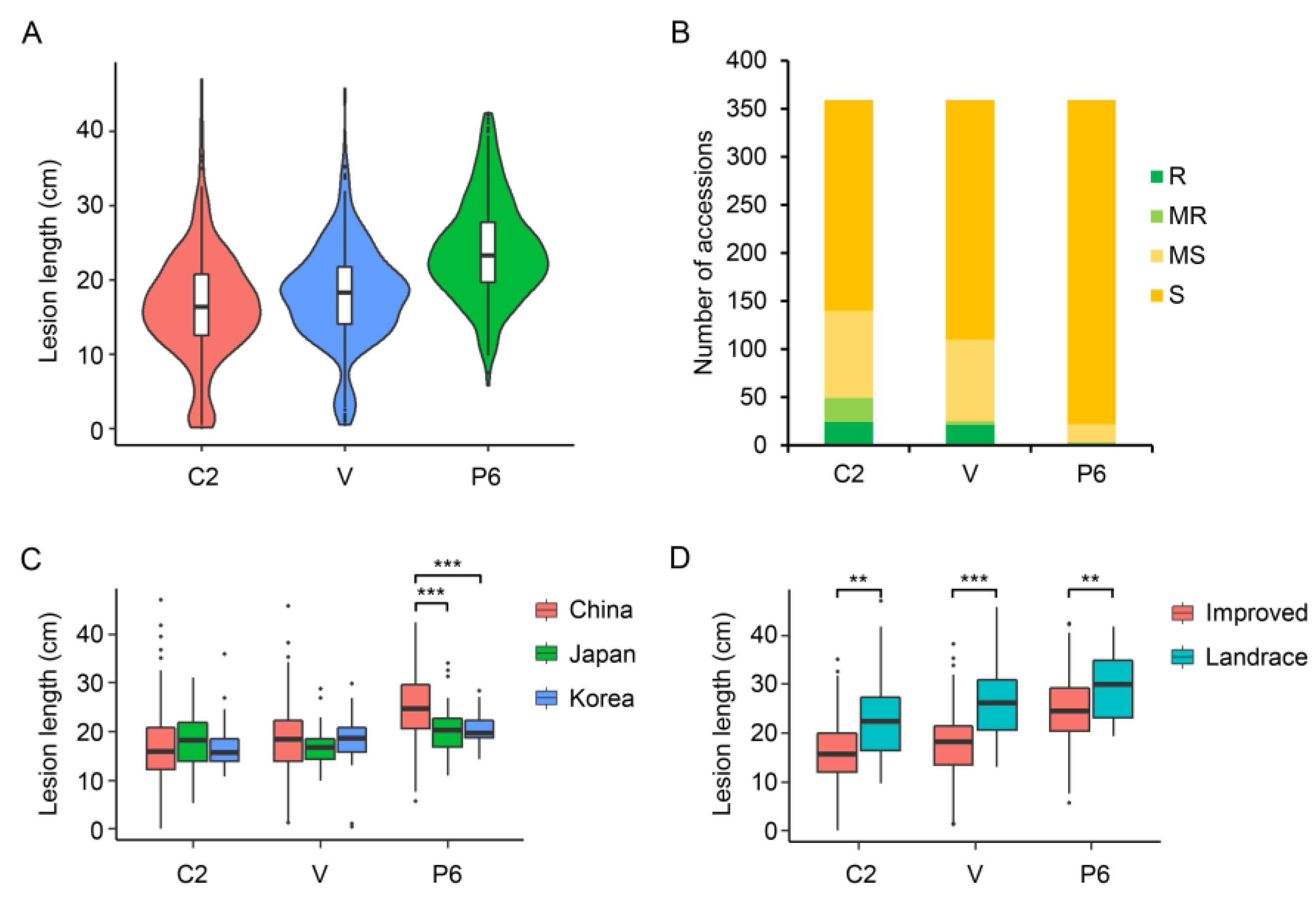
2.2. Resistance of Rice Accessions to Three Xoo Strains
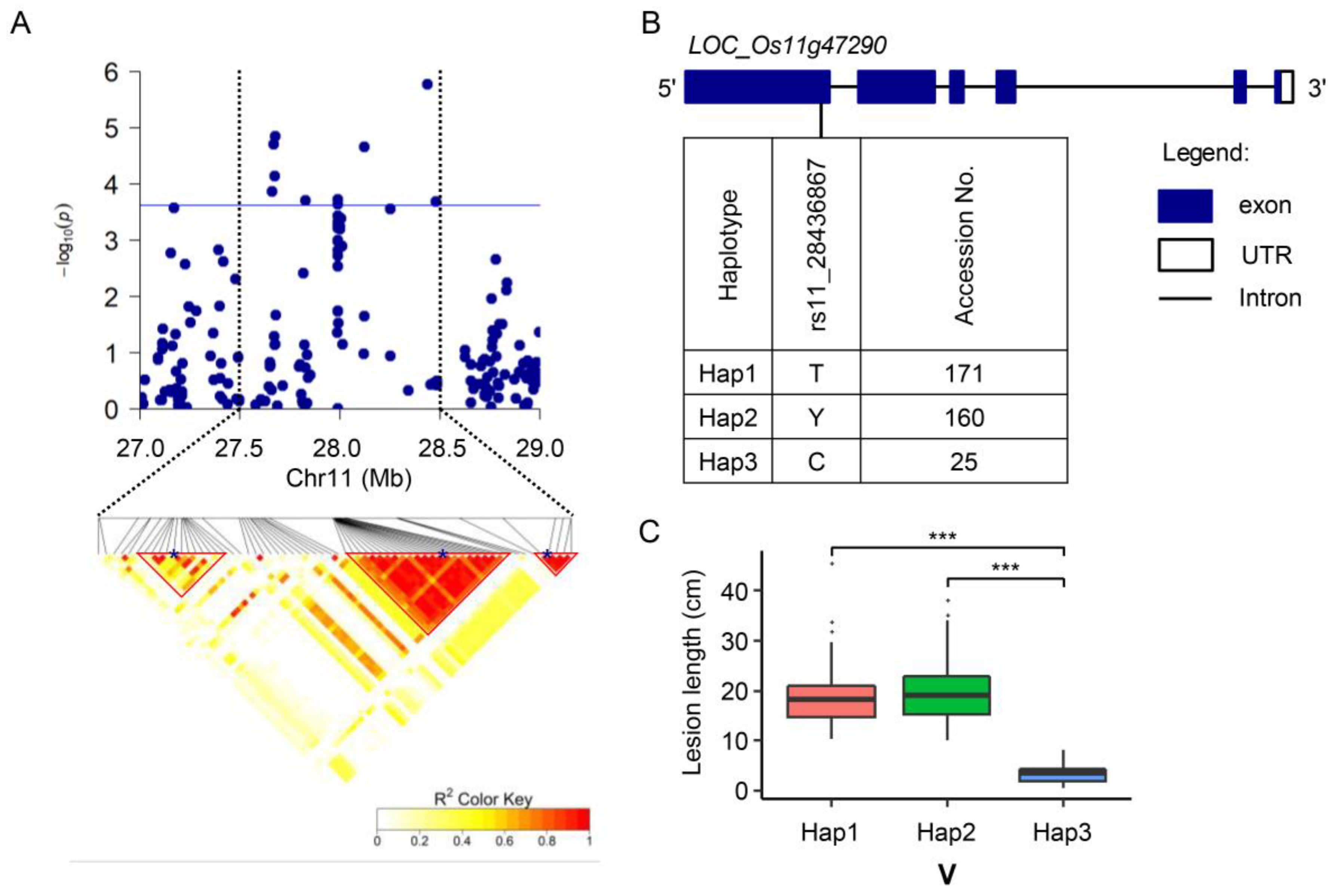
2.3. Identification of Resistance Loci against BB through GWAS
2.4. Candidate Gene Prediction
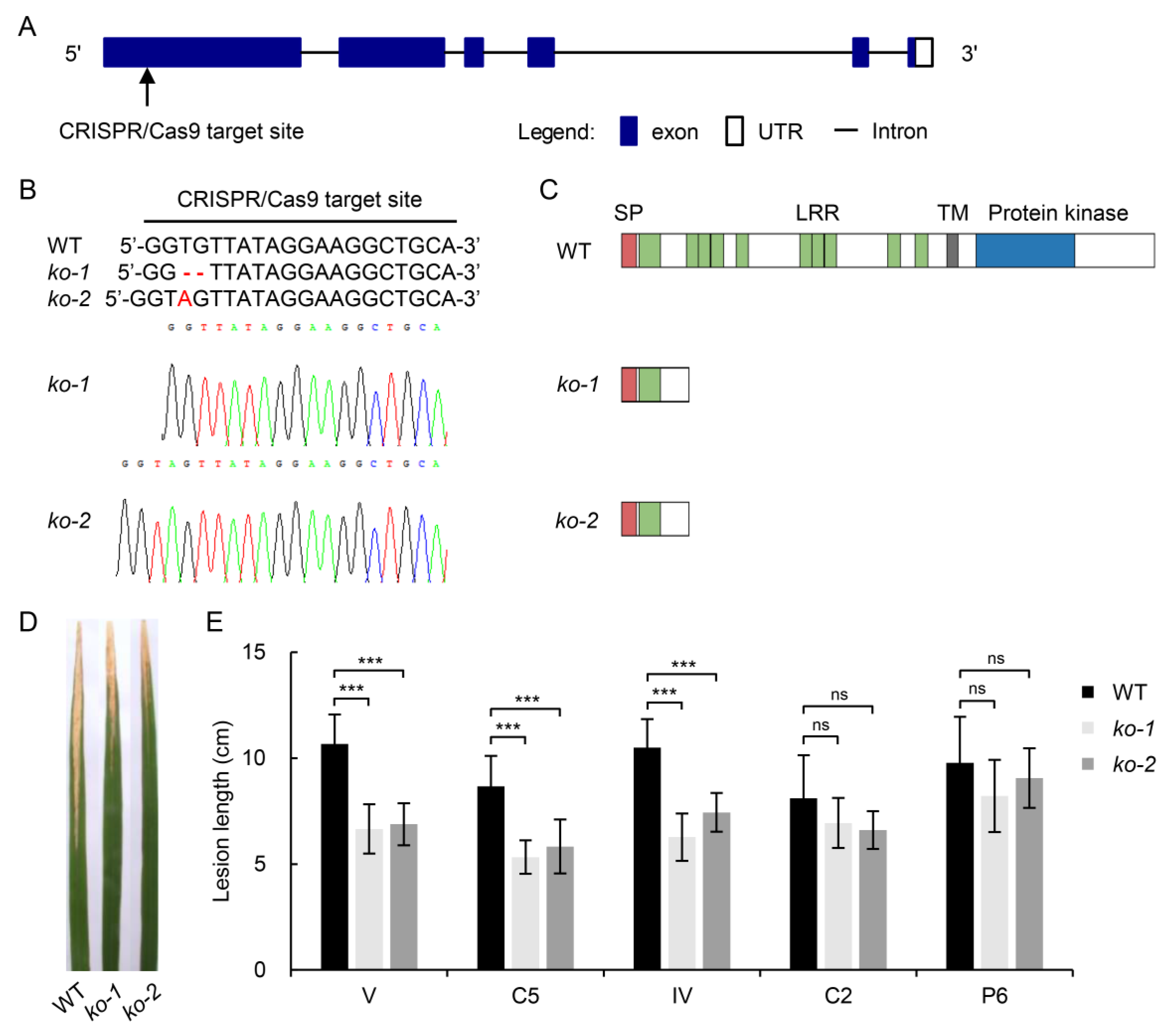
2.5. Involvement of LOC_Os11g47290 Strain-Specific Resistance to BB
3. Discussion
3.1. Variation in Resistance of the Temperate Japonica Rice Accessions
3.2. QTL and Candidate Genes Associated with BB Resistance
3.3. Improving BB Resistance in Rice through Editing LOC_Os11g47290
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Xoo Strains and Artificial Inoculation
4.3. Genotyping with the 55K SNP Array
4.4. Population Structure Analysis
4.5. Genome-Wide Association Analysis
4.6. QTL Identification and Analysis of Candidate Genes
4.7. Vector Construction and Rice Transformation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Niño-Liu, D.O.; Ronald, P.C.; Bogdanove, A.J. Xanthomonas oryzae pathovars: Model pathogens of a model crop. Mol. Plant Pathol. 2006, 7, 303–324. [Google Scholar] [CrossRef]
- Chukwu, S.C.; Rafii, M.Y.; Ramlee, S.I.; Ismail, S.I.; Hasan, M.M.; Oladosu, Y.A.; Magaji, U.G.; Akos, I.; Olalekan, K.K. Bacterial leaf blight resistance in rice: A review of conventional breeding to molecular approach. Mol. Biol. Rep. 2019, 46, 1519–1532. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Nayak, D.K.; Mohanty, S.; Behera, L.; Barik, S.R.; Pandit, E.; Lenka, S.; Anandan, A. Pyramiding of three bacterial blight resistance genes for broad-spectrum resistance in deepwater rice variety, Jalmagna. Rice 2015, 8, 51. [Google Scholar] [CrossRef]
- Dossa, G.S.; Sparks, A.; Cruz, C.V.; Oliva, R. Decision tools for bacterial blight resistance gene deployment in rice-based agricultural ecosystems. Front. Plant Sci. 2015, 6, 305. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, S. Linkage studies on the resistance to bacterial leaf blight, Xanthomonas oryzae (Uyeda et Ishiyama) Dowson, in rice. Bull. Nat. Inst. Agric. Sci. 1967, 16, 1–18. [Google Scholar]
- Yang, Y.; Zhou, Y.; Sun, J.; Liang, W.; Chen, X.; Wang, X.; Zhou, J.; Yu, C.; Wang, J.; Wu, S.; et al. Research progress on cloning and function of Xa genes against rice bacterial blight. Front. Plant Sci. 2022, 13, 847199. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhong, Q.; Xiao, S.; Wang, B.; Ke, X.; Zhang, Y.; Yin, F.; Zhang, D.; Jiang, C.; Liu, L.; et al. A new NLR disease resistance gene Xa47 confers durable and broad-spectrum resistance to bacterial blight in rice. Front. Plant Sci. 2022, 13, 1037901. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q. Genetics and improvement of bacterial blight resistance of hybrid rice in China. Rice Sci. 2009, 16, 83–92. [Google Scholar] [CrossRef]
- Khan, M.A.; Naeem, M.; Iqbal, M. Breeding approaches for bacterial leaf blight resistance in rice (Oryza sativa L.), current status and future directions. Eur. J. Plant Pathol. 2014, 139, 27–37. [Google Scholar] [CrossRef]
- Xu, Z.; Xu, X.; Wang, Y.; Liu, L.; Li, Y.; Yang, Y.; Liu, L.; Zou, L.; Chen, G. A varied AvrXa23-like TALE enables the bacterial blight pathogen to avoid being trapped by Xa23 resistance gene in rice. J. Adv. Res. 2022, 42, 263–272. [Google Scholar] [CrossRef]
- Lu, J.; Li, Q.; Wang, C.; Wang, M.; Zeng, D.; Zhang, F.; Zhai, W.; Zhou, Y.J. Identification of quantitative trait loci associated with resistance to Xanthomonas oryzae pv. oryzae pathotypes prevalent in South China. Crop J. 2022, 10, 498–507. [Google Scholar] [CrossRef]
- Zhao, K.; Tung, C.W.; Eizenga, G.C.; Wright, M.H.; Ali, M.L.; Price, A.H.; Norton, G.J.; Islam, M.R.; Reynolds, A.; Mezey, J.; et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2011, 2, 467. [Google Scholar] [CrossRef] [PubMed]
- Burghardt, L.T.; Young, N.D.; Tiffin, P. A guide to genome-wide association mapping in plants. Curr. Protoc. Plant Biol. 2017, 2, 22–38. [Google Scholar] [CrossRef] [PubMed]
- Demirjian, C.; Vailleau, F.; Berthomé, R.; Roux, F. Genome-wide association studies in plant pathosystems: Success or failure? Trends Plant Sci. 2023, 28, 471–485. [Google Scholar] [CrossRef] [PubMed]
- Dhatt, B.K.; Paul, P.; Sandhu, J.; Hussain, W.; Irvin, L.; Zhu, F.; Adviento-Borbe, M.A.; Lorence, A.; Staswick, P.; Yu, H.; et al. Allelic variation in rice Fertilization Independent Endosperm 1 contributes to grain width under high night temperature stress. New Phytol. 2021, 229, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Dilla-Ermita, C.J.; Tandayu, E.; Juanillas, V.M.; Detras, J.; Lozada, D.N.; Dwiyanti, M.S.; Vera Cruz, C.; Mbanjo, E.G.N.; Ardales, E.; Diaz, M.G.; et al. Genome-wide association analysis tracks bacterial leaf blight resistance loci in rice diverse germplasm. Rice 2017, 10, 8. [Google Scholar] [CrossRef]
- Zhang, F.; Wu, Z.C.; Wang, M.M.; Zhang, F.; Dingkuhn, M.; Xu, J.L.; Zhou, Y.L.; Li, Z.K. Genome-wide association analysis identifies resistance loci for bacterial blight in a diverse collection of indica rice germplasm. PLoS ONE 2017, 12, e0174598. [Google Scholar] [CrossRef]
- Alexandrov, N.; Tai, S.; Wang, W.; Mansueto, L.; Palis, K.; Fuentes, R.R.; Ulat, V.J.; Chebotarov, D.; Zhang, G.; Li, Z.; et al. SNP-Seek database of SNPs derived from 3000 rice genomes. Nucleic Acids Res. 2015, 43, D1023–D1027. [Google Scholar] [CrossRef]
- Wang, W.; Mauleon, R.; Hu, Z.; Chebotarov, D.; Tai, S.; Wu, Z.; Li, M.; Zheng, T.; Fuentes, R.R.; Zhang, F.; et al. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 2018, 557, 43–49. [Google Scholar] [CrossRef]
- Lu, J.; Wang, C.; Zeng, D.; Li, J.; Shi, X.; Shi, Y.; Zhou, Y. Genome-wide association study dissects resistance loci against bacterial blight in a diverse rice panel from the 3000 Rice Genomes Project. Rice 2021, 14, 22. [Google Scholar] [CrossRef]
- Zhang, F.; Hu, Z.; Wu, Z.; Lu, J.; Shi, Y.; Xu, J.; Wang, X.; Wang, J.; Zhang, F.; Wang, M.; et al. Reciprocal adaptation of rice and Xanthomonas oryzae pv. oryzae: Cross-species 2D GWAS reveals the underlying genetics. Plant Cell 2021, 33, 2538–2561. [Google Scholar] [CrossRef]
- Zaidi, S.S.; Mukhtar, M.S.; Mansoor, S. Genome editing: Targeting susceptibility genes for plant disease resistance. Trends Biotechnol. 2018, 36, 898–906. [Google Scholar] [CrossRef] [PubMed]
- Derry, W.B. CRISPR: Development of a technology and its applications. FEBS J. 2021, 288, 358–359. [Google Scholar] [CrossRef] [PubMed]
- Oliva, R.; Ji, C.; Atienza-Grande, G.; Huguet-Tapia, J.C.; Perez-Quintero, A.; Li, T.; Eom, J.S.; Li, C.; Nguyen, H.; Liu, B.; et al. Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 2019, 37, 1344–1350. [Google Scholar] [CrossRef] [PubMed]
- Blanvillain-Baufumé, S.; Reschke, M.; Solé, M.; Auguy, F.; Doucoure, H.; Szurek, B.; Meynard, D.; Portefaix, M.; Cunnac, S.; Guiderdoni, E.; et al. Targeted promoter editing for rice resistance to Xanthomonas oryzae pv. oryzae reveals differential activities for SWEET14-inducing TAL effectors. Plant Biotechnol. J. 2017, 15, 306–317. [Google Scholar] [CrossRef]
- Xu, Z.; Xu, X.; Gong, Q.; Li, Z.; Li, Y.; Wang, S.; Yang, Y.; Ma, W.; Liu, L.; Zhu, B.; et al. Engineering broad-spectrum bacterial blight resistance by simultaneously disrupting variable TALE-binding elements of multiple susceptibility genes in rice. Mol. Plant 2019, 12, 1434–1446. [Google Scholar] [CrossRef]
- Kim, Y.A.; Moon, H.; Park, C.J. CRISPR/Cas9-targeted mutagenesis of Os8N3 in rice to confer resistance to Xanthomonas oryzae pv. oryzae. Rice 2019, 12, 67. [Google Scholar] [CrossRef]
- Chen, Z.; Bu, Q.; Liu, G.; Wang, M.; Wang, H.; Liu, H.; Li, X.; Li, H.; Fang, J.; Liang, Y.; et al. Genomic decoding of breeding history to guide breeding-by-design in rice. Natl. Sci. Rev. 2023, 10, nwad029. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Wang, B.; Zhao, X.; Ponce, K.; Qian, Q.; Ye, G. Association mapping of ferrous, zinc, and aluminum tolerance at the seedling stage in indica rice using MAGIC populations. Front. Plant Sci. 2017, 8, 1822. [Google Scholar] [CrossRef]
- Descalsota, G.I.L.; Swamy, B.P.M.; Zaw, H.; Inabangan-Asilo, M.A.; Amparado, A.; Mauleon, R.; Chadha-Mohanty, P.; Arocena, E.C.; Raghavan, C.; Leung, H.; et al. Genome-wide association mapping in a rice MAGIC plus population detects QTLs and genes useful for biofortification. Front. Plant Sci. 2018, 9, 1347. [Google Scholar] [CrossRef]
- Chen, T.X.; Zhu, Y.J.; Mi, X.F.; Chen, K.; Meng, L.J.; Zuo, S.M.; Xu, J.L. Mapping of QTLs for bacterial blight resistance and screening of resistant materials using MAGIC populations of rice. Acta Agron. Sin. 2016, 42, 1437. [Google Scholar] [CrossRef]
- Kim, S.M.; Reinke, R.F. A novel resistance gene for bacterial blight in rice, Xa43(t) identified by GWAS, confirmed by QTL mapping using a bi-parental population. PLoS ONE 2019, 14, e0211775. [Google Scholar] [CrossRef]
- Wang, Y.S.; Pi, L.Y.; Chen, X.; Chakrabarty, P.K.; Jiang, J.; De Leon, A.L.; Liu, G.Z.; Li, L.; Benny, U.; Oard, J.; et al. Rice XA21 binding protein 3 is a ubiquitin ligase required for full Xa21-mediated disease resistance. Plant Cell 2006, 18, 3635–3646. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ye, B.; Yin, J.; Yuan, C.; Zhou, X.; Li, W.; He, M.; Wang, J.; Chen, W.; Qin, P.; et al. Characterization and fine mapping of a light-dependent leaf lesion mimic mutant 1 in rice. Plant Physiol. Biochem. 2015, 97, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Man, J.; Gallagher, J.P.; Bartlett, M. Structural evolution drives diversification of the large LRR-RLK gene family. New Phytol. 2020, 226, 1492–1505. [Google Scholar] [CrossRef]
- Lee, L.Y.; Hou, X.; Fang, L.; Fan, S.; Kumar, P.P.; Yu, H. STUNTED mediates the control of cell proliferation by GA in Arabidopsis. Development 2012, 139, 1568–1576. [Google Scholar] [CrossRef]
- Morillo, S.A.; Tax, F.E. Functional analysis of receptor-like kinases in monocots and dicots. Curr. Opin. Plant Biol. 2006, 9, 460–469. [Google Scholar] [CrossRef]
- Fang, Z.D.; Xu, Z.G.; Guo, C.J.; Yin, S.Z.; Wu, S.Z.; Xu, X.M.; Zhang, Q. Studies on pathotypes of Xanthomonas campestris pv. oryzae in China. Acta Phytopathol. Sin. 1990, 20, 81–88. [Google Scholar]
- Zeng, L. Resistance of IRBB21 (Xa21) to five races of bacterial blight in Guangdong. Acta Phytophy. Sin. 2002, 29, 97–100. [Google Scholar]
- Ichimaru, K.; Yamaguchi, K.; Harada, K.; Nishio, Y.; Hori, M.; Ishikawa, K.; Inoue, H.; Shigeta, S.; Inoue, K.; Shimada, K.; et al. Cooperative regulation of PBI1 and MAPKs controls WRKY45 transcription factor in rice immunity. Nat. Commun. 2022, 13, 2397. [Google Scholar] [CrossRef]
- Sett, S.; Prasad, A.; Prasad, M. Resistance genes on the verge of plant-virus interaction. Trends Plant Sci. 2022, 27, 1242–1252. [Google Scholar] [CrossRef] [PubMed]
- Uji, Y.; Kashihara, K.; Kiyama, H.; Mochizuki, S.; Akimitsu, K.; Gomi, K. Jasmonic acid-induced VQ-Motif-Containing protein OsVQ13 influences the OsWRKY45 signaling pathway and grain size by associating with OsMPK6 in Rice. Int. J. Mol. Sci. 2019, 20, 2917. [Google Scholar] [CrossRef] [PubMed]
- Nelson, R.; Wiesner-Hanks, T.; Wisser, R.; Balint-Kurti, P. Navigating complexity to breed disease-resistant crops. Nat. Rev. Genet. 2018, 19, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Nowack, M.K.; Holmes, D.R.; Lahaye, T. TALE-induced cell death executors: An origin outside immunity? Trends Plant Sci. 2022, 27, 536–548. [Google Scholar] [CrossRef]
- Li, N.; Lin, B.; Wang, H.; Li, X.; Yang, F.; Ding, X.; Yan, J.; Chu, Z. Natural variation in ZmFBL41 confers banded leaf and sheath blight resistance in maize. Nat. Genet. 2019, 51, 1540–1548. [Google Scholar] [CrossRef]
- Kim, H.S.; Delaney, T.P. Arabidopsis SON1 is an F-box protein that regulates a novel induced defense response independent of both salicylic acid and systemic acquired resistance. Plant Cell 2002, 14, 1469–1482. [Google Scholar] [CrossRef]
- Chen, X.; Chen, H.; Yuan, J.S.; Köllner, T.G.; Chen, Y.; Guo, Y.; Zhuang, X.; Chen, X.; Zhang, Y.J.; Fu, J.; et al. The rice terpene synthase gene OsTPS19 functions as an (S)-limonene synthase in planta, and its overexpression leads to enhanced resistance to the blast fungus Magnaporthe oryzae. Plant Biotechnol. J. 2018, 16, 1778–1787. [Google Scholar] [CrossRef]
- Gish, L.A.; Clark, S.E. The RLK/Pelle family of kinases. Plant J. 2011, 66, 117–127. [Google Scholar] [CrossRef]
- Coleman, A.D.; Maroschek, J.; Raasch, L.; Takken, F.L.W.; Ranf, S.; Hückelhoven, R. The Arabidopsis leucine-rich repeat receptor-like kinase MIK2 is a crucial component of early immune responses to a fungal-derived elicitor. New Phytol. 2021, 229, 3453–3466. [Google Scholar] [CrossRef]
- Chen, X.; Zuo, S.; Schwessinger, B.; Chern, M.; Canlas, P.E.; Ruan, D.; Zhou, X.; Wang, J.; Daudi, A.; Petzold, C.J.; et al. An XA21-associated kinase (OsSERK2) regulates immunity mediated by the XA21 and XA3 immune receptors. Mol. Plant 2014, 7, 874–892. [Google Scholar] [CrossRef]
- Eom, J.S.; Luo, D.; Atienza-Grande, G.; Yang, J.; Ji, C.; Thi Luu, V.; Huguet-Tapia, J.C.; Char, S.N.; Liu, B.; Nguyen, H.; et al. Diagnostic kit for rice blight resistance. Nat. Biotechnol. 2019, 37, 1372–1379. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Matsuoka, M. Genetic approaches to crop improvement: Responding to environmental and population changes. Nat. Rev. Genet. 2008, 9, 444–457. [Google Scholar] [CrossRef] [PubMed]
- Kauffman, H.E.; Reddy, A.; Hsieh, S.; Merca, S.D. An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis. Rep. 1973, 57, 537–541. [Google Scholar]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Galinsky, K.J.; Bhatia, G.; Loh, P.R.; Georgiev, S.; Mukherjee, S.; Patterson, N.J.; Price, A.L. Fast principal-component analysis reveals convergent evolution of ADH1B in Europe and East Asia. Am. J. Hum. Genet. 2016, 98, 456–472. [Google Scholar] [CrossRef]
- Balding, D.J.; Nichols, R.A. A method for quantifying differentiation between populations at multi-allelic loci and its implications for investigating identity and paternity. Genetica 1995, 96, 3–12. [Google Scholar] [CrossRef]
- Kang, H.M.; Sul, J.H.; Service, S.K.; Zaitlen, N.A.; Kong, S.Y.; Freimer, N.B.; Sabatti, C.; Eskin, E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010, 42, 348–354. [Google Scholar] [CrossRef]
- Li, M.X.; Yeung, J.M.; Cherny, S.S.; Sham, P.C. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum. Genet. 2012, 131, 747–756. [Google Scholar] [CrossRef]
- Lu, K.; Wei, L.; Li, X.; Wang, Y.; Wu, J.; Liu, M.; Zhang, C.; Chen, Z.; Xiao, Z.; Jian, H.; et al. Whole-genome resequencing reveals Brassica napus origin and genetic loci involved in its improvement. Nat. Commun. 2019, 10, 1154. [Google Scholar] [CrossRef]
- Turner, S.D. qqman: An R package for visualizing GWAS results using Q-Q and manhattan plots. bioRxiv 2014. [CrossRef]
- Li, C.; Su, P.; Dan, W.; Peng, S.; Yong, L. Dissection of the genetic architecture of rice resistance to Xanthomonas oryzae pv. oryzae using a genome wide association study. J. Phytopathol. 2018, 166, 470–476. [Google Scholar] [CrossRef]
- Guo, H.; Zeng, Y.; Li, J.; Ma, X.; Zhang, Z.; Lou, Q.; Li, J.; Gu, Y.; Zhang, H.; Li, J.; et al. Differentiation, evolution and utilization of natural alleles for cold adaptability at the reproductive stage in rice. Plant Biotechnol. J. 2020, 18, 2491–2503. [Google Scholar] [CrossRef]
- Zhang, F.; Zeng, D.; Zhang, C.S.; Lu, J.L.; Chen, T.J.; Xie, J.P.; Zhou, Y.L. Genome-wide association analysis of the genetic basis for sheath blight resistance in rice. Rice 2019, 12, 93. [Google Scholar] [CrossRef]
- Yano, K.; Yamamoto, E.; Aya, K.; Takeuchi, H.; Lo, P.C.; Hu, L.; Yamasaki, M.; Yoshida, S.; Kitano, H.; Hirano, K.; et al. Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nat. Genet. 2016, 48, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.H.; Blay, S.; Mcneney, B.; Graham, J. LD heatmap: An R function for graphical display of pairwise linkage disequilibriam between single nucleotide polymorphisms. J. Stat. Softw. 2006, 16, 9. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Ma, X.; Zhu, Q.; Zeng, D.; Li, G.; Liu, Y.G. CRISPR-GE: A convenient software toolkit for CRISPR-based genome editing. Mol. Plant 2017, 10, 1246–1249. [Google Scholar] [CrossRef]
- Nishimura, A.; Aichi, I.; Matsuoka, M. A protocol for Agrobacterium-mediated transformation in rice. Nat. Protoc. 2006, 1, 2796–2802. [Google Scholar] [CrossRef]

| QTL Name a | Xoo Strains | Chromosome | LD Block Interval (bp) | Number of Significant SNP | Lead SNP | p-Value | PVE b (%) | Effect | Reported QTL/Genes |
|---|---|---|---|---|---|---|---|---|---|
| qBBC2-1 | C2 | 1 | 4,747,337–4,961,779 | 2 | rs1_4762032 | 1.03 × 10−4 | 3.8 | 6.01 | Novel |
| qBBC2-2 | C2 | 2 | 1,614,380–1,778,296 | 3 | rs2_1651016 | 1.28 × 10−6 | 6.1 | −6.08 | Novel |
| qBBC2-10.1 | C2 | 10 | 58,063–850,339 | 6 | rs10_187309 | 1.26 × 10−4 | 3.5 | −3.36 | Novel |
| qBBC2-10.2 | C2 | 10 | 2,375,502–2,569,504 | 10 | rs10_2508049 | 6.82 × 10−6 | 4.4 | −4.51 | Novel |
| qBBV-11.1 | V | 11 | 27,648,838–27,675,916 | 4 | rs11_27660271 | 1.36 × 10−4 | 4.6 | 3.80 | Xa35(t) [6], Xa36(t) [6], L11 [17], qC4-11 [20], qBB11.4 [11], qBLB11:1 [30] |
| qBBV-11.2 | V | 11 | 27,987,854–28,010,037 | 2 | rs11_27988643 | 1.88 × 10−4 | 4.0 | −1.59 | Xa22(t) [6], Xa35(t) [6], Xa36(t) [6], xa44(t) [6], Xa47 [7], qC5-11.1 [20], qBB11.5 [11], QBbr11-2 [31], qBLB11:1 [30] |
| qBBV-11.3 | V | 11 | 28,436,867–28,484,909 | 2 | rs11_28436867 | 1.69 × 10−6 | 22.8 | 3.23 | Xa3/Xa26 [6], Xa43(t) [32], qC5-11.2 [20], qBB11.6 [11], qBLB11:1 [30] |
| qBBP6-4 | P6 | 4 | 30,856,230–31,068,491 | 2 | rs4_31010527 | 1.70 × 10−4 | 3.8 | 2.62 | qC5-4 [20] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Shi, X.; Wang, C.; Li, Q.; Lu, J.; Zeng, D.; Xie, J.; Shi, Y.; Zhai, W.; Zhou, Y. Genome-Wide Association Study Identifies Resistance Loci for Bacterial Blight in a Collection of Asian Temperate Japonica Rice Germplasm. Int. J. Mol. Sci. 2023, 24, 8810. https://doi.org/10.3390/ijms24108810
Li J, Shi X, Wang C, Li Q, Lu J, Zeng D, Xie J, Shi Y, Zhai W, Zhou Y. Genome-Wide Association Study Identifies Resistance Loci for Bacterial Blight in a Collection of Asian Temperate Japonica Rice Germplasm. International Journal of Molecular Sciences. 2023; 24(10):8810. https://doi.org/10.3390/ijms24108810
Chicago/Turabian StyleLi, Jianmin, Xiaorong Shi, Chunchao Wang, Quanlin Li, Jialing Lu, Dan Zeng, Junping Xie, Yingyao Shi, Wenxue Zhai, and Yongli Zhou. 2023. "Genome-Wide Association Study Identifies Resistance Loci for Bacterial Blight in a Collection of Asian Temperate Japonica Rice Germplasm" International Journal of Molecular Sciences 24, no. 10: 8810. https://doi.org/10.3390/ijms24108810
APA StyleLi, J., Shi, X., Wang, C., Li, Q., Lu, J., Zeng, D., Xie, J., Shi, Y., Zhai, W., & Zhou, Y. (2023). Genome-Wide Association Study Identifies Resistance Loci for Bacterial Blight in a Collection of Asian Temperate Japonica Rice Germplasm. International Journal of Molecular Sciences, 24(10), 8810. https://doi.org/10.3390/ijms24108810





