Supervised Learning and Multi-Omics Integration Reveals Clinical Significance of Inner Membrane Mitochondrial Protein (IMMT) in Prognostic Prediction, Tumor Immune Microenvironment and Precision Medicine for Kidney Renal Clear Cell Carcinoma
Abstract
1. Introduction
2. Results
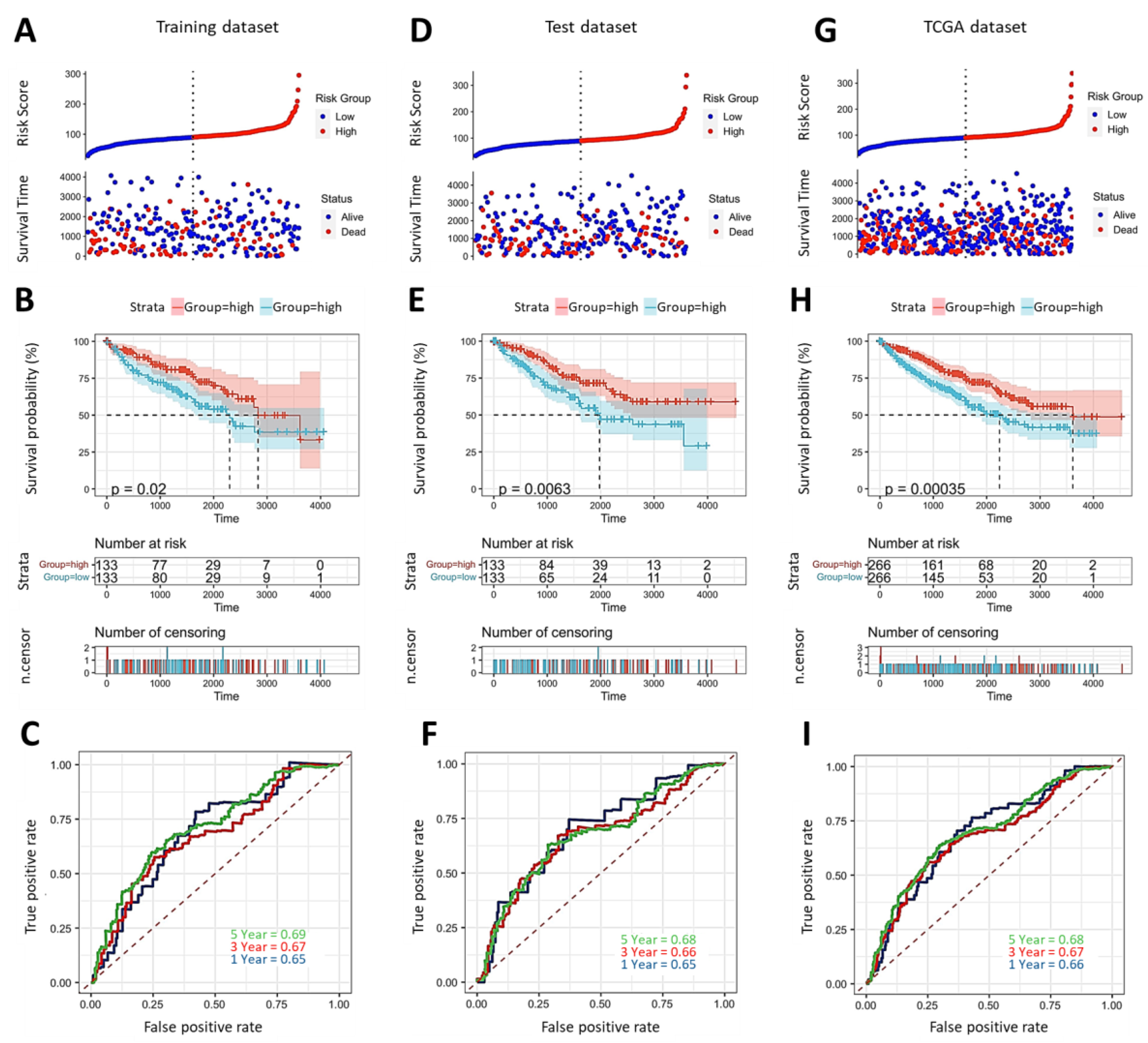
2.1. Evaluation and Verification of Prognostic Significance of IMMT in KIRC
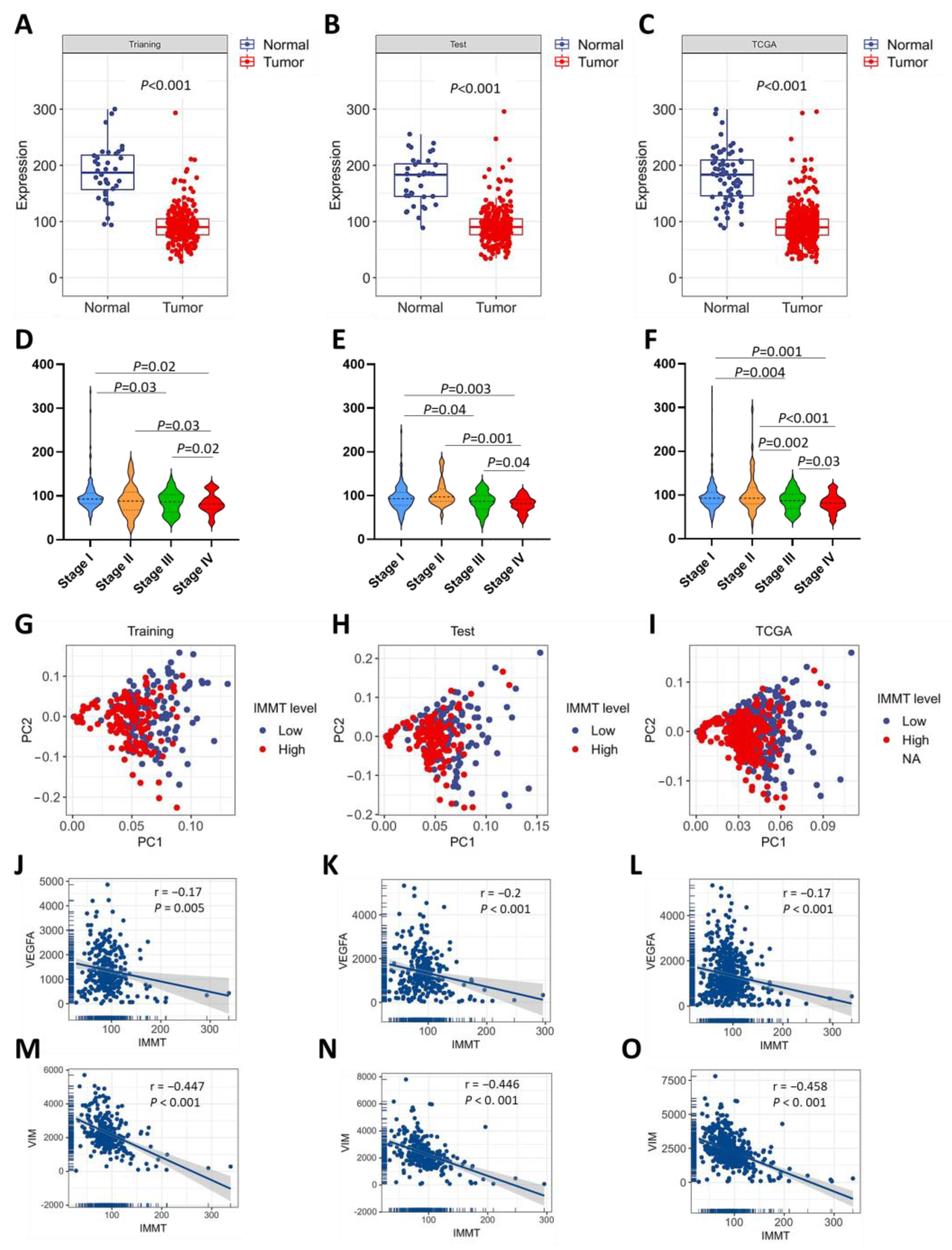
2.2. Low IMMT Expressions Correlate with KIRC Progression
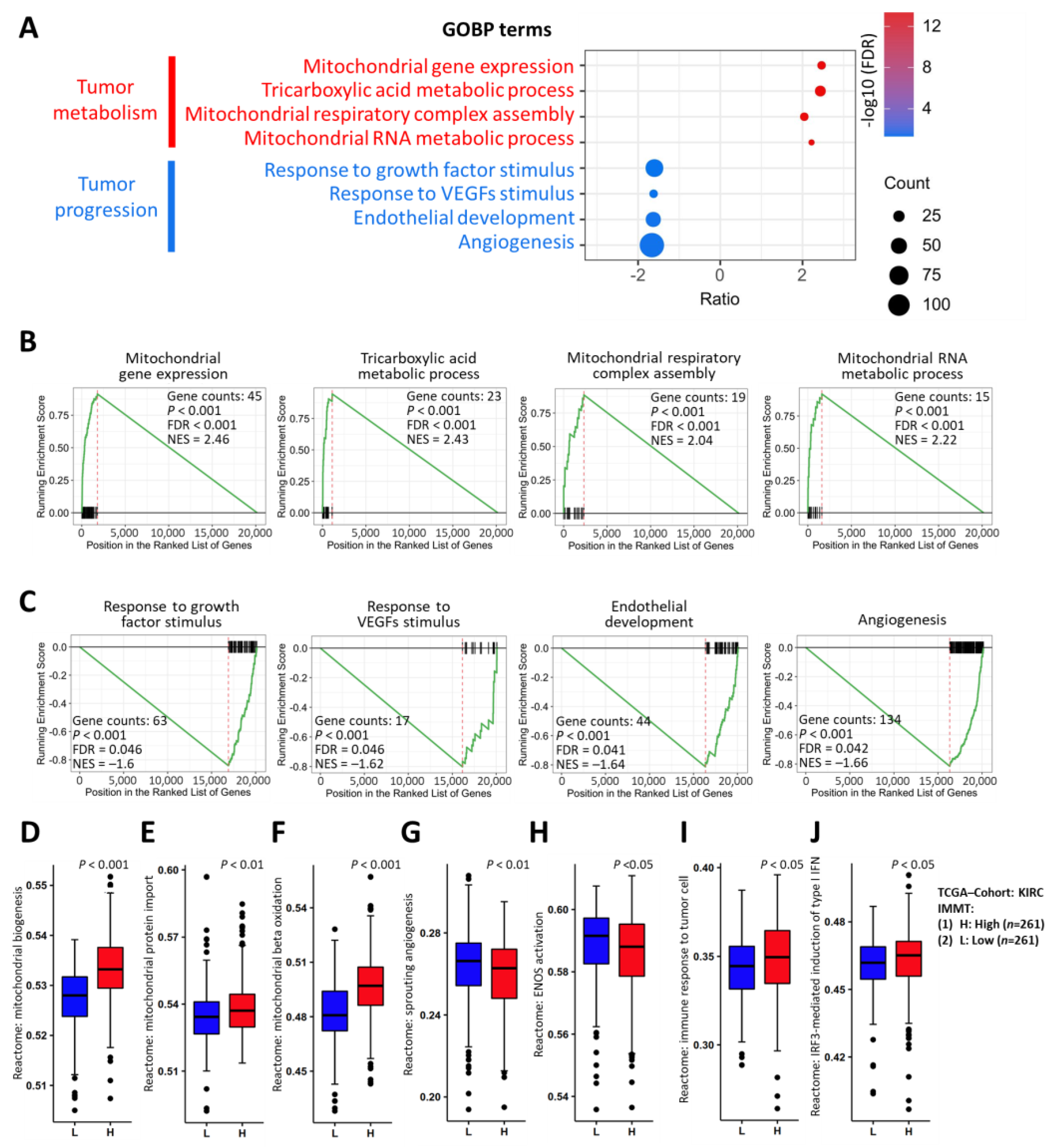
2.3. Implications of Low IMMT in Mitochondrial Inhibition and Angiogenetic Activation
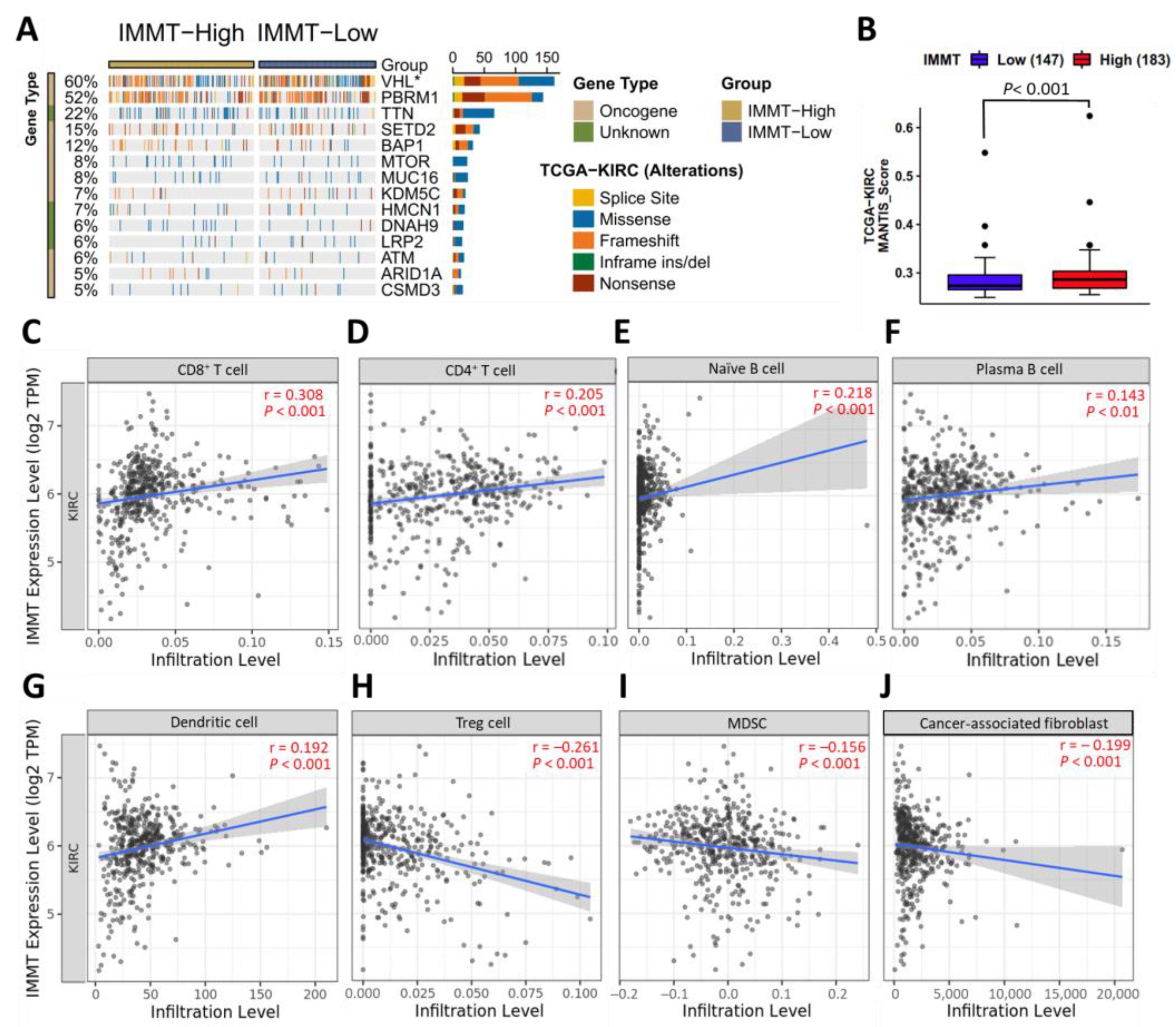
2.4. Low IMMT Expressions Are Implicated in Reduced Immunogenicity and an Immunosuppressive TIME
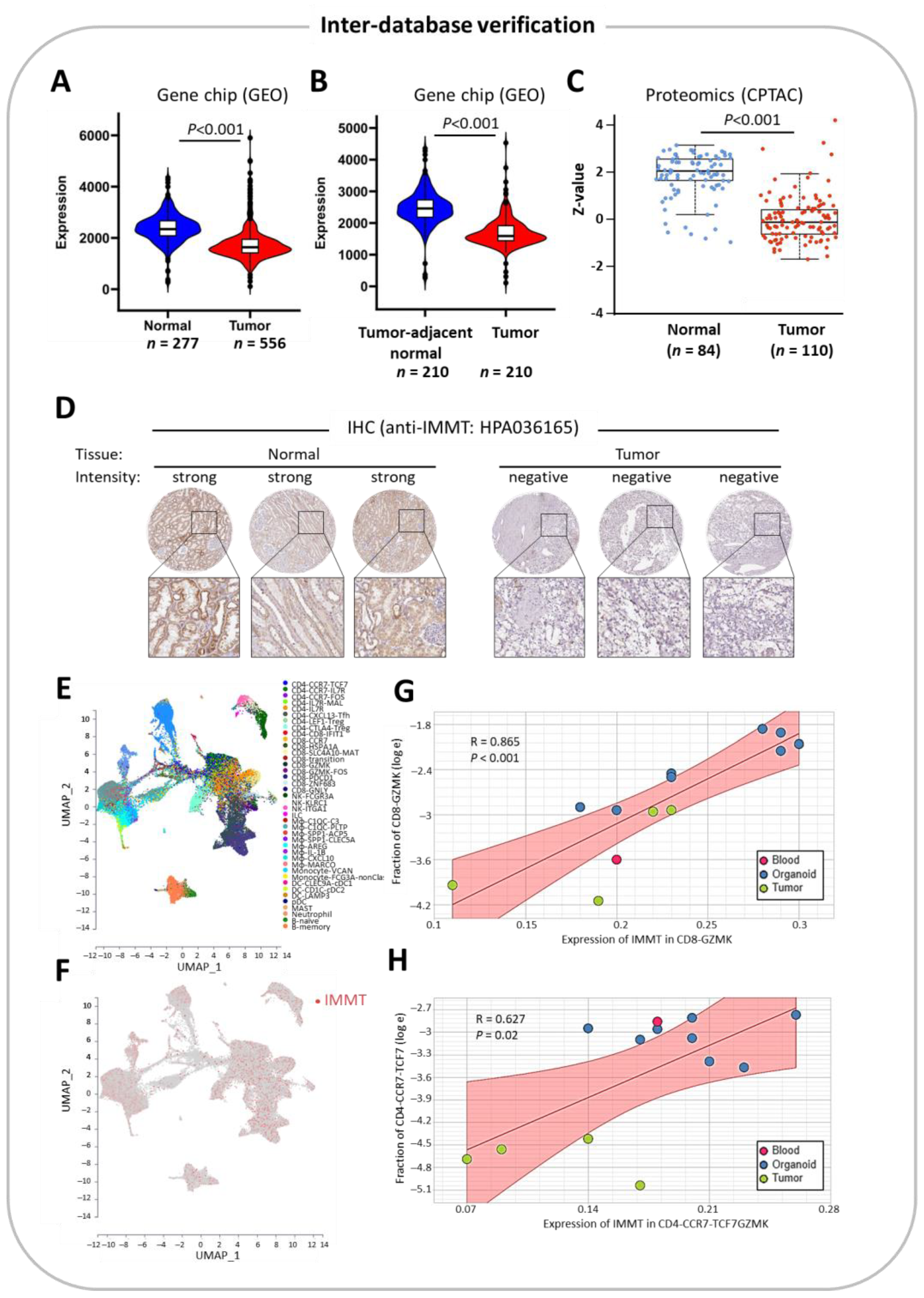
2.5. Inter-Database Verification
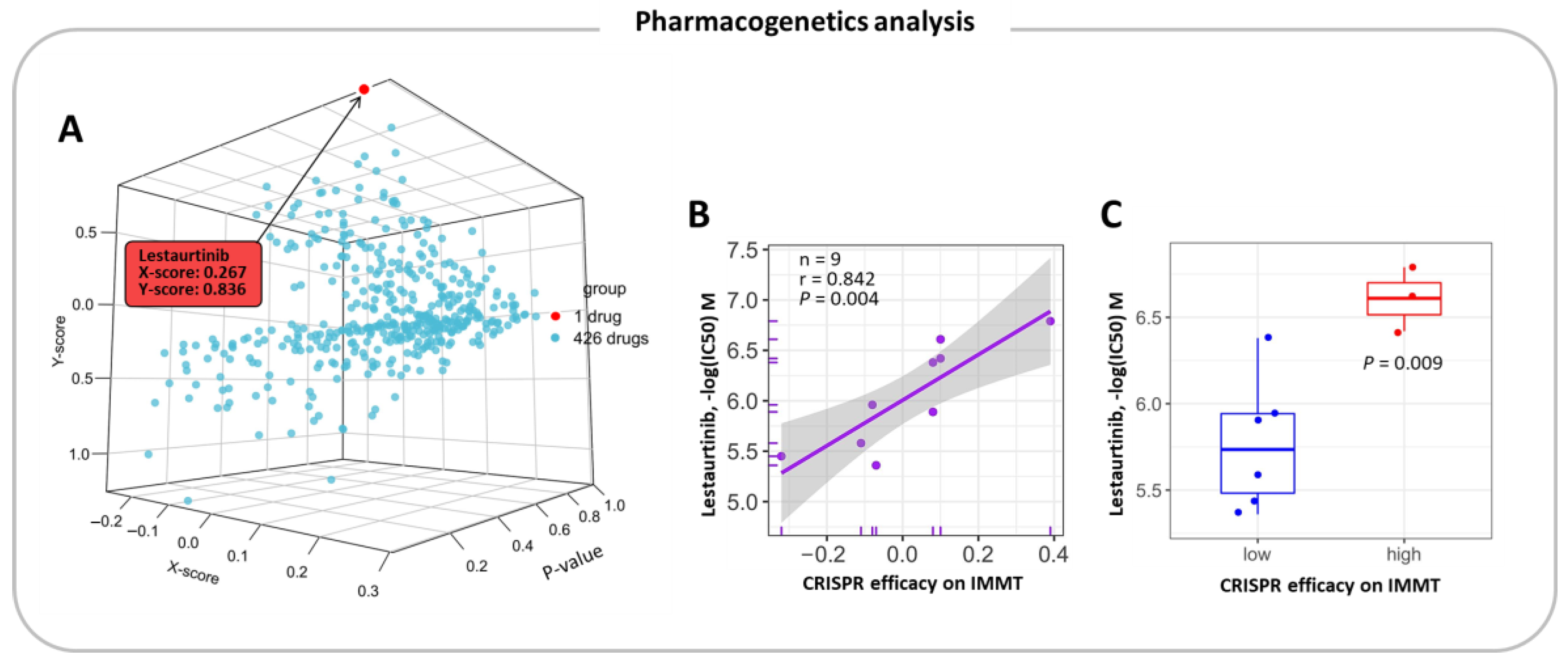
2.6. Pharmacogenetic Prediction for Potent Drugs
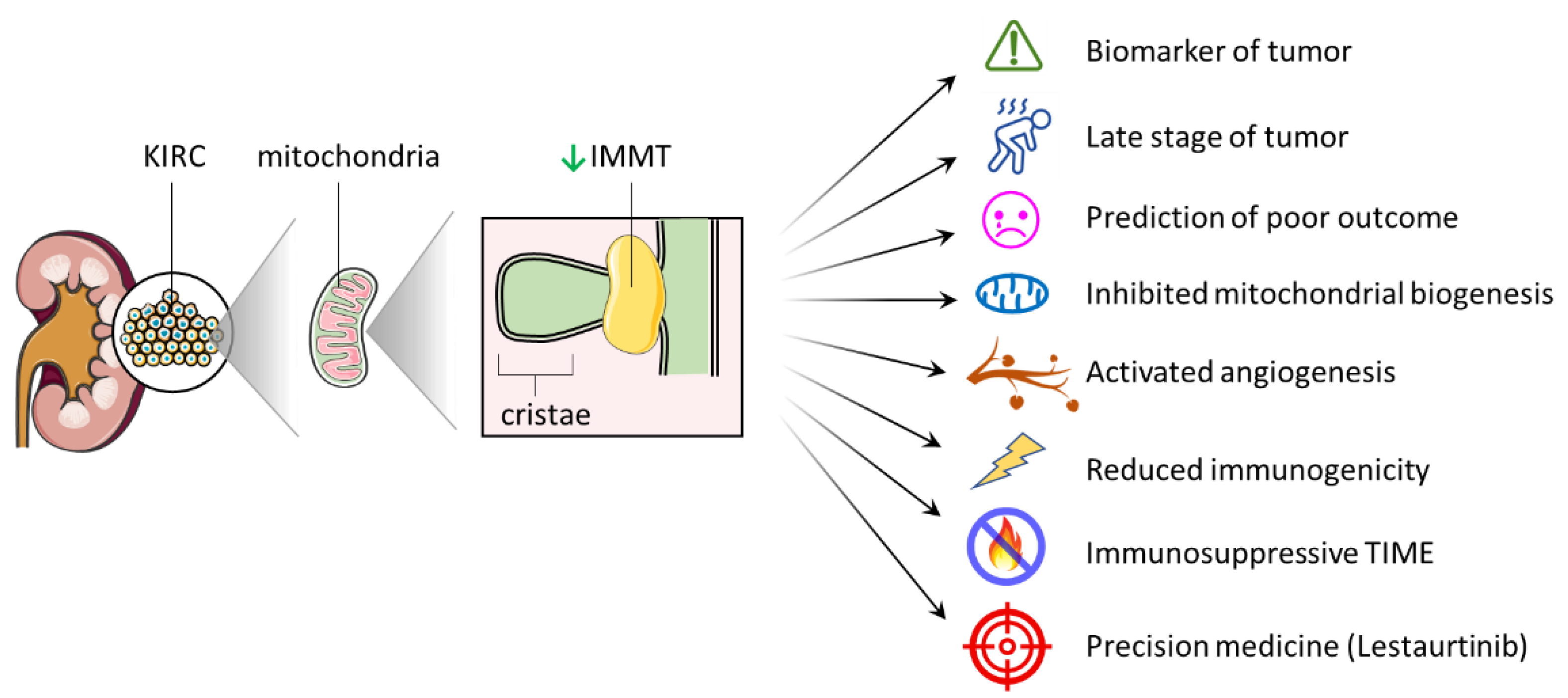
3. Discussion
4. Materials and Methods
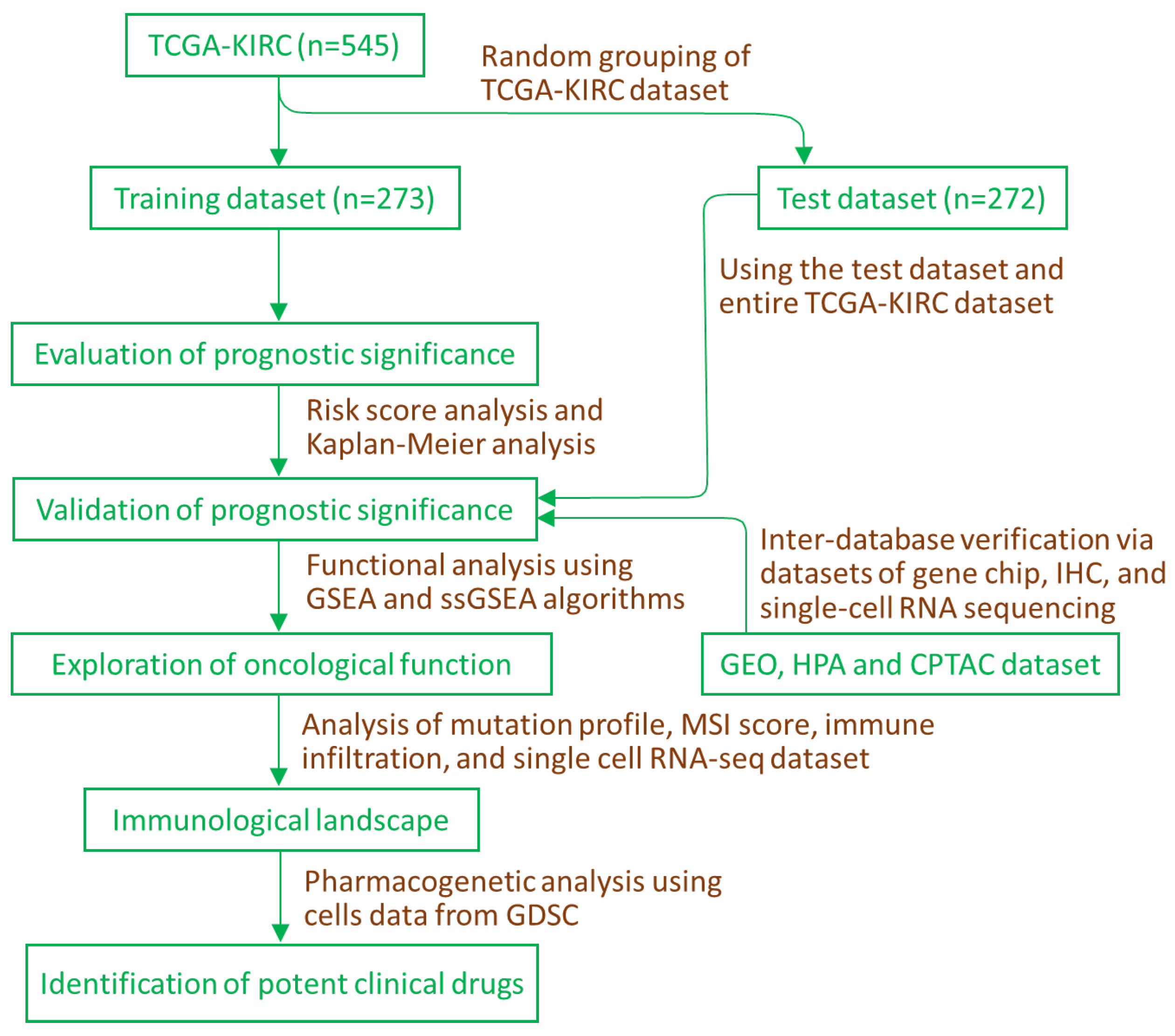
4.1. Data Sources and Supervised Learning Approach
4.2. Analysis of Predictive Significance
4.3. Functional Enrichment Analysis
4.4. Immunogenicity, Immunological Landscape and Single-Cell Analysis
4.5. Pharmacogenetic Prediction
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Owens, B. Kidney cancer. Nature 2016, 537, S97. [Google Scholar] [CrossRef] [PubMed]
- Turajlic, S.; Swanton, C.; Boshoff, C. Kidney cancer: The next decade. J. Exp. Med. 2018, 215, 2477–2479. [Google Scholar] [CrossRef]
- Odgren, P.R.; Toukatly, G.; Bangs, P.L.; Gilmore, R.; Fey, E.G. Molecular characterization of mitofilin (HMP), a mitochondria-associated protein with predicted coiled coil and intermembrane space targeting domains. J. Cell Sci. 1996, 109 Pt 9, 2253–2264. [Google Scholar] [CrossRef] [PubMed]
- Kondadi, A.K.; Anand, R.; Reichert, A.S. Cristae Membrane Dynamics—A Paradigm Change. Trends Cell Biol. 2020, 30, 923–936. [Google Scholar] [CrossRef]
- Feng, Y.; Imam Aliagan, A.; Tombo, N.; Draeger, D.; Bopassa, J.C. RIP3 Translocation into Mitochondria Promotes Mitofilin Degradation to Increase Inflammation and Kidney Injury after Renal Ischemia-Reperfusion. Cells 2022, 11, 1894. [Google Scholar] [CrossRef]
- Ghosh, J.C.; Perego, M.; Agarwal, E.; Bertolini, I.; Wang, Y.; Goldman, A.R.; Tang, H.Y.; Kossenkov, A.V.; Landis, C.J.; Languino, L.R.; et al. Ghost mitochondria drive metastasis through adaptive GCN2/Akt therapeutic vulnerability. Proc. Natl. Acad. Sci. USA 2022, 119, e2115624119. [Google Scholar] [CrossRef]
- Lin, H.Y.; Wu, H.J.; Chu, P.Y. Multi-omics and experimental analysis unveil theragnostic value and immunological roles of inner membrane mitochondrial protein (IMMT) in breast cancer. J. Transl. Med. 2023, 21, 189. [Google Scholar] [CrossRef]
- Petitprez, F.; Ayadi, M.; de Reynies, A.; Fridman, W.H.; Sautes-Fridman, C.; Job, S. Review of Prognostic Expression Markers for Clear Cell Renal Cell Carcinoma. Front. Oncol. 2021, 11, 643065. [Google Scholar] [CrossRef]
- Bonneville, R.; Krook, M.A.; Kautto, E.A.; Miya, J.; Wing, M.R.; Chen, H.Z.; Reeser, J.W.; Yu, L.; Roychowdhury, S. Landscape of Microsatellite Instability Across 39 Cancer Types. JCO Precis. Oncol. 2017, 2017, 1–15. [Google Scholar] [CrossRef]
- Casuscelli, J.; Vano, Y.A.; Fridman, W.H.; Hsieh, J.J. Molecular Classification of Renal Cell Carcinoma and Its Implication in Future Clinical Practice. Kidney Cancer 2017, 1, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Kossenkov, A.V.; Milcarek, A.; Notta, F.; Jang, G.H.; Wilson, J.M.; Gallinger, S.; Zhou, D.C.; Ding, L.; Ghosh, J.C.; Perego, M.; et al. Mitochondrial fitness and cancer risk. PLoS ONE 2022, 17, e0273520. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Cho, M.; Wang, X. OncoDB: An interactive online database for analysis of gene expression and viral infection in cancer. Nucleic Acids Res. 2022, 50, D1334–D1339. [Google Scholar] [CrossRef] [PubMed]
- Thul, P.J.; Akesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Bjork, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356, eaal3321. [Google Scholar] [CrossRef]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjostedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, 2507. [Google Scholar] [CrossRef]
- Bartha, A.; Gyorffy, B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef]
- Vasaikar, S.V.; Straub, P.; Wang, J.; Zhang, B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018, 46, D956–D963. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Vasaikar, S.; Shi, Z.; Greer, M.; Zhang, B. WebGestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017, 45, W130–W137. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Severson, E.; Pignon, J.C.; Zhao, H.; Li, T.; Novak, J.; Jiang, P.; Shen, H.; Aster, J.C.; Rodig, S.; et al. Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol. 2016, 17, 174. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef] [PubMed]
- Hong, F.; Meng, Q.; Zhang, W.; Zheng, R.; Li, X.; Cheng, T.; Hu, D.; Gao, X. Single-Cell Analysis of the Pan-Cancer Immune Microenvironment and scTIME Portal. Cancer Immunol. Res. 2021, 9, 939–951. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kim, Y.; Jin, S.; Yoo, H.; Jeong, S.; Jeong, E.; Yoon, S. Q-omics: Smart Software for Assisting Oncology and Cancer Research. Mol. Cells 2021, 44, 843–850. [Google Scholar] [CrossRef]
- Chang, K.T.; Wu, H.J.; Liu, C.W.; Li, C.Y.; Lin, H.Y. A Novel Role of Arrhythmia-Related Gene KCNQ1 Revealed by Multi-Omic Analysis: Theragnostic Value and Potential Mechanisms in Lung Adenocarcinoma. Int. J. Mol. Sci. 2022, 23, 2279. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.-C.; Chu, P.-Y.; Lin, H.-Y. Supervised Learning and Multi-Omics Integration Reveals Clinical Significance of Inner Membrane Mitochondrial Protein (IMMT) in Prognostic Prediction, Tumor Immune Microenvironment and Precision Medicine for Kidney Renal Clear Cell Carcinoma. Int. J. Mol. Sci. 2023, 24, 8807. https://doi.org/10.3390/ijms24108807
Chen C-C, Chu P-Y, Lin H-Y. Supervised Learning and Multi-Omics Integration Reveals Clinical Significance of Inner Membrane Mitochondrial Protein (IMMT) in Prognostic Prediction, Tumor Immune Microenvironment and Precision Medicine for Kidney Renal Clear Cell Carcinoma. International Journal of Molecular Sciences. 2023; 24(10):8807. https://doi.org/10.3390/ijms24108807
Chicago/Turabian StyleChen, Chun-Chi, Pei-Yi Chu, and Hung-Yu Lin. 2023. "Supervised Learning and Multi-Omics Integration Reveals Clinical Significance of Inner Membrane Mitochondrial Protein (IMMT) in Prognostic Prediction, Tumor Immune Microenvironment and Precision Medicine for Kidney Renal Clear Cell Carcinoma" International Journal of Molecular Sciences 24, no. 10: 8807. https://doi.org/10.3390/ijms24108807
APA StyleChen, C.-C., Chu, P.-Y., & Lin, H.-Y. (2023). Supervised Learning and Multi-Omics Integration Reveals Clinical Significance of Inner Membrane Mitochondrial Protein (IMMT) in Prognostic Prediction, Tumor Immune Microenvironment and Precision Medicine for Kidney Renal Clear Cell Carcinoma. International Journal of Molecular Sciences, 24(10), 8807. https://doi.org/10.3390/ijms24108807





