Abstract
The erythropoietin receptor (EPOR) is a transmembrane type I receptor with an essential role in the proliferation and differentiation of erythroid progenitors. Besides its function during erythropoiesis, EPOR is expressed and has protective effect in various non-hematopoietic tissues, including tumors. Currently, the advantageous aspect of EPOR related to different cellular events is still under scientific investigation. Besides its well-known effect on cell proliferation, apoptosis and differentiation, our integrative functional study revealed its possible associations with metabolic processes, transport of small molecules, signal transduction and tumorigenesis. Comparative transcriptome analysis (RNA-seq) identified 233 differentially expressed genes (DEGs) in EPOR overexpressed RAMA 37-28 cells compared to parental RAMA 37 cells, whereas 145 genes were downregulated and 88 upregulated. Of these, for example, GPC4, RAP2C, STK26, ZFP955A, KIT, GAS6, PTPRF and CXCR4 were downregulated and CDH13, NR0B1, OCM2, GPM6B, TM7SF3, PARVB, VEGFD and STAT5A were upregulated. Surprisingly, two ephrin receptors, EPHA4 and EPHB3, and EFNB1 ligand were found to be upregulated as well. Our study is the first demonstrating robust differentially expressed genes evoked by simple EPOR overexpression without the addition of erythropoietin ligand in a manner which remains to be elucidated.
1. Introduction
EPOR cytokine receptor occurs in diverse types of cells, mainly erythropoietic progenitors and several types of non-hematopoietic tissues as well as cancer cells. In early erythroblasts, EPOR holds the position of regulator of cell size and cell cycle duration [1], and induces transcriptional reprogramming throughout their maturation [2]. EPOR and its tissue-protective effect was described in nervous system, retina, heart, kidneys, endothelium, muscle and bone tissues [3,4,5,6,7,8,9]. EPO/EPOR axis is also implied in regulation of energy metabolism, obesity and insulin response [10,11]. As a typical member of the type I cytokine receptor superfamily with no intrinsic kinase activity, the EPOR molecule consists of extracellular, transmembrane and intracellular regions. Part of the EPOR intracellular region named cytoplasmic box1 is constitutively associated with JAK2 kinases [12] and after binding of the EPO ligand, a cascade of phosphorylations is activated. Activation of the EPO/EPOR complex triggers signaling cascades JAK2/STAT5, PI3K/AKT, RAS/MAP and PKC signaling pathways [13]. It was reported that for successful signal transmission, EPOR translocates to membrane lipid microdomains along with JAK2, STAT5 and LYN kinase [14]. After these events, the signal translocates to the nucleus, where transcription of EPO responsive target genes is turned on. To date, the overall understanding of EPOR effects on the biological processes in the cells is still missing. Global transcriptome and proteome studies were formerly applied to examine EPO/EPOR effects during embryonic and adult erythropoiesis [15,16]. Lately, phospho-proteome analysis of erythroid progenitors detected 121 novel EPO/EPOR target proteins, including core regulators of metabolic processes aldolase and pyruvate dehydrogenase-α1, and also phosphorylation of 19 novel cytoskeletal targets [17]. Although EPOR signaling is well described in a variety of tissues and cells [13,14] studies dealing with EPOR-induced DEGs in malignant cells are completely missing. EPOR plays an important role in the progression of numerous cancers and can be used as a prognostic marker [18], but the precise mechanism of EPOR action on cancer cells needs to be further investigated. In breast cancer, its expression on the surface of tumor cells is associated with better survival, proliferation and invasiveness [19].
The objective of the present study was to assess the effects of EPOR on the whole transcriptome and proteome changes and interpret these results with regard to cell biological processes. For our purpose, we used rat mammary adenocarcinoma RAMA 37-28 cell line with stable EPOR overexpression, which is a subclone of benign non-invasive mammary rat cell line RAMA 37 [3]. Using RNA-seq and proteomics, a global picture of signaling events in RAMA 37-28 vs. RAMA 37 cells has been obtained, in which 233 DEGs were affected. Subsequently, the accuracy of the expression profile of 13 DEGs from RNA-seq result was verified using qRT-PCR. The main goal of our study was to reveal new candidate genes associated with EPOR expression and to categorize them according to their biological processes.
2. Results
2.1. Differentially Expressed Genes and Validation
We used RNA-seq analysis to understand the molecular changes occurring after the introduction and overexpression of human EPOR in the mammary adenocarcinoma cell line RAMA 37. RNA isolated from both the EPOR overexpressed clone RAMA 37-28 and the parental line RAMA 37 was evaluated for its quality. cDNA libraries were prepared from biological replicates that had an optimal fragment size between 150–300 nt. Sequencing yielded 10 million raw reads per sample of both the RAMA 37 and RAMA 37-28 lines. A total of 15,177 genes were mapped for each sample. Genes with a minimum mean logCPM (counts per million) of 3 were considered differentially expressed and included in the differential expression analysis. Genes with logFC (fold change) in the range above ±1.2 were included in the final list of differentially expressed genes (DEGs). Additionally, we checked the p-value for shortlisted DEGs and removed any gene with p-value > 0.01 (Supplementary Figure S1).
A total of 233 genes were differentially expressed in RAMA 37-28 versus RAMA 37 cells (Supplementary Table S1). Among them, 145 were downregulated and 88 were upregulated. To validate the results obtained from RNA-seq, 13 DEGs were analyzed by qRT-PCR. The correlation of both results was carried out by calculating the Pearson correlation coefficient, the value of which was equal 0.943 (p-value = 1.3 × 10−7) (Figure 1). DEGs were divided based on GO biological processes using the peer-reviewed server Omnalysis (http://lbmi.uvlf.sk/omnalysis.html, accessed on 15 February 2023) (Figure 2).
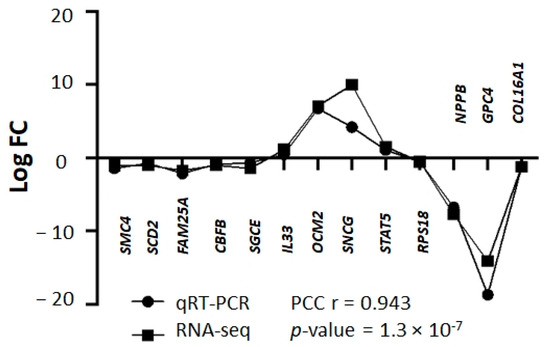
Figure 1.
Validation of RNA-seq data using qRT-PCR. Squares bar—logFC values of RNA-seq, bullets bar—logFC values of qRT-PCR. Pearson correlation coefficient (PCC) r = 0.943; p-value = 1.3 × 10−7. Please note that standard deviation (SD) is not shown here as logFC values of DEGs and qRT-PCR were calculated based on the average CT values of triplicates. The representative results of 13 DEGs are shown.
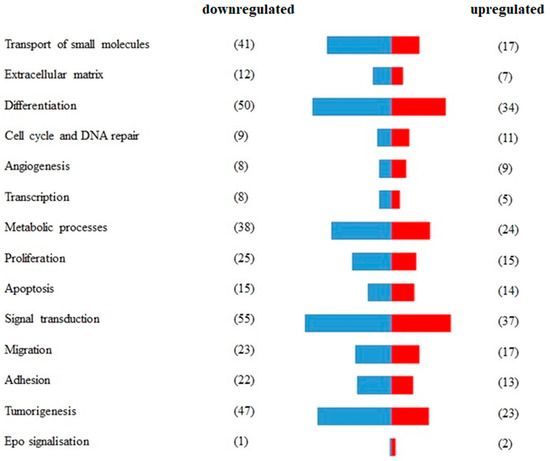
Figure 2.
Segregation of differentially expressed genes (DEGs) according to the GO biological processes. A total of 233 DEGs expressed in RAMA 37-28 versus RAMA 37 cells were validated and divided according to GO biological processes and using the peer-reviewed server Omnalysis. Blue bars—downregulated DEGs. Red bars—upregulated DEGs. Number of DEGs is displayed in parenthesis.
2.2. DEGs Involved in Erythropoietin Signaling
Three genes are associated with erythropoietin signaling, of which two are upregulated and one is downregulated (GO biological processes “erythropoietin signaling”). EPOR overexpression evoked the upregulation of STAT5A and LYN and downregulation of KIT.
2.3. DEGs Involved in Proliferation
Forty genes associate with cell proliferation (GO biological processes “cell proliferation”). In this regard, 25 genes were downregulated and 15 genes were upregulated as a result of EPOR overexpression in RAMA 37-28 cells. Twelve downregulated genes KIT (logFC −7.93), NPPB (logFC −7.71), SFRP2 (logFC −5.07), SLURP1 (logFC −2.28), XDH (logFC −2.71), NR4A1 (logFC −2.45), CLDN1 (logFC −2.07), SMAD3, IFT80, CDH3, ECM1 and RGCC and seven upregulated genes CDH13 (logFC 8.01), CXCL12 (logFC 2.28), TGFB2 (logFC 2.04), FST, FGFR2, STAT5A and SIX1 associate with negative regulation of epithelial cell proliferation. Furthermore, upregulated CDKN1A and PAK1 can associate with negative regulation of vascular-associated smooth muscle cell proliferation and/or negative regulation of cell proliferation involved in contact inhibition (Figure 3a).
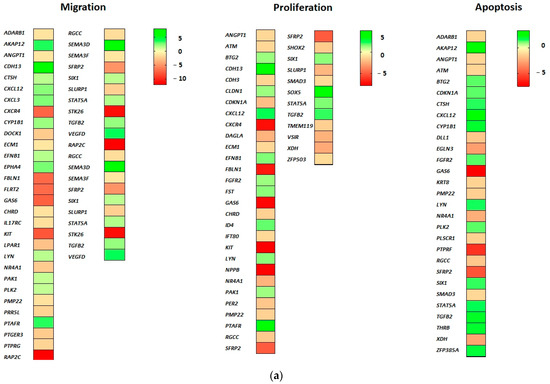
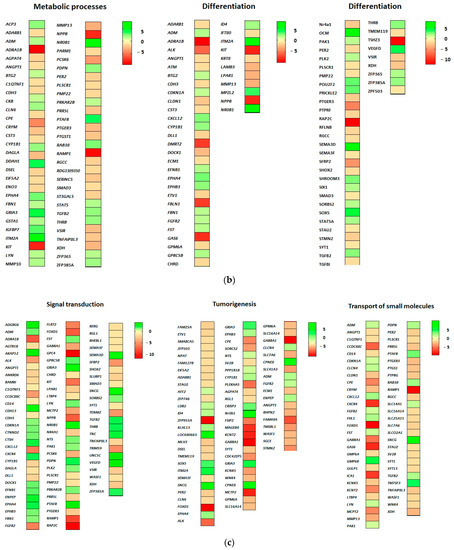
Figure 3.
(a) DEGs related to proliferation, apoptosis and cell migration. (b) DEGs related to metabolic processes and cell differentiation. (c) DEGs related to signal transduction, cell transport and tumorigenesis. Green-shaded genes—upregulated, red-shaded genes—downregulated. Shading intensity indicates the degree of upregulation or downregulation. Range of the fold change (logFC values) is presented in the scale.
2.4. DEGs Involved in Apoptosis
Twenty-nine genes were categorized either in the regulation of apoptotic signaling pathway or simply involved in the apoptotic process. Of these, 15 were downregulated and 14 were upregulated (GO biological process “apoptosis”). The most downregulated PTPRF (logFC −5.67), SFRP2 (logFC −5.07), EGLN3 (logFC −2.91), XDH (logFC −2.71) and NR4A1 (logFC −2.45) associate with negative regulation of both intrinsic and extrinsic apoptotic signaling pathways. Moreover, downregulated EGLN3, NR4A1 and SMAD3 can participate also in the inhibition of cysteine-type endopeptidase activity (involved in apoptotic process) and downregulated ANGPT1 and RGCC correlate with diminishing of both epithelial and endothelial apoptosis. In spite of the proapoptotic (tumor suppressor) potential of upregulated genes AKAP12 (logFC 3.36), TGFB2 and BTG2, the group of overexpressed genes CXCL12 (logFC 2.28), ZFP385A and CDKN1A associate with negative regulation of the intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator (Figure 3a).
2.5. DEGs Involved in Cell Migration
Forty genes associate with migration, of which 23 are downregulated and 17 upregulated (GO biological processes “cell migration”). Despite EPOR-induced upregulated genes such as TGFB2, PLK2, CTSH, AKAP12, PAK1, VEGFD and CXCL12, the presence of downregulated genes RAP2C (logFC −12.01), STK26 (logFC −11.04), KIT (logFC −7.93) and SFRP2 (logFC −5.07) implies an association with negative regulation of RAMA 37-28 cell migration (Figure 3a).
2.6. DEGs Involved in Metabolic Processes
Sixty-two genes associate with metabolic processes, of which 38 are downregulated and 24 upregulated (GO biological processes “metabolic processes”). Many metabolic processes could be regulated as a result of EPOR-induced differentially expressed genes in RAMA 37-28 cells. The most significant seems to be altered lipid metabolism, in which downregulated KIT (logFC −7.93), PMP22 and CLN6 and upregulated NR0B1 (logFC 7.70), IGFBP7, THRB, STAT5A, ADM and CYP1B1 associate with steroid metabolic process (GO biological processes “metabolic processes”). While upregulated LYN and RAB38 associate with the regulation of phospholipid and lipid metabolic processes, upregulated THRB could be directly involved in the regulation of triglyceride or cholesterol metabolic processes. In addition, PTGS1, GSTA1, PDPN, DAGLA and CYP1B1 genes associate with icosanoid, unsaturated fatty acid and long-chain fatty acid metabolism. In relation to apoptosis, the following associations between the metabolic processes and DEGs appear to be important. The first one is between the metabolism of reactive oxygen species and both downregulated XDH and SMAD3 and upregulated DDAH1 (logFC 3.67), CDKN1A and CYP1B1 genes. The second is between the metabolism of both xenobiotic and glutathione and the upregulated GSTA1 gene, and the last one is between diminished phosphatidylserine metabolism and downregulated PLSCR1 and SERINC5 genes (Figure 3b).
2.7. DEGs Involved in Cell Differentiation
Eighty-four genes associate with cell differentiation, of which 50 are downregulated and 34 upregulated (GO biological processes “cell differentiation”). A large group of genes associates with the regulation of cell morphogenesis involved in cell differentiation including upregulated OCM2 (logFC 7.08), SEMA3D (logFC 6.08), EPHA4, CXCL12, PDPN, PAK1, EPHB3, LYN and GPRC5B and downregulated CXCR4 (logFC −7.30), FBLN1 (logFC −7.08), PTPRF (logFC −5.97), ALK (logFC −5.57), LPAR1, DOCK1, STMN2, SHOX2, STAU2 and SYT1 and SEMA2F (GO biological processes “cell differentiation”). Upregulated genes CDKN1A, SHROOM3 and FGFR2 associate also with columnar and/or cuboidal epithelial cell differentiation. Indeed, many genes evoked by EPOR overexpression correlate with differentiation of muscle, neuronal and other cells, e.g., TSHZ3 (logFC −11.34), ADRA1B (logFC −9.27), TMEM119, KIT, PMP2, NPPB, KRT8, SORBS2, RAMP2, SIX1, ADM and DLL1, but their role in the differentiation of RAMA 37-28 cells remains unclear and requires additional studies (Figure 3b).
2.8. DEGs Involved in Signal Transduction
Ninety-two genes associate with signal transduction, of which 55 are downregulated and 37 upregulated (GO biological processes “signal transduction”). Most signaling pathways in RAMA 37-28 cells appear to be downregulated based on changes in the expression of categorized genes (GO biological processes “signal transduction”). In this case, both small GTPase-mediated signal transduction via downregulated genes RAP2C (logFC −12.01), ADRA1B (logFC −9.27), CCDC88C (logFC −5.63), RGL1, WASF1, RHEBL1, LPAR1, DOCK1 and RERG and Ras signal transduction via downregulated SFRP2 (logFC −5.07), ANKRD6, SMAD3, BAMBI and CDH3 are inhibited. Except for upregulated STAT5A, cytokine-mediated signaling is also downregulated via reduced expression of KIT (logFC −7.93), GAS6 (logFC −7.53), CXCR4 (logFC −7.30), PTPRF (logFC −5.97), IL17RC, KRT8, ANGPT1 and ECM1 genes. Finally, the intrinsic apoptotic signaling pathway in response to DNA damage is also negatively regulated based on downregulated PLSCR and ATM and upregulated CDKN1A, CYP1B1, CXCL12 and ZFP385A genes. On the contrary, the combination of upregulated CTNND2, GPRC5B and FGFR2 and downregulated CCDC88C and ANKRD6 associate with positive regulation of Wnt signaling. Interestingly, ephrin receptor signaling pathway with three upregulated EFNB, EPHA, EPHB3 is also positively regulated in RAMA 37-28 cells (Figure 3c).
2.9. DEGs Involved in Cell Transport
According to GO biological processes “transport of small molecules”, 58 genes were found, of which 41 are downregulated and 17 upregulated. The majority of EPOR-evoked genes in RAMA 37-28 cells associate with the regulation of transmembrane protein, hormone and ion transport. While in protein transport, PRR5L, MMP13, CD14, RGCC and GAS6 were downregulated and TM7SF3, TGFB2 and PAK1 were upregulated; in hormone transport, C1QTNF1, ICA1, PTGER3, PER2, LTBP4, FOXD1, CPE and CRYM were downregulated and LYN, FST and ADM upregulated. The most significantly downregulated genes associated with protein and hormone transport were FOXD1 (logFC −9.13) and GAS6 (logFC −7.53), respectively, whereas the TM7SF3 gene was significantly upregulated (logFC 4.34) in both transports. In the case of ion transmembrane transporter activity regulation, downregulated PTGER3, GPM6B, KCNT2, WNK4, KCNK5 and SYT1 with most significant RAMP1 (logFC −8.68) and CXCR4 (logFC −7.31) and upregulated FHL1, PTAFR, CXCL12, TGFB2 and LYN genes were categorised. The four abovementioned TGFB2, GPM6B, PTGER3 and WNK4 genes associate also with negative regulation of calcium transmembrane transport (Figure 3c).
2.10. DEGs Involved in Tumorigenesis
Seventy genes associate with tumorigenesis, of which 47 are downregulated and 23 upregulated (GO biological processes “tumorigenesis”). Among them are 15 oncogenes and 10 tumor suppressors which can contribute to the development of a malignant phenotype of RAMA 37-28 cells. Moreover, overexpression of subset of genes was found to be associated with breast cancer tumorigenesis (ID4, SOX5, ITM2A, SNCG, EPHA4, NTS, CYP1B1, FGFR2, ENPEP), whereas ID4, SEMA3F, FOXD, KCNK5, WNK4 and ECM1 genes may play a role in the chemoresistance of cancer cells. Interestingly, upregulated DEGs like SNCG (logFC 9.95), CPNE8 (logFC 7.18), GRIA3 (logFC 4.08), SOX5 (logFC 3.94) and CRISP3 (logFC 3.44) representing the highest log fold changes were involved in tumor cell metastasis and invasiveness. Transcriptome analysis revealed also upregulated RADX (logFC 9.92) and downregulated MLH6 (logFC −2.00), NPAT, ATM and SMARCA5 genes associated with DNA damage response (Figure 3c).
2.11. Proteomic Analysis and Validation by Western Blot
Proteome analysis of more than 5000 peptides confirmed the altered expression of OCM2 (logFC 7.43) and SNCG (logFC 2.08) genes from the transcriptome analysis, but also revealed new DEGs and/or proteins such as ALDH6 (logFC 5.51), SPRR1B (logFC 2.64), ZYX (logFC 2.28), CTPB (logFC 4.62), HPRT (logFC −5.31) and hypothetical protein MJ0443 (logFC −3.51) (Supplementary Figure S2). To validate the results obtained from proteomic analysis, five representative proteins were analyzed using Western blot. Results obtained from both techniques were consistent (Supplementary Figure S3). EPOR-induced changes were confirmed by using specific siRNA designed against EPOR mRNA. In addition, 48 h silencing of EPOR did not abolish the expression of selected STAT5A and OCM2 genes; however, it significantly reduced their protein levels. On the contrary, EPOR silencing did not increase the expression of downregulated HPRT (Figure 4).
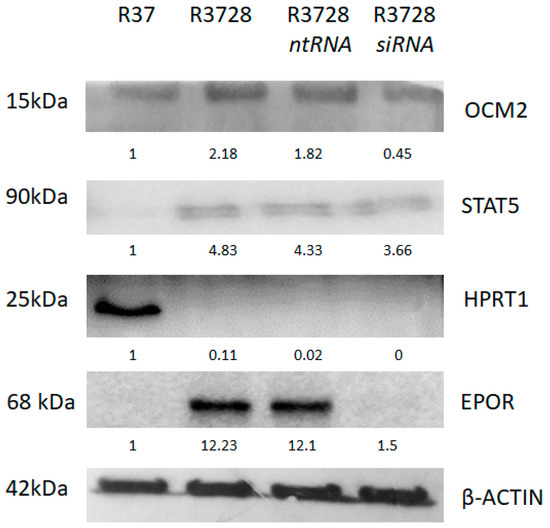
Figure 4.
Western blot analysis of representative proteins (OCM2, STAT5A, HPRT1, EPOR) as validation of proteomic data. EPOR-induced changes were confirmed by specific siRNA designed against EPOR mRNA.
3. Discussion
EPOR was discovered in 1989 [20] and has been intensively studied for over 30 years. Activation of EPO-EPOR cascades during erythropoiesis results in transcriptional activation of proliferation and differentiation [2], while in non-hematopoietic tissues, anti-apoptotic and protective mechanism of EPOR signaling is described [21,22,23]. The expression of EPOR has been demonstrated in a panel of 29 tumor cell lines [24] and may be related to increased resistance of cancer cells to various therapies. In the current study, we describe EPOR as a causative factor of extensive expression changes referred to 233 DEGs in RAMA 37-28 cell line, affecting numerous biological functions and resulting in the origin of a new cell identity. The genes obtained from RNA-seq have been processed and clustered into particular biological categories (GO biological processes). In our previous studies, we have shown that RAMA 37-28 cells have lower proliferation capacity compared to RAMA 37 cells, as a result of EPOR overexpression [25,26] (Supplementary Figure S4). Currently, we used xCelligence screening to show faster proliferation of RAMA 37-28 cells under conditions of paclitaxel added, whereas EPOR silencing suppressed such an effect (Supplementary Figure S5).
According to our transcriptome analysis, EPOR overexpression in RAMA 37-28 cell line results in 40 DEGs associated with cell proliferation. Indeed, 12 downregulated genes such as KIT, NPPB, SFRP2, SLURP1, XDH, NR4A1, CLDN1, SMAD3, IFT80, CDH3, ECM1 and RGCC and seven upregulated genes, CDH13, CXCL12, TGFB2, FST, FGFR2, STAT5A, SIX1, are involved in the inhibition of epithelial cell proliferation. In this regard, STAT5A, as a member of JAK2/STAT5 signaling cascade regulated by EPO/EPOR activation, is identified as a chemoresistance inducer and the regulator of ABCB1 transporter protein [27] with the ability to stabilize also heterochromatin structure followed by the inhibition of cell growth [28]. We demonstrate EPOR-induced overexpression of STAT5A, whose direct relationship (without EPO addition) using siRNA against EPOR was confirmed. Upregulated TGFB itself encodes the transforming growth factor beta family of cytokines which functions in proliferation, differentiation, adhesion and migration in many cell types [29]. TGFB receptors, moreover, activate both SMAD-dependent and independent pathways that not only regulate SMAD signaling, but also allow SMAD-independent TGFB responses [30]. The data of Livitsanou et al. [31] demonstrated a novel mechanism of TGFB/SMAD signaling modulation by the small GTPase RHOB and show that TGFB/RHOB signaling cross talk affects the nuclear and cytoplasmic responses to TGFB in opposite ways. The slower proliferation of our RAMA 37-28 cells could therefore be explained by the downregulation of SMAD3 and the upregulation of TGBF2 and RHOB genes. Furthermore, Marlow et al. [32] showed that RHOB has divergent downstream signaling partners, which are dependent on the HDAC isoform that is inhibited. When RHOB upregulates only P21 using a class I HDACi (romidepsin), cells undergo cytostasis. When RHOB upregulates BIM using class II/(I) HDACi (belinostat or vorinostat), apoptosis occurs. Combinatorial synergy with paclitaxel is dependent upon RHOB and BIM while upregulation of RHOB and only P21 blocks synergy. Interestingly, RAMA 37-28 cells which demonstrate paclitaxel resistance compared to RAMA 37 cells [26] reveal overexpression of both RHOB and CDKN1A genes. In search of mechanisms responsible for the proliferative changes of RAMA 37-28 cells, we emphasize the overexpression of IGFBP7 and simultaneous downregulation of NPPB genes. Indeed, it has been reported that IGFBP7, involved in p53-dependent growth suppression of lung and colorectal tumors [33,34], decreased the production of NPPB during viral tumorigenesis [35], which points to the conditional expression of some DEGs. Moreover, overexpressed P21 activated PAK1 may also be involved in the negative regulation of cell proliferation involved in the contact inhibition together with downregulated CLDN-1, whose lower expression is either associated with cancer progression (invasion) or improved survival of cancer patients [36]. Highly downregulated proto-oncogene and receptor tyrosine kinase KIT itself might be another important factor contributing to both neoplastic breast epithelium transformation and inhibition of cell migration which were already demonstrated by Janostiak et al. [37].
Furthermore, we demonstrate the upregulation of ephrin receptors (EPHA4, EPHB3) and ligand (EFNB1) which can associate with a slowdown in RAMA 37-28 proliferation. While EPHA4 signaling promotes mesenchymal-to-epithelial transition during morphogenesis in zebra fish embryo development [38], it was reported that overexpression of EPHB3 enhances cell–cell contacts and inhibits the growth in HT-29 human colon cancer cells [39]. EFNB1 plays important role in cell adhesion in healthy tissues [40], while in tumors, it inhibits cell proliferation and adhesion [41].
Interestingly, overexpression of EPOR is associated with the emergence of a malignant phenotype in RAMA 37-28 cell line [42]. In our previous studies, overexpression of EPOR in RAMA 37-28 breast cancer cells altered the sensitivity of these cells to tamoxifen and paclitaxel via mechanism related to prolonged activation of AKT and ERK1/2 pathways, respectively [25,26]. Acquisition of chemotherapy-resistant cancer phenotype of RAMA 37-28 cells may also associate with the overexpression of widely described cell cycle regulator CDKN1A (P21Waf1/Cip1) [43], which can induce cell cycle arrest, followed by reduction of cancer cell growth rate. Indeed, cytoplasmic P21 phosphorylated by AKT increases cell survival and contributes to taxol resistance in glioblastoma cells [44], cisplatin resistance in testicular and ovarian cancer [45,46], doxorubicin resistance in triple-negative breast cancer cells SUM159 [47] and failure of paclitaxel treatment in human nasal squamous carcinoma RPMI-2650 [48]. In addition, P21 mediates 5-fluorouracil resistance in colorectal cancer cells [49] and is associated with poor response to tamoxifen in MCF7 breast cancer cells [50]. CDKN1A overexpression drives cells to acquire a more aggressive phenotype that is capable of escaping cell block, senescence and apoptosis [43]. Equally important in the resistance of RAMA 37-28 cells to tamoxifen alone is PAK1, whose overexpression and the role in tamoxifen resistance of breast cancer patients has already been described [51]. Besides this, ID4, SEMA3F, FOXD, KCNK5, WNK4 and ECM1 associate with chemoresistance origin and SOX5, ITM2A, SNCG, EPHA4, NTS, FGFR2 and ENPEP associate moreover with breast cancer tumorigenesis. The most significantly upregulated DEGs such as SNCG, CPNE8, GRIA3, SOX5 and CRISP3 can be involved in metastasis and invasivity of cancer cells as well. In this regard, SNCG is overexpressed in human infiltrating breast carcinomas and promotes metastasis [52], while it is undetectable in normal and benign breast tissues [53]. Tian et al. [54] indicated a possible relationship between SNCG and P21 and their relation to radioresistance. We show that EPOR evoked upregulation of SNCG (both mRNA and protein levels) in RAMA 37-28 cells and suppose that the overexpression of both SNCG and P21 genes could play a significant role in the chemoresistance phenotype of RAMA 37-28 cells.
The sensitivity of a tumor cell to therapy can also be affected by changes in the cell’s metabolism [55]. Our transcriptome analysis reveals 62 DEGs associated with such a biological property and genes such as XDH, CYP1B1, GSTA1, PLSCR1 and DDAH1 are definitely genes worth mentioning. In addition, proteomic analysis enriched this category with ALDH6 and HPRT1 proteins. The expression of XDH inversely correlates with the expression of cancer stem cell–related genes, such as CD44 or CD133, and their downregulation promotes TGFβ signaling in hepatocellular carcinoma [56]. It is noteworthy that knocking-down or inhibiting XDH results in development and progression of hepatocellular HCC and promotes migration and invasion but not proliferation of these cells, while the changes themselves are dependent on increased activation of TGFB2/SMAD2/3 signaling pathway [56]. It seems that low activity of XDH provides a selective privilege to cancer cells and may also contribute to neoplastic differentiation of RAMA 37-28 cells. However, the main biological effects of TGFβ are inhibition or promotion of proliferation, differentiation, apoptosis, cell dormancy, autophagy and cellular senescence; reduced levels of SMAD3 in RAMA 37-28 cells can suppress the function of TGF-β-induced expression of tumor suppressor genes. So, downregulation of TGFB2 results in the expression of anti-apoptotic proteins BCL2 and BCL-W, and enhanced cancer cell survival, which confers platinum resistance in NSCLC and 5-FU resistance in CRC cells, respectively [57,58]. A similar relationship to resistance to cisplatin and paclitaxel chemotherapy was described in the case of GSTA1 and CYP1B1 genes, respectively [59,60]. Although both genes are overexpressed in RAMA 37-28 cells, the CYP1B1 could be one of the mechanisms explaining the resistance phenotype of RAMA 37-28 cells to paclitaxel [26]. The sensitivity and/or proliferative capacity of tumor cells can also be affected by phospholipid scramblase 1 (PLSCR1) which is involved in several processes including phosphatidylserine exposure on an apoptotic cell surface. Its upregulation promoted cell proliferation, invasion and migration, while its downregulation inhibited these effects [61]. On the contrary, downregulation of PLSCR1 expression, which is also seen in RAMA 37-28 cells, significantly inhibited the proliferation, adhesion, migration and invasion of cancer cells [62]. Another important gene that is involved in cellular metabolism and at the same time plays an important role in the altered phenotype of the tumor cell is DDAH1. The encoded enzyme plays a role in NO generation by regulating cellular concentrations of methylarginines, which in turn inhibits NO synthase activity. While endothelium-derived NO stimulates angiogenesis through the inhibition of apoptosis [63], in cancer cells, the roles of NO are diverse, and might have dual pro- and anti-tumor effects depending on local concentration [64]. Finally, the upregulated ALDH6 enzyme confirmed in RAMA 37-28 cells by proteome analysis may also play a significant role in the resistance [65] of our cells, while the role of our downregulated HPRT1 in relation to the published results [66] is questionable.
It is well known that the characteristic attribute of cancer cells is alteration of cell adhesion and/or extracellular matrix organization (ECM). According to DEGs identified by transcriptome analysis of RAMA 37-28 cells compared to RAMA 37 control, we suppose the loss of cell adhesion ability of RAMA 37-28 cells via plasma membrane adhesion molecules which are closely related to strong downregulation of morphogen and tumor suppressor glypican 4 (GPC4). The effect of GPC4 downregulation was earlier demonstrated in relation to breast tumor progression [67], as well as in healthy tissues, to disruption of epithelial integrity and tight junction organization [68]. GPCs have been shown as well-known and accepted cell surface coreceptors for growth factors such as Wnt/β-catenin, FGF, IGF, VEGF and TGFB and matrix modifying enzymes in many cancer cells, thus being involved in control of tumor growth, metastasis, angiogenesis and ECM [69,70,71]. Moreover, Varma et al. [72] showed that GPC4 mRNA level was downregulated in oxaliplatin-resistant ovarian carcinoma cell line A2780/C10. We demonstrate expression changes also in Frizzled-related protein 2 (SFRP2), which is secreted and incorporated into ECM of the normal and tumor cells. Its association with the fibronectin–integrin protein complex, promotion of cell adhesion and/or transformation of normal mammary epithelial cells into a tumor was already described earlier [73]. SFRP2 was downregulated in radiotherapy-treated glioma patients, and low SFRP2 expression was correlated with advanced tumor stage and poor prognosis. CRISP/Cas9-mediated SFRP2 knockdown promoted soft agar colony formation, cancer stemness and radioresistance of glioma cells, while increased SFRP2 expression exhibited opposite effects. Moreover, SFRP2 knockdown activated Wnt/β-catenin signaling in glioma cell lines, while overexpression of SFRP2 inhibited Wnt/β-catenin activation [74]. In the case of ECM assembly, downregulated LAMB3, SMAD3, RGCC and GAS6 and upregulated GPM6B were categorized. On the other hand, downregulated MMP13 and CST3 and upregulated TGFB2 and PDPN correlate with ECM disassembly. Interestingly, upregulated FBN1 associates with sequestering of TGFB in ECM. Many more DEGs of RAMA 37-28 cells are related to ECM remodeling and changes of cell adhesion, such as upregulated CYR61, MMP10, NOV, CTGF, TMEM47, CDH13, CTNND2, PAK1 and LOXL1 and downregulated FBLN1, CLDN1, ECM1, TENM2, RGD1561161, COL16a1, COLA3, SGCE, THBS2, PTPRF, MEPE, C1QTNF1, PLEKHA2, PCSK6, FLRT2 and GPM6A.
Not only alterations in cell growth and proliferation but also different morphology in RAMA 37-28 cells compared to parental RAMA 37 ones was observed [25,26]. Although the molecular basis of such difference has not been fully described, actually, 84 genes out of 233 associate with cell differentiation of RAMA 37-28 cells. The most significantly overexpressed gene in this category, both at the level of mRNA and the protein itself, is high-affinity calcium ion-binding protein (OCM2), which belongs to the superfamily of calmodulin proteins. We conclude and show for the first time that the expression of the OCM2 gene which associates with positive regulation of cell morphogenesis involved in differentiation (GO biological processes “cell differentiation”) is directly related to the expression of EPOR. This effect was confirmed by siRNA against EPOR. Indeed, high expression of OCM2 was demonstrated earlier in rat cancer cell lines undergoing neoplastic transformation [75,76]. Besides OCM2, 19 DEGs participate directly in the morphogenesis process. Thus, we demonstrate several upregulated DEGs related to columnar or cuboidal epithelial differentiation, namely, P21, SHROOM3 and FGFR2. A large body of evidence suggests that P21 also plays an important role in the differentiation of various normal or malignant cells and tissues, but its effect is dependent on the cell type and the stage of differentiation. Interestingly, P21 has a positive role in differentiation in most studies, partly by inhibiting apoptosis, which promotes cell survival [77]. The STAT5A protein itself plays a key role both in the positive regulation of myeloid cell differentiation and gamma-delta T cell differentiation, as well as in the negative regulation of erythrocyte differentiation, while its role in the differentiation of RAMA 37-28 cells remains unclear.
4. Materials and Methods
4.1. Culture of RAMA Cells
Rat mammary adenocarcinoma cell lines RAMA 37 and RAMA 37-28 were kindly provided by Dr. Mohamed El-Tanani (Center of Cancer Research and Cell Biology, Queens University Belfast, Belfast, UK). RAMA 37-28 cell line is a clone of parental RAMA 37 cells, with stabile overexpression of human EPOR using an expression vector pcDNA3.1/V5-His-TOPO and selection antibiotic Geneticin (Invitrogen, Carlsbad, CA, USA) (1 mg/mL). RAMA 37 and RAMA 37-28 cells were cultured in RPMI-1640 medium (Biosera, Cholet, France) supplemented by antibiotics (antibiotic-antimycotic solution 100×, Sigma-Aldrich, Darmstadt, Germany) and 10% of FCS (fetal calf serum) (Gibco, Carlsbad, CA, USA) in the presence of 5% CO2 in a humidified atmosphere at 37 °C. The 5 × 106 cells were plated onto a 75 cm2 flask; the culture reached 70–90% confluency in 2–3 days and was ready to split or harvest for experiments. To determine the linear range of each assay, six cell densities ranging from 50–10,000 cells/well were plated into sterile 96-well plates and incubated for 24, 48 or 72 h.
4.2. Preparation of the Library
A quantity of 250 ng of RNA was reverse transcribed to first strand cDNA using oligodT primers and QuantSeq. 3′ mRNA-Seq Library Prep Kit (Lexogen, Wien, Austria). RNA template was removed using RNA removal solution (RS buffer) (Lexogen, Wien, Austria) and the second strand was synthesized using random hexamer primer with Illumina-compatible linker sequences at its 5′ end. The dsDNA libraries were purified using magnetic beads and then amplified by PCR using specific single indexing i7 primers with the aim of adding adapter sequences important for cluster generation and to generate DNA for quality control and sequencing. PCR Add-on kit for Illumina (Lexogen, Wien, Austria) was used for the determination of 20 PCR cycles required for RAMA cells. Later, magnetic beads of the kit were used for purification of amplified libraries, which were checked for the length of the fragments by fragment analyzer.
4.3. Sequencing and Data Analysis
cDNA libraries were sequenced using Illumina NextSeq, single-end 75 bp, with 8 million reads per sample. Fastq files were processed and aligned to reference genome (Rattus norvegicus, mRatBN7.2) using STAR aligner (STAR V 2.5.2b, (https://github.com/alexdobin/STAR/releases/tag/2.5.2b, accessed on 20 February 2023). The pre-processing included adaptor trimming and removal of recommended initial 10 bases. Reads were counted in STAR V 2.5.2b. to perform differential gene expression analysis edgeR (open source R package version 3.12 was used: https://bioconductor.org/packages/release/bioc/html/edgeR.html, accessed on 20 February 2023). The low read count with less than 3 CPM (count per million) was filtered out using the filterByExp function of edgeR package. The identification of differentially expressed gene (DEGs) was accomplished by using glmTreat and glmQLFit (quasi-likelihood, QL) functions of edgeR in R package considering log fold change (logFC) values beyond ±1.2 and FDR less than 0.05. The logical relation of DEGs between the challenged RAMA cells (RAMA 37-28 vs. RAMA 37) was calculated using Excel (MS office) and Venn diagrams were constructed. To categorize DEGs groups into GO biological processes and to construct heat maps https://reactome.org/ (accessed on 20 February 2023) and http://www.heatmapper.ca/expression/ (accessed on 20 February 2023), respectively, were used.
4.4. Validation of DEGs by qRT-PCR
Reverse transcription (RT) was performed at 37 °C in a 20 μL volume using 1 μg of the total RNA, 10 mM random hexamer primer and 200U M-MLV reverse transcriptase (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Briefly, 1 μg of RNA and 100 pM of random hexamers were mixed and incubated 5 min at 65 °C. Subsequently, 4 μL of 5× reaction buffer, 2 μL dNTP (10 mM), 1 μL RevertAid reverse transcriptase (200 U) (Thermo Fisher Scientific, Waltham, MA, USA) and 0.5 μL RiboLock RNase inhibitor (20U) (Thermo Fisher Scientific, Waltham, MA, USA) were added. The reaction mixture was incubated for 10 min at 25 °C and 1 h at 42 °C, followed by 70 °C for 10 min. A set of DEGs significantly up and downregulated in RNA-seq (23 genes–upregulated and downregulated) were selected for qRT-PCR. Primers used in qRT-PCR were designed using Geneious Pro software 2020 (Biomatters, San Francisco, CA, USA) (Supplementary Table S2). Reaction mix of qRT-PCR was composed of 1 µg of cDNA, 1× Maxima SYBR Green/ROX qPCR Master Mix (2x) (Thermo Fisher Scientific, Waltham, MA, USA), gene-specific primers (10 pM each) and RNase free water up to total volume of 20 μL. Each reaction was performed in triplicate. Amplification cycles were as follows: 10 min at 95 °C, 40 cycles [15 s at 95 °C, 30 s at 60 °C, 30 s at 72 °C], 5 min at 72 °C; melting curve 54 °C to 95 °C–0.3% temperature increment/s (Eco™ qRT-PCR System, CA; Eco™ Software v4.1.2.0). The gene expression (ΔΔCt) was normalized to β–actin (house-keeping gene). ΔΔCt values were converted to logFC (https://goldbio.com/qpcr-and-rtqpcr-analysis-tool, accessed on 27 February 2023). The correlation of DEGs values obtained from RNA-seq and qRT-PCR was determined by calculating the Pearson correlation coefficient (r2). Correlation plots and Pearson correlation were performed by Prism 9 software (Graphpad, Boston, MA, USA).
4.5. RNA Isolation, Reverse Transcription and PCR Analysis
After incubation of RAMA cells in 6-well plates, mRNA was isolated using TRIzol® reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. DNaseI treatment was essentially incorporated during RNA isolation. Total RNA concentration and purity (OD 260/280 and 260/230 ratio) was measured using Nanodrop (Thermo Fisher Scientific, Waltham, MA, USA). The RNA was stored at −80 °C. Reverse transcription was performed according to the qRT-PCR method mentioned above.
4.6. Isolation, Purification of the Proteins and Western Blot Analysis
RAMA cells cultivated in 6-well plates were lysed in (lysis buffer FNN0011, Thermo Fisher Scientific, Waltham, MA, USA) and 1× protease inhibitor cocktail (PIC P8340, Sigma-Aldrich, Darmstadt, Germany), followed by sonication on ice (2 cycles; 30-s pulses, 100% amplitude). The cell lysates were sonicated for 15 s at 30% power of the BANDELIN SONOPULS HD2070 (BANDELIN electronic, Berlin, Germany) on ice and centrifuged for 10 min/13,000× g at 4 °C. Protein concentration was determined using a detergent-compatible protein assay (Bio-Rad Laboratories, Hercules, CA, USA). Equal protein amounts (30 μg) supplemented with 0.01% bromphenol blue, 1% 2-mercaptoethanol, 0.4% SDS and 5% glycerol were then separated with 10% SDS-polyacrylamide gel (Acrylamide/Bis Solution, 37.5:1) and transferred (dry transfer, 10 min) onto a NC membrane (Bio-Rad Laboratories, Hercules, CA, USA) by dry transfer (iBlot 2 Gel Transfer Device, Thermo Fisher Scientific, Waltham, MA, USA). After 1 h of membrane blocking at RT in 5% non-fat milk (20 mM Tris–HCl, pH 7.6, 150 mM NaCl, 0.1 % TWEEN 20, pH 7.4), the NC membrane blots were incubated overnight at 4 °C with primary antibody anti–Oncomodulin (Polyclonal Antibody PA5-115689 R&D); anti–Glypican 4 (Polyclonal Antibody, PA5-115301); anti–HPRT1 (Recombinant Rabbit Monoclonal Antibody, JU03-26); anti–gamma Synuclein (Recombinant Rabbit Monoclonal Antibody (JM90-32, MA5-32748); anti-STAT5 (ab 230670). After 20 min washing in wash buffer, the membranes were incubated with appropriate horseradish peroxidase-conjugated secondary antibody for 1 h at RT (Goat anti-rabbit IgG 1:2000, 31461 Thermo Fisher Scientific, Waltham, MA, USA Rabbit anti-mouse IgG 1:5000, Santa Cruz, Dallas, TX, USA). Detection of antibody reactivity was performed using a Pierce ECL Western Blotting Substrate (Pierce, Thermo Fisher Scientific, Waltham, MA, USA) and visualised by bio-imaging system (DNR MF Chemibis 2.0, Neve Yamin, Israel). Equal sample loading was verified by immunodetection of anti- β-actin Monoclonal Antibody AC-15 (MA1-91399). The pictures were scanned with GS-800 Calibrated Densitometer and the quantification was performed using Image J software version 1.52 (NIH; National Institutes of Health, Bethesda, MD, USA).
4.7. Proteomics Analysis
Total proteins (100 μg) were reduced with 10 mM dithiothreitol in 100 mM ammonium bicarbonate at 56 °C for 30 min. Alkylation was performed with 100 μL of 50 mM iodoacetamide in 100 mM ammonium bicarbonate in the dark for 30 min. Proteins were digested with trypsin in ratio 1:100 at 37 °C, overnight. Aliquots of purified complex peptide mixtures of 100 ng were separated using Acquity M-Class UHPLC (Waters, Etten-Leur, The Netherlands). Samples were loaded onto the nanoEase Symmetry C18 trap column (25 mm length, 180 μm diameter, 5 μm particles size). After 2 min of desalting/concentration by 1% acetonitrile containing 0.1% formic acid at a flow rate 8 μL/min, peptides were introduced to the nanoEase HSS T3 C18 analytical column (100 mm length, 75 μm diameter, 1.8 μm particle size). For the thorough separation, a 90 min gradient of 5–35% acetonitrile with 0.1% formic acid was applied at a flow rate of 300 nL/min. The samples were nanosprayed (3.1 kV capillary voltage) to the quadrupole time-of-flight mass spectrometer Synapt G2-Si with ion mobility option (Waters, Etten-Leur, The Netherlands). Spectra were recorded in a data-independent manner in high definition MSE mode. Ions with 50–2000 m/z were detected in both channels, with a 1 s spectral acquisition scan rate. Spectra were preprocessed with the Compression and Archival Tool 1.0 (Waters, Etten-Leur, The Netherlands) to reduce noise, removing ion counts below 15.
4.8. Silencing of EPOR Gene Expression by siRNA
For silence of EPOR expression in RAMA 37-28 cells, we used siRNAs (siRNA Dharma SMARTpool ON-TARGETplus human EPOR siRNA, GE HealthCare, PerkinElmer Holdings Inc., Lafayette, CO, USA), ntRNAs (negative controls to siRNA Dharma ON-TARGETplus–non-targeting) and transfect reagent DharmaFECT 1 (GE HealthCare, PerkinElmer Holdings Inc., Lafayette, CO, USA) following the manufacturer’s instructions. Transfection reagents were stable and none of the tested conditions significantly affected cell toxicity. RAMA 37-28 cells were cultured in 6-well plates (500,000 cells/well) in RPMI media without antibiotics, supplemented by 10% of FBS and incubated for 24 h at 37 °C in the presence of 5% CO2. Subsequently, cells were cultured in plates with changed fresh media without antibiotics, and were supplemented by 10% of FCS; the experimental groups were enriched by siRNA (14 µM) or ntRNA (5 µM) for the next 48 h. After that, the media was changed and followed by isolation and purification of proteins. Subsequently, Western blot analysis was carried out according to the protocol described above.
4.9. Agilent xCELLigence Real-Time Cell Analysis
RAMA 38-28 cells (8 × 103 cells/well) were seeded in 96-well plates (RTCA E-Plates 96) in xCELLigence RTCA systems (Agilent Technologies, Santa Clara, CA, USA) for 24 h followed by the treatment using siRNA (14 nM), or ntRNA (5 nM) for 48 h. Subsequently, cells were treated by PTX (200 nM) for 60 h. The cell adhesion and spread of the cells were continuously monitored in 60 min intervals over the course of monitoring period using the xCELLigence RTCA system.
4.10. Statistical Analysis of xCELLigence Analysis
Experiments under all conditions were performed in at least three independent measurements. Mean value and standard deviation were calculated using descriptive statistics. The data were analyzed by using the RTCA software Pro 1.2.1 (ACEA Bioscience, Santa Clara, CA, USA). Statistical analysis was carried out by a non-parametric method, one-way ANOVA using SigmaPlot (Ver. 12.0); p < 0.05 was considered significant (see Supplementary Figure S5).
5. Conclusions
We demonstrate for the first time robust 233 DEGs evoked by human EPOR overexpression in rat mammary adenocarcinoma RAMA 37-28 cells. Identified DEGs are associated with many biological processes such as proliferation, apoptosis, tumorigenesis, cellular metabolism, differentiation and others. It is obvious that EPOR overexpression affects also signaling pathways either by attenuation of RAS signaling, small GTPase, cytokine-mediated transduction, intrinsic apoptotic pathway in response to DNA damage and cAMP-mediated signaling or through the activation of STAT5, NOTCH signaling and certain Ephrin and WNT signaling mediators. We are aware of certain limitations of this work, which is based on the comparison of rat EPOR overexpressed RAMA 37-28 cells and their parental control only. However, we believe that our newly prepared EPOR-overexpressed human cell lines will soon confirm EPOR-induced gene expression and its potential tissue-specific origin.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24108482/s1.
Author Contributions
Conceptualization, P.S.; Methodology, M.Š., K.B., M.B., A.K., P.M., M.K., J.Š. and P.S.; Software, M.B.; Validation, K.B., M.B., A.K. and P.S.; Formal analysis, Z.T., M.Š., M.B., A.K., P.M., M.K. and J.Š.; Investigation, P.S.; Data curation, M.Š. and M.B.; Writing—original draft, Z.T. and P.S.; Writing—review & editing, Z.S. and P.S.; Visualization, M.B.; Supervision, P.S.; Project administration, Z.S. and P.S.; Funding acquisition, P.S. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Scientific Grant Agency of the Ministry of Education of the Slovak Republic by grant No. VEGA1/0500/23.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hidalgo, D.; Bejder, J.; Pop, R.; Gellatly, K.; Hwang, Y.; Scalf, S.M.; Eastman, A.E.; Chen, J.J.; Zhu, L.J.; Heuberger, J.A.A.C.; et al. EpoR stimulates rapid cycling and larger red cells during mouse and human erythropoiesis. Nat. Commun. 2021, 12, 7334. [Google Scholar] [CrossRef] [PubMed]
- Bhoopalan, S.V.; Huang, L.J.; Weiss, M.J. Erythropoietin regulation of red blood cell production: From bench to bedside and back. F1000Research 2020, 9, 1153. [Google Scholar] [CrossRef]
- Ostrowski, D.; Heinrich, R. Alternative erythropoietin receptors in the nervous system. J. Clin. Med. 2018, 7, 24. [Google Scholar] [CrossRef]
- Sanchis-Gomar, F.; Garcia-Gimenez, J.L.; Pareja-Galeano, H.; Romagnoli, M.; Perez-Quilis, C.; Lippi, G. Erythropoietin and the heart: Physiological effects and the therapeutic perspective. Int. J. Cardiol. 2014, 171, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Hiram-Bab, S.; Neumann, D.; Gabet, Y. Erythropoietin in bone—Controversies and consensus. Cytokine 2017, 89, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhao, D.; Na, N.; Li, H.; Miao, B.; Hong, L.; Huang, Z. Renoprotective effect of erythropoietin via modulation of the STAT6/MAPK/NF-kappaB pathway in ischemia/reperfusion injury after renal transplantation. Int. J. Mol. Med. 2018, 41, 25–32. [Google Scholar]
- Wang, K.; Wang, C.; Xiao, F.; Wang, H.; Wu, Z. JAK2/STAT2/STAT3 are required for myogenic differentiation. J. Biol. Chem. 2008, 283, 34029–34036. [Google Scholar] [CrossRef]
- Anagnostou, A.; Liu, Z.Y.; Steiner, M.; Chin, K.; Lee, E.S.; Kessimian, N.; Noguchi, C.T. Erythropoietin receptor messenger-RNA expression in human endothelial-cells. Proc. Natl. Acad. Sci. USA 1994, 91, 3974–3978. [Google Scholar] [CrossRef]
- Bretz, C.A.; Ramshekar, A.; Kunz, E.; Wang, H.B.; Hartnett, M.E. Signaling through the erythropoietin receptor affects angiogenesis in retinovascular disease. Investig. Ophthalmol. Vis. Sci. 2020, 61, 23. [Google Scholar] [CrossRef]
- Debeljak, N.; Sytkowski, A.J. Erythropoietin: New approaches to improved molecular designs and therapeutic alternatives. Curr. Pharm. Des. 2008, 14, 1302–1310. [Google Scholar] [CrossRef]
- McGraw, K.L.; Fuhler, G.M.; Johnson, J.O.; Clark, J.A.; Caceres, G.C.; Sokol, L.; List, A.F. Erythropoietin receptor signaling is membrane raft dependent. PLoS ONE 2012, 7, e34477. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Y.; Wang, L.; Dey, S.; Alnaeeli, M.; Suresh, S.; Rogers, H.; Teng, R.F.; Noguchi, C.T. Erythropoietin action in stress response, tissue maintenance and metabolism. Int. J. Mol. Sci. 2014, 15, 10296–10333. [Google Scholar] [CrossRef]
- Tóthová, Z.; Tomc, J.; Debeljak, N.; Solár, P. STAT5 as a key protein of erythropoietin signalization. Int. J. Mol. Sci. 2021, 22, 7109. [Google Scholar] [CrossRef] [PubMed]
- Tothova, Z.; Semelakova, M.; Solarova, Z.; Tomc, J.; Debeljak, N.; Solar, P. The Role of PI3K/AKT and MAPK signaling pathways in erythropoietin signalization. Int. J. Mol. Sci. 2021, 22, 7682. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Dev, A.; Verma, R.; Pradeep, A.; Sathyanarayana, P.; Green, J.M.; Narayanan, A.; Wojchowski, D.M. Defining an EPOR-regulated transcriptome for primary progenitors, including Tnfr-sf13c as a novel mediator of EPO-dependent erythroblast formation. PLoS ONE 2012, 7, e38530. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Andrieux, G.; Kastl, P.; Adlung, L.; Altamura, S.; Boehm, M.E.; Schwarzmueller, L.E.; Abdullah, Y.; Wagner, M.C.; Helm, B.; et al. Erythropoietin-driven dynamic proteome adaptations during erythropoiesis prevent iron overload in the developing embryo. Cell Rep. 2022, 40, 111360. [Google Scholar] [CrossRef] [PubMed]
- Held, M.A.; Greenfest-Allen, E.; Jachimowicz, E.; Stoeckert, C.J.; Stokes, M.P.; Wood, A.W.; Wojchowski, D.M. Phospho-proteomic discovery of novel signal transducers including thioredoxin-interacting protein as mediators of erythropoietin-dependent human erythropoiesis. Exp. Hematol. 2020, 84, 29–44. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Wang, S.Y.; Han, S.T.; Feng, Y.C. Pan-cancer analysis based on EPOR expression with potential value in prognosis and tumor immunity in 33 tumors. Front. Oncol. 2022, 12, 844794. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.K.; Matchett, K.B.; Coulter, J.A.; Yuen, H.F.; McCrudden, C.M.; Zhang, S.D.; Irwin, G.W.; Davidson, M.A.; Rulicke, T.; Schober, S.; et al. Erythropoietin drives breast cancer progression by activation of its receptor EPOR. Oncotarget 2017, 8, 38251–38263. [Google Scholar] [CrossRef]
- Dandrea, A.D.; Lodish, H.F.; Wong, G.G. Expression cloning of the murine erythropoietin receptor. Cell 1989, 57, 277–285. [Google Scholar] [CrossRef]
- Zafeiriou, M.P. The erythropoietin system protects the heart upon injury by cardiac progenitor cell activation. Vitam. Horm. 2017, 105, 233–248. [Google Scholar] [PubMed]
- Siren, A.L.; Fratelli, M.; Brines, M.; Goemans, C.; Casagrande, S.; Lewczuk, P.; Keenan, S.; Gleiter, C.; Pasquali, C.; Capobianco, A.; et al. Erythropoietin prevents neuronal apoptosis after cerebral ischemia and metabolic stress. Proc. Natl. Acad. Sci. USA 2001, 98, 4044–4049. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.Y.; Lai, Y.F.; Chen, Y.H.; Lu, D.W. The latest evidence of erythropoietin in the treatment of glaucoma. Int. J. Mol. Sci. 2022, 23, 16038. [Google Scholar] [CrossRef]
- Miller, C.P.; Lowe, K.A.; Valliant-Saunders, K.; Kaiser, J.F.; Mattern, D.; Urban, N.; Henke, M.; Blau, C.A. Evaluating erythropoietin-associated tumor progression using archival tissues from a phase III clinical trial. Stem Cells 2009, 27, 2353–2361. [Google Scholar] [CrossRef] [PubMed]
- Ilkovicova, L.; Trost, N.; Szentpeteriova, E.; Solar, P.; Komel, R.; Debeljak, N. Overexpression of the erythropoietin receptor in RAMA 37 breast cancer cells alters cell growth and sensitivity to tamoxifen. Int. J. Oncol. 2017, 51, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Zsokova, E.; Ilkovicova, L.; Kimakova, P.; Feckova, B.; Solar, P. Erythropoietin receptor induces a paclitaxel resistance phenotype in mammary adenocarcinoma cells. Oncol. Rep. 2019, 42, 1149–1160. [Google Scholar] [PubMed]
- Li, Z.Q.; Chen, C.; Chen, L.N.; Hu, D.D.; Yang, X.Q.; Zhuo, W.Y.; Chen, Y.X.; Yang, J.J.; Zhou, Y.L.; Mao, M.S.; et al. STAT5a confers Doxorubicin resistance to breast cancer by regulating ABCB1. Front. Oncol. 2021, 11, 697950. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.Y.; Dutta, P.; Tsurumi, A.; Li, J.H.; Wang, J.T.; Land, H.; Li, W.X. Unphosphorylated STAT5A stabilizes heterochromatin and suppresses tumor growth. Proc. Natl. Acad. Sci. USA 2013, 110, 10213–10218. [Google Scholar] [CrossRef]
- Lu, R.J.; Ji, Z.L.; Li, X.Q.; Qin, J.; Cui, G.H.; Chen, J.; Zhai, Q.N.; Zhao, C.J.; Zhang, W.; Yu, Z.D. Tumor suppressive microRNA-200a inhibits renal cell carcinoma development by directly targeting TGFB2. Tumor Biol. 2015, 36, 6691–6700. [Google Scholar] [CrossRef]
- Derynck, R.; Zhang, Y.E. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 2003, 425, 577–584. [Google Scholar] [CrossRef]
- Livitsanou, M.; Vasilaki, E.; Stournaras, C.; Kardassis, D. Modulation of TGF beta/Smad signaling by the small GTPase RhoB. Cell Signal. 2018, 48, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Marlow, L.A.; Bok, I.; Smallridge, R.C.; Copland, J.A. RhoB upregulation leads to either apoptosis or cytostasis through differential target selection. Endocr.-Relat. Cancer 2015, 22, 777–792. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Igarashi, S.; Nojima, M.; Maruyama, R.; Yamamoto, E.; Kai, M.; Akashi, H.; Watanabe, Y.; Yamamoto, H.; Sasaki, Y.; et al. IGFBP7 is a p53-responsive gene specifically silenced in colorectal cancer with CpG island methylator phenotype. Carcinogenesis 2010, 31, 342–349. [Google Scholar] [CrossRef]
- Chen, Y.; Cui, T.T.; Knosel, T.; Yang, L.L.; Zoller, K.; Petersen, I. IGFBP7 is a p53 target gene inactivated in human lung cancer by DNA hypermethylation. Lung Cancer 2011, 73, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Dang, W.; Cao, P.F.; Yan, Q.J.; Yang, L.; Wang, Y.W.; Yang, J.; Xin, S.Y.; Zhang, J.; Li, J.; Long, S.J.; et al. IGFBP7-AS1 is a p53-responsive long noncoding RNA downregulated by Epstein-Barr virus that contributes to viral tumorigenesis. Cancer Lett. 2021, 523, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.A.; Syed, N.; Therachiyil, L.; Nisar, S.; Hashem, S.; Macha, M.A.; Yadav, S.K.; Krishnankutty, R.; Muralitharan, S.; Al-Naemi, H.; et al. Claudin-1, a double-edged sword in cancer. Int. J. Mol. Sci. 2020, 21, 569. [Google Scholar] [CrossRef] [PubMed]
- Janostiak, R.; Vyas, M.; Cicek, A.F.; Wajapeyee, N.; Harigopal, M. Loss of of c-KIT expression in breast cancer correlates with malignant transformation of breast epithelium and is mediated by KIT gene promoter DNA hypermethylation. Exp. Mol. Pathol. 2018, 105, 41–49. [Google Scholar] [CrossRef]
- Barrios, A.; Poole, R.J.; Durbin, L.; Brennan, C.; Holder, N.; Wilson, S.W. Eph/Ephrin signaling regulates the mesenchymal-to-epithelial transition of the paraxial mesoderm during somite morphogenesis. Curr. Biol. 2003, 13, 1571–1582. [Google Scholar] [CrossRef]
- Chiu, S.T.; Chang, K.J.; Ting, C.H.; Shen, H.C.; Li, H.; Hsieh, F.J. Over-expression of EphB3 enhances cell-cell contacts and suppresses tumor growth in HT-29 human colon cancer cells. Carcinogenesis 2009, 30, 1475–1486. [Google Scholar] [CrossRef]
- Klein, R. Eph/ephrin signaling in morphogenesis, neural development and plasticity. Curr. Opin. Cell Biol. 2004, 16, 580–589. [Google Scholar] [CrossRef]
- Shi, Y.H.; Sun, Y.Y.; Cheng, H.Y.; Wang, C. EFNB1 acts as a novel prognosis marker in glioblastoma through bioinformatics methods and experimental validation. J. Oncol. 2021, 2021, 4701680. [Google Scholar] [CrossRef]
- Shi, Z.Z.; Hodges, V.M.; Dunlop, E.A.; Percy, M.J.; Maxwell, A.P.; El-Tanani, M.; Lappin, T.R.J. Erythropoietin-induced activation of the JAK2/STAT5, PI3K/Akt, and Ras/ERK pathways promotes malignant cell behavior in a modified breast cancer cell line. Mol. Cancer Res. 2010, 8, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Georgakilas, A.G.; Martin, O.A.; Bonner, W.M. p21: A two-faced genome guardian. Trends Mol. Med. 2017, 23, 310–319. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Dowbenko, D.; Lasky, L.A. AKT/PKB phosphorylation of p21(Cip/WAF1) enhances protein stability of p21(Cip/WAF1) and promotes cell survival. J. Biol. Chem. 2002, 277, 11352–11361. [Google Scholar] [CrossRef]
- Koster, R.; di Pietro, A.; Timmer-Bosscha, H.; Gibcus, J.H.; van den Berg, A.; Suurmeijer, A.J.; Bischoff, R.; Gietema, J.A.; de Jong, S. Cytoplasmic p21 expression levels determine cisplatin resistance in human testicular cancer. J. Clin. Investig. 2010, 120, 3594–3605. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Ma, Q.F.; Li, X.; Ji, T.; Chen, P.B.; Xu, H.B.; Li, K.Z.; Fang, Y.; Weng, D.H.; Weng, Y.J.; et al. Cytoplasmic p21 is a potential predictor for cisplatin sensitivity in ovarian cancer. BMC Cancer 2011, 11, 399. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.J.; Ren, S.P.; Harris, L.G.; Devine, D.J.; Samant, R.S.; Fodstad, O.; Shevde, L.A. Cytoplasmic translocation of p21 mediates NUPR1-induced chemoresistance NUPR1 and p21 in chemoresistance. FEBS Lett. 2012, 586, 3429–3434. [Google Scholar] [CrossRef]
- Heliez, C.; Baricault, L.; Barboule, N.; Valette, A. Paclitaxel increases p21 synthesis and accumulation of its AKT-phosphorylated form in the cytoplasm of cancer cells. Oncogene 2003, 22, 3260–3268. [Google Scholar] [CrossRef]
- Maiuthed, A.; Ninsontia, C.; Erlenbach-Wuensch, K.; Ndreshkjana, B.; Muenzner, J.K.; Caliskan, A.; Ahmed, P.H.; Chaotham, C.; Hartmann, A.; Roehe, A.V.; et al. Cytoplasmic p21 mediates 5-Fluorouracil resistance by inhibiting pro-apoptotic Chk2. Cancers 2018, 10, 373. [Google Scholar] [CrossRef]
- Perez-Tenorio, G.; Berglund, F.; Merca, A.E.; Nordenskjold, B.; Rutqvist, L.E.; Skoog, L.; Stal, O. Cytoplasmic p21(WAF1/CIP1) correlates with Akt activation and poor response to tamoxifen in breast cancer. Int. J. Oncol. 2006, 28, 1031–1042. [Google Scholar]
- Holm, C.; Rayala, S.; Jirstrom, K.; Stal, O.; Kumar, R.; Landberg, G. Association between Pak1 expression and subcellular localization and tamoxifen resistance in breast cancer patients. J. Natl. Cancer Inst. 2006, 98, 671–680. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.W.; Spence, M.J.; Zhang, Y.L.; Jiang, Y.F.; Liu, Y.L.E.; Shi, Y.E. Transcriptional suppression of synuclein gamma (SNCG) expression in human breast cancer cells by the growth inhibitory cytokine oncostatin M. Breast Cancer Res. Treat. 2000, 62, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.J.; Weng, Z.Y.; Tao, Q.H.; Lin, G.F.; Wu, X.R.; Qian, H.Q.; Zhang, Y.C.; Ding, X.Y.; Jiang, Y.F.; Shi, Y.E. Stage-specific expression of breast cancer-specific gene gamma-synuclein. Cancer Epidemiol. Biomark. 2003, 12, 920–925. [Google Scholar]
- Tian, L.; Zhao, Y.; Truong, M.J.; Lagadec, C.; Bourette, R.P. Synuclein gamma expression enhances radiation resistance of breast cancer cells. Oncotarget 2018, 8, 27435–27447. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, S.W.; Yu, D.S. Metabolic reprogramming of chemoresistant cancer cells and the potential significance of metabolic regulation in the reversal of cancer chemoresistance. Metabolites 2020, 10, 289. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.L.; Ye, T.; Chen, H.L.; Zhao, Z.Y.; Tang, W.Q.; Wang, L.S.; Xia, J.L. Xanthine dehydrogenase downregulation promotes TGF beta signaling and cancer stem cell-related gene expression in hepatocellular carcinoma. Oncogenesis 2017, 6, e382. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Yang, H.H.; Qiu, Y.; Deng, Q.H.; Liu, J.; Zhao, M.L.; He, P.; Mo, M.C.; Zou, X.S.; He, J.X. Association between epidermal growth factor receptor gene copy number and ERCC1, BRCA1 protein expression in Chinese patients with non-small cell lung cancer. Med. Oncol. 2014, 31, 803. [Google Scholar] [CrossRef]
- Vu, J.V.; Sheetz, K.H.; De Roo, A.C.; Hiatt, T.; Hendren, S. Variation in colectomy rates for benign polyp and colorectal cancer. Surg. Endosc. 2021, 35, 802–808. [Google Scholar] [CrossRef]
- Zou, M.Y.; Hu, X.L.; Xu, B.T.; Tong, T.T.; Jing, Y.X.; Xi, L.; Zhou, W.; Lu, J.P.; Wang, X.; Yang, X.L.; et al. Glutathione S-transferase isozyme alpha 1 is predominantly involved in the cisplatin resistance of common types of solid cancer. Oncol. Rep. 2019, 41, 989–998. [Google Scholar] [CrossRef]
- Zhu, Z.Y.; Mu, Y.Q.; Qi, C.X.; Wang, J.; Xi, G.P.; Guo, J.C.; Mi, R.R.; Zhao, F.X. CYP1B1 enhances the resistance of epithelial ovarian cancer cells to paclitaxel in vivo and in vitro. Int. J. Mol. Med. 2015, 35, 340–348. [Google Scholar] [CrossRef]
- Behuria, H.G.; Dash, S.; Sahu, S.K. Phospholipid Scramblases: Role in cancer progression and anticancer therapeutics. Front. Genet. 2022, 13, 875894. [Google Scholar] [CrossRef] [PubMed]
- Gui, L.; Zhu, Y.W.; Xu, Q.; Huang, J.J.; Hua, P.; Wu, G.J.; Lu, J.; Ni, J.B.; Tang, H.; Zhang, L.L. RNA interference-mediated downregulation of phospholipid scramblase 1 expression in primary liver cancer in vitro. Oncol. Lett. 2020, 20, 361. [Google Scholar] [CrossRef] [PubMed]
- Dimmeler, S.; Zeiher, A.M. Nitric oxide—An endothelial cell survival factor. Cell Death Differ. 1999, 6, 964–968. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, D.C.; Charles, I.G.; Thomsen, L.L.; Moss, D.W.; Holmes, L.S.; Baylis, S.A.; Rhodes, P.; Westmore, K.; Emson, P.C.; Moncada, S. Roles of nitric-oxide in tumor growth. Proc. Natl. Acad. Sci. USA 1995, 92, 4392–4396. [Google Scholar] [CrossRef]
- Januchowski, R.; Wojtowicz, K.; Zabel, M. The role of aldehyde dehydrogenase (ALDH) in cancer drug resistance. Biomed. Pharmacother. 2013, 67, 669–680. [Google Scholar] [CrossRef]
- Sedano, M.J.; Ramos, E.I.; Choudhari, R.; Harrison, A.L.; Subramani, R.; Lakshmanaswamy, R.; Zilaie, M.; Gadad, S.S. Hypoxanthine phosphoribosyl transferase 1 is upregulated, predicts clinical outcome and controls gene expression in breast cancer. Cancers 2020, 12, 1522. [Google Scholar] [CrossRef]
- Munir, J.; Ngu, T.V.; Ayudthaya, P.D.N.; Ryu, S. Downregulation of glypican-4 facilitates breast cancer progression by inducing cell migration and proliferation. Biochem. Biophys. Res. Commun. 2020, 526, 91–97. [Google Scholar] [CrossRef]
- Legier, T.; Rattier, D.; Llewellyn, J. Epithelial disruption drives mesendoderm differentiation in human pluripotent stem cells by enabling TGF-β protein sensing. Nat. Commun. 2023, 14, 349. [Google Scholar] [CrossRef]
- Wang, S.; Qiu, Y.D.; Bai, B. The expression, regulation, and biomarker potential of glypican-1 in Cancer. Front. Oncol. 2019, 9, 614. [Google Scholar] [CrossRef]
- Fico, A.; Maina, F.; Dono, R. Fine-tuning of cell signaling by glypicans. Cell Mol. Life Sci. 2011, 68, 923–929. [Google Scholar] [CrossRef]
- Filmus, J.; Capurro, M.; Rast, J. Glypicans. Genome Biol. 2008, 9, 224. [Google Scholar] [CrossRef] [PubMed]
- Varma, R.R.; Hector, S.M.; Clark, K.; Greco, W.R.; Hawthorn, L.; Pendyala, L. Gene expression profiling of a clonal isolate of oxaliplatin resistant ovarian carcinoma cell line A2780/C10. Oncol. Rep. 2005, 14, 925–932. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.L.; Lin, C.T.; Chueh, L.L.; Chang, C.J. Autocrine/paracrine secreted Frizzled-related protein 2 induces cellular resistance to apoptosis—A possible mechanism of mammary tumorigenesis. J. Biol. Chem. 2004, 279, 14602–14609. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.S.; Yin, X.F.; Zhao, W.B.; Xu, W.L.; Chen, L.Z. Downregulation of SFRP2 facilitates cancer stemness and radioresistance of glioma cells via activating Wnt/beta-catenin signaling. PLoS ONE 2021, 16, e0260864. [Google Scholar] [CrossRef]
- Macmanus, J.P.; Whitfield, J.F.; Boynton, A.L.; Durkin, J.P.; Swierenga, S.H.H. Oncomodulin—A widely distributed, tumor-specific, calcium-binding protein. Oncodev. Biol. Med. 1982, 3, 79–90. [Google Scholar]
- Sommer, E.W.; Heizmann, C.W. Expression of the tumor-specific and calcium-binding protein oncomodulin during chemical Transformation of Rat Fibroblasts. Cancer Res. 1989, 49, 899–905. [Google Scholar]
- Kreis, N.N.; Louwen, F.; Yuan, J. The multifaceted p21 (Cip1/Waf1/CDKN1A) in cell differentiation, migration and cancer therapy. Cancers 2019, 11, 1220. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).