The Antimicrobial Peptide Cathelicidin Exerts Immunomodulatory Effects via Scavenger Receptors
Abstract
1. Introduction
2. Results
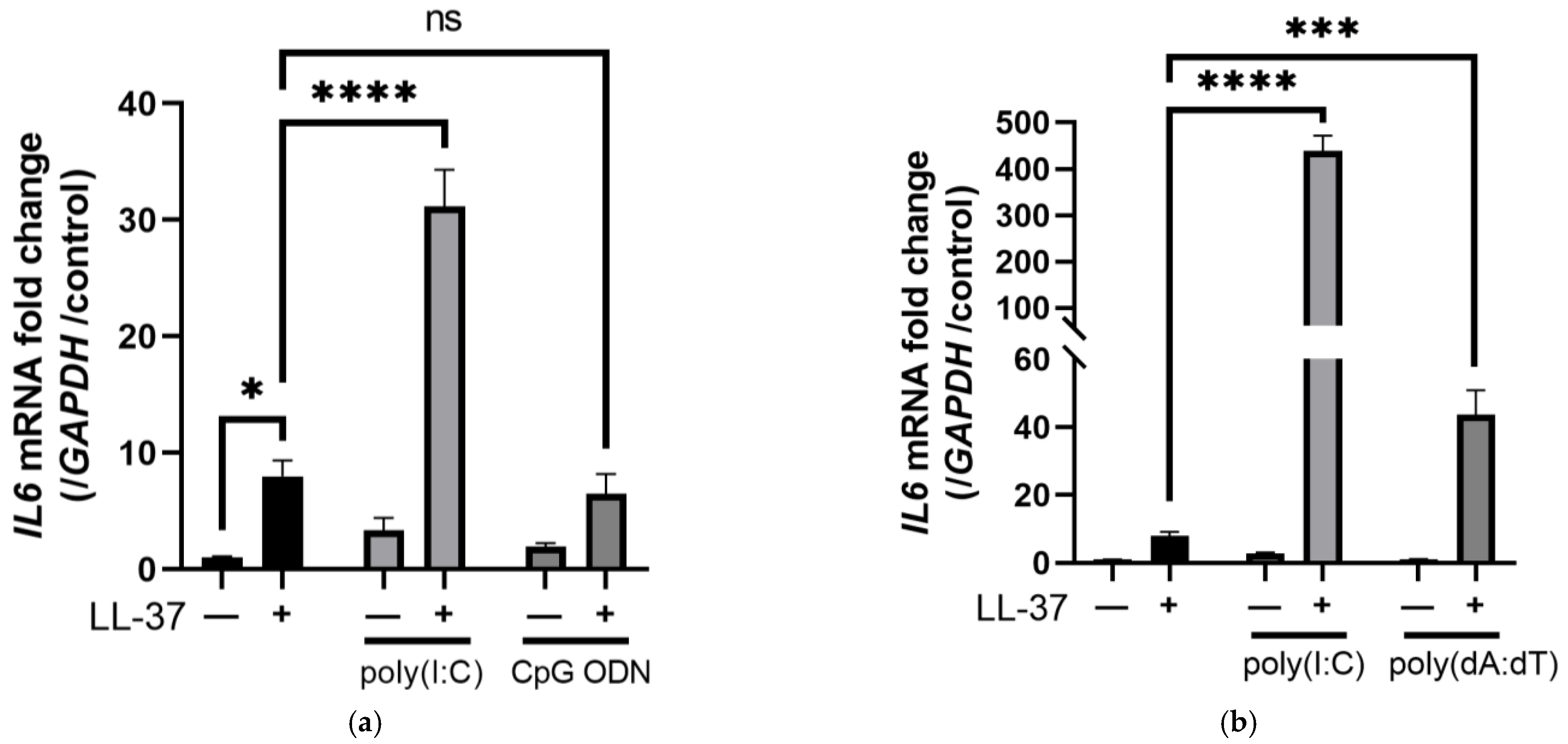
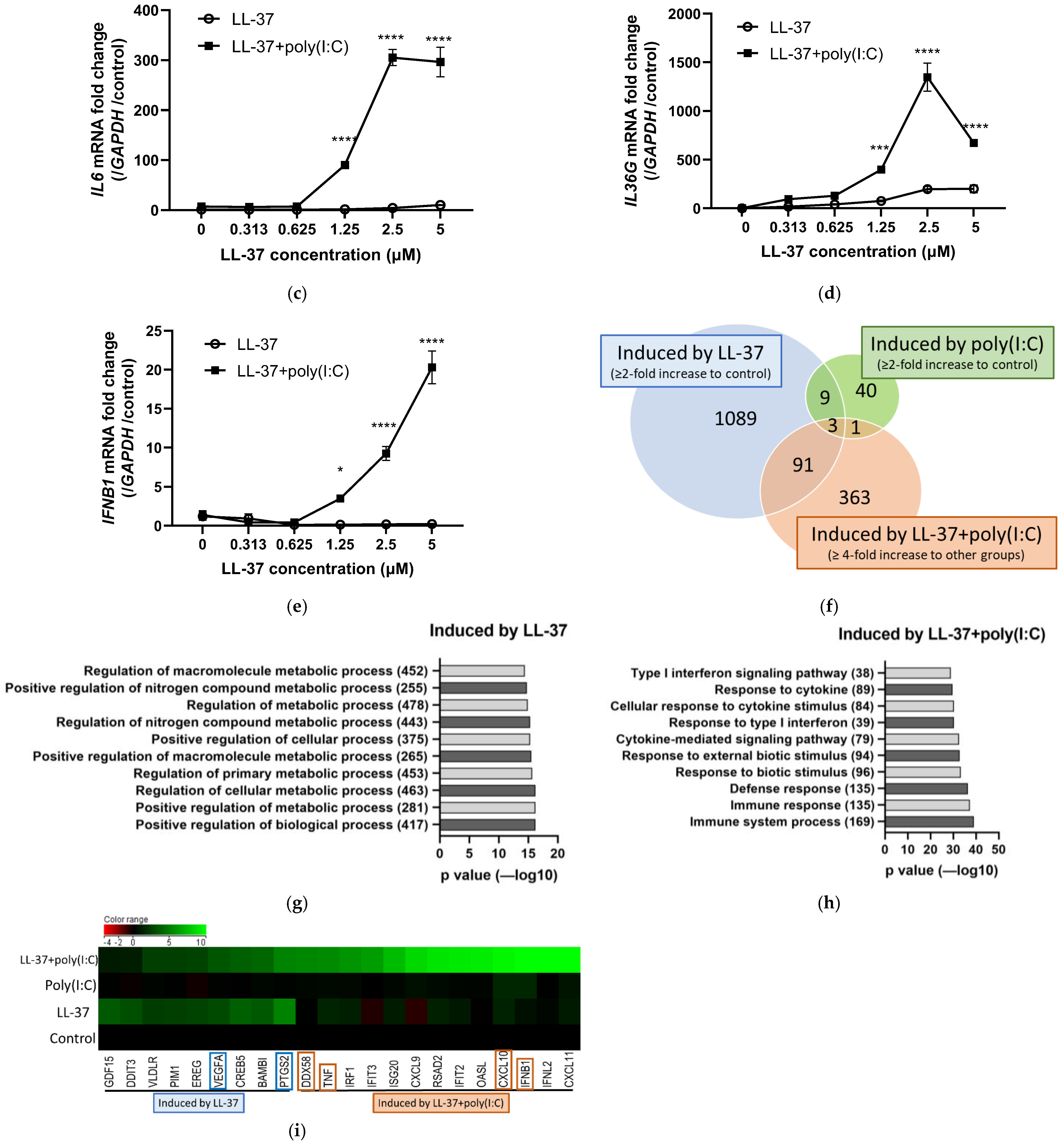
2.1. Unlike the Combination of LL-37 and dsRNA, LL-37 alone Induces a Set of Genes Mainly Related to Biological Metabolic Processes in Keratinocytes
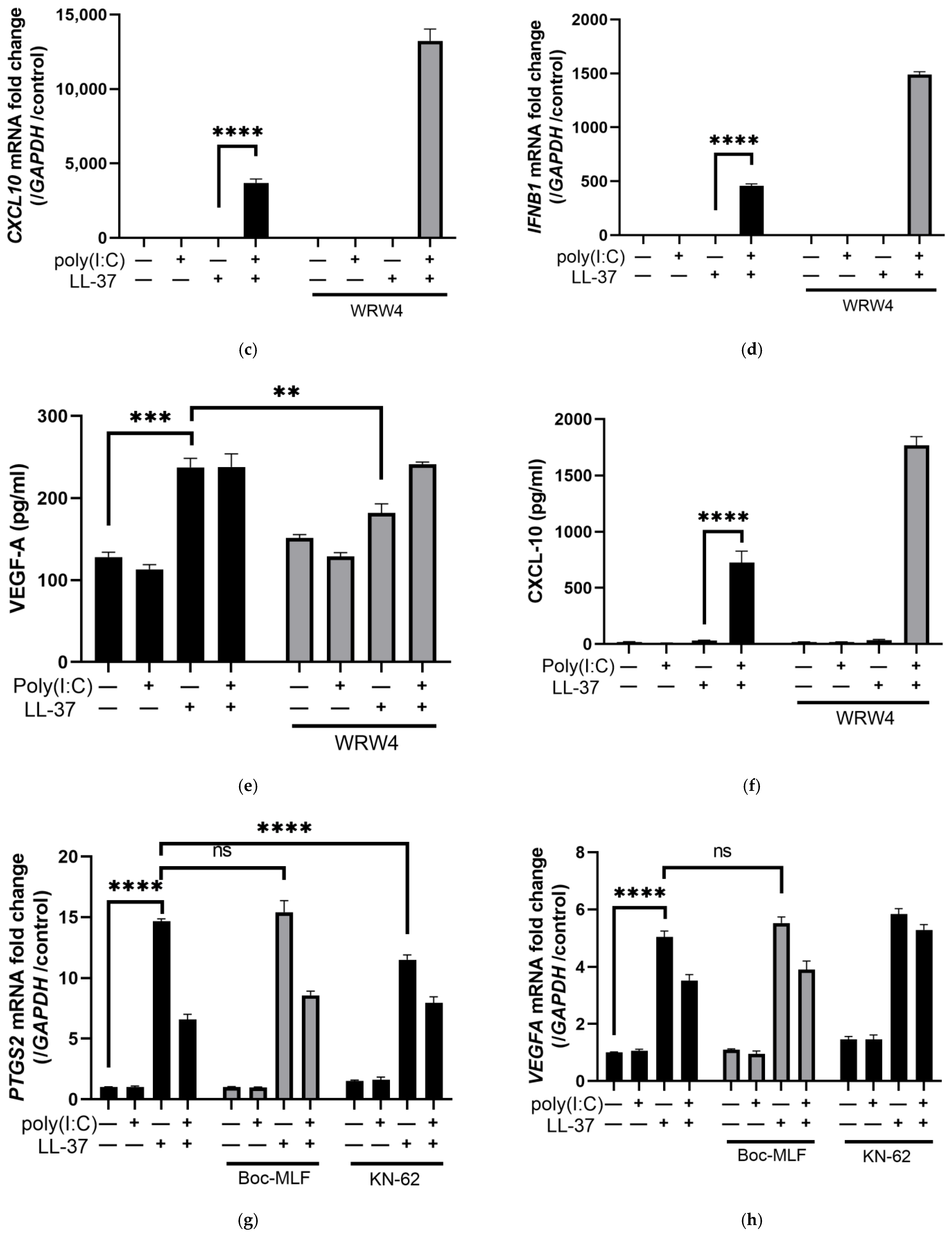
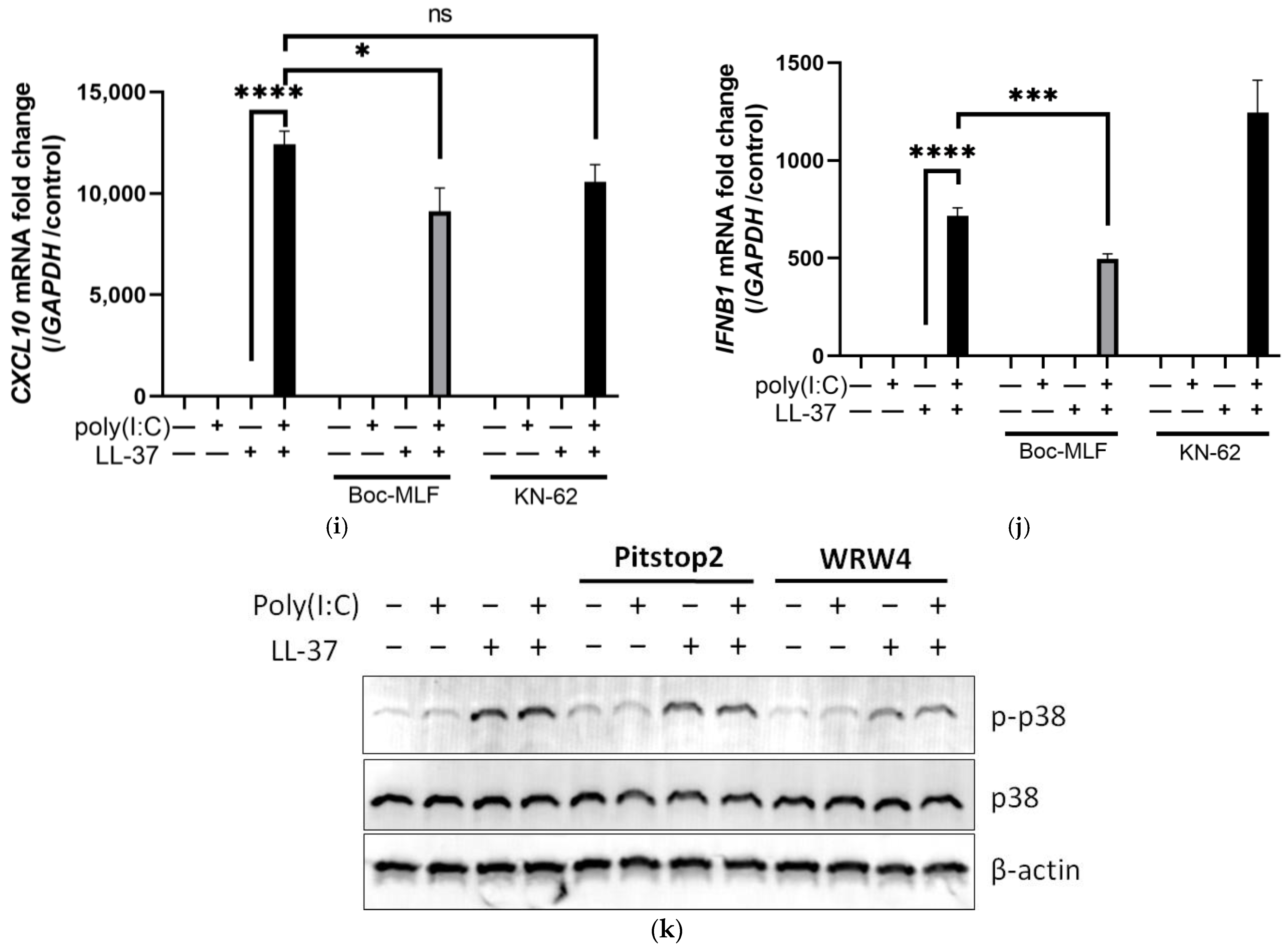
2.2. FPR2 Is a Receptor for LL-37 alone in Keratinocytes, but Is Not Involved in Signal Transmission of the Combination of LL-37 and poly(I:C)
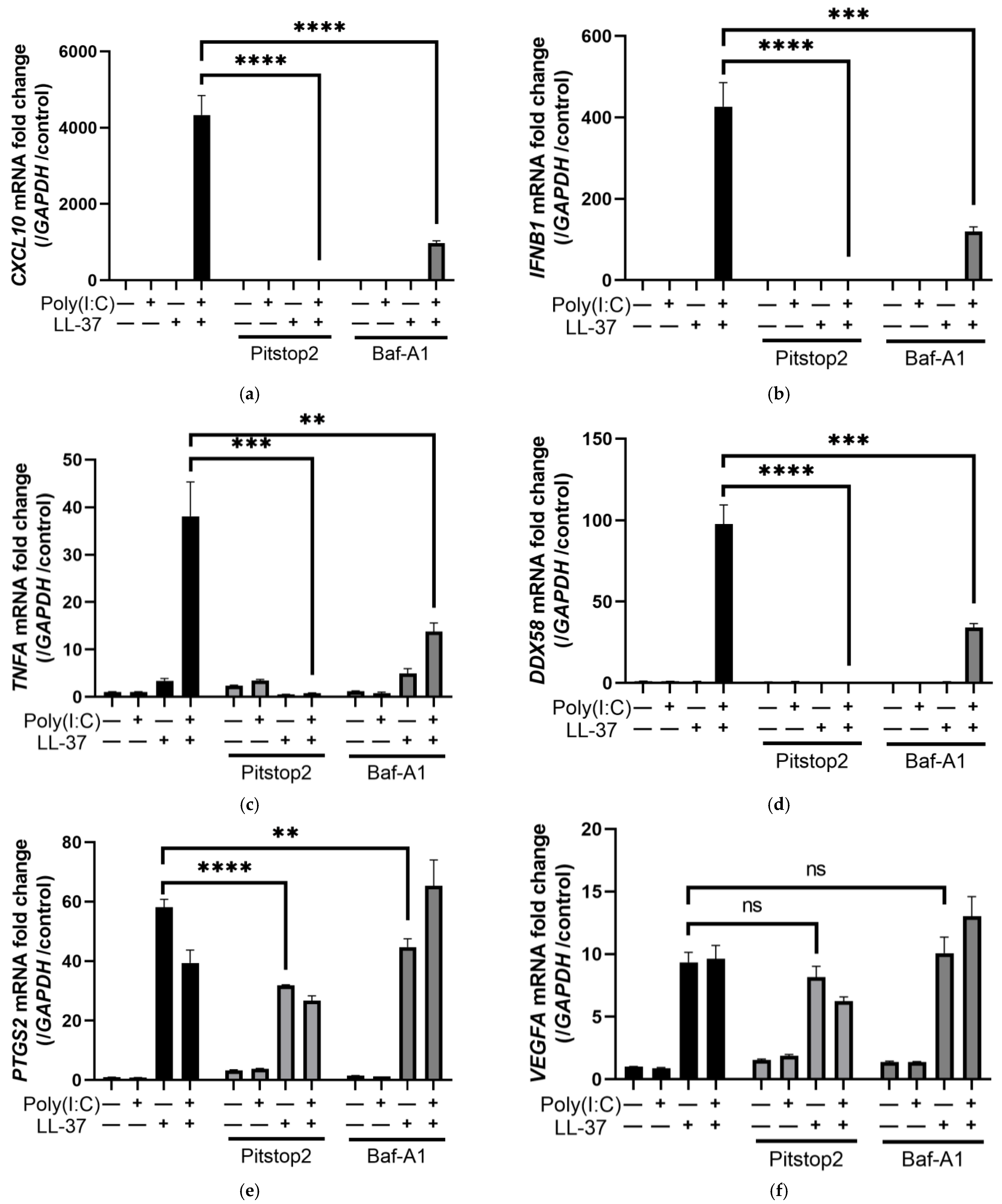
2.3. LL-37 alone and in Combination with poly(I:C) Is Transported by Scavenger Receptors in Keratinocytes
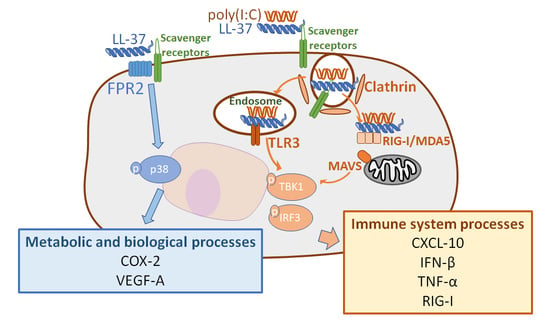
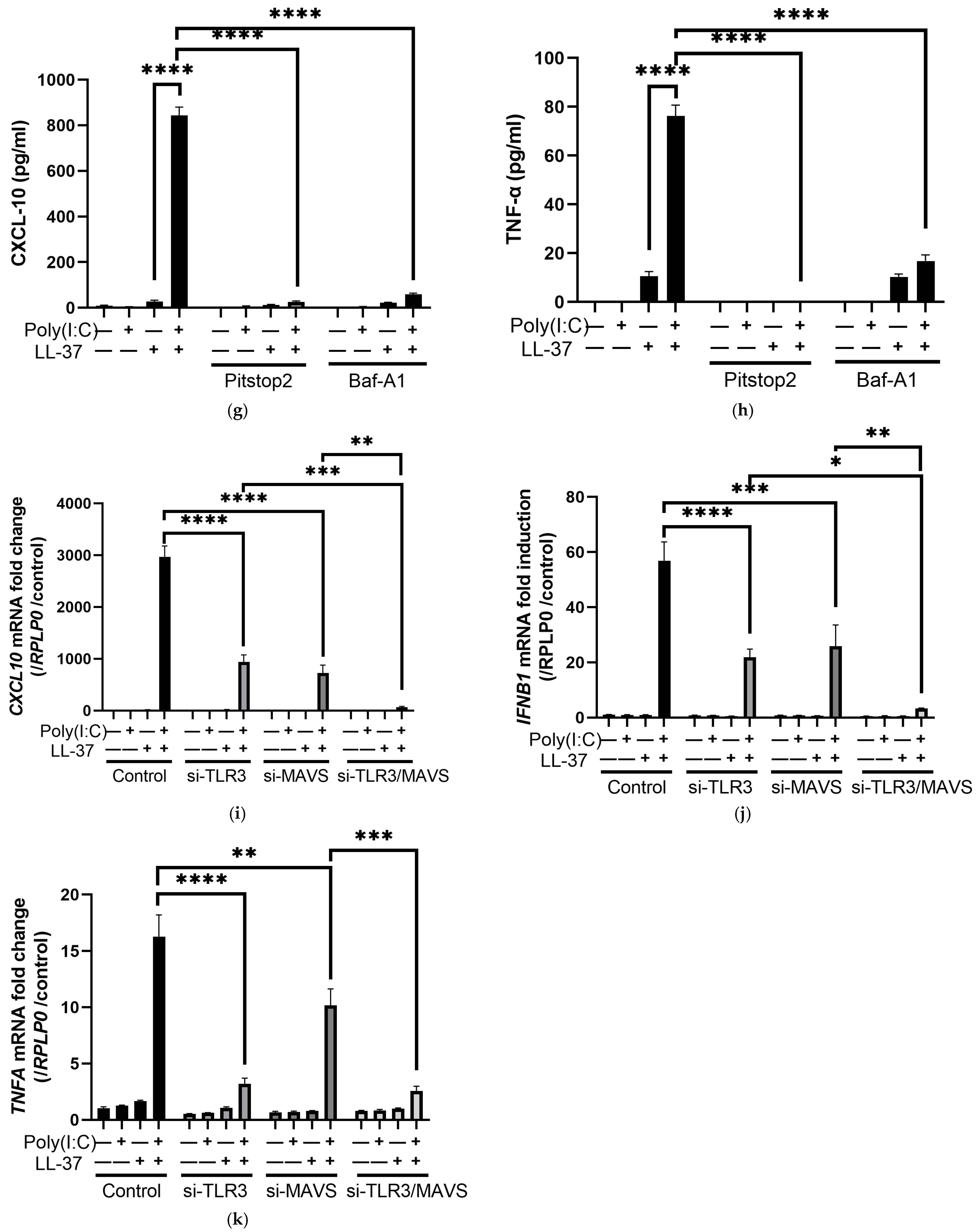
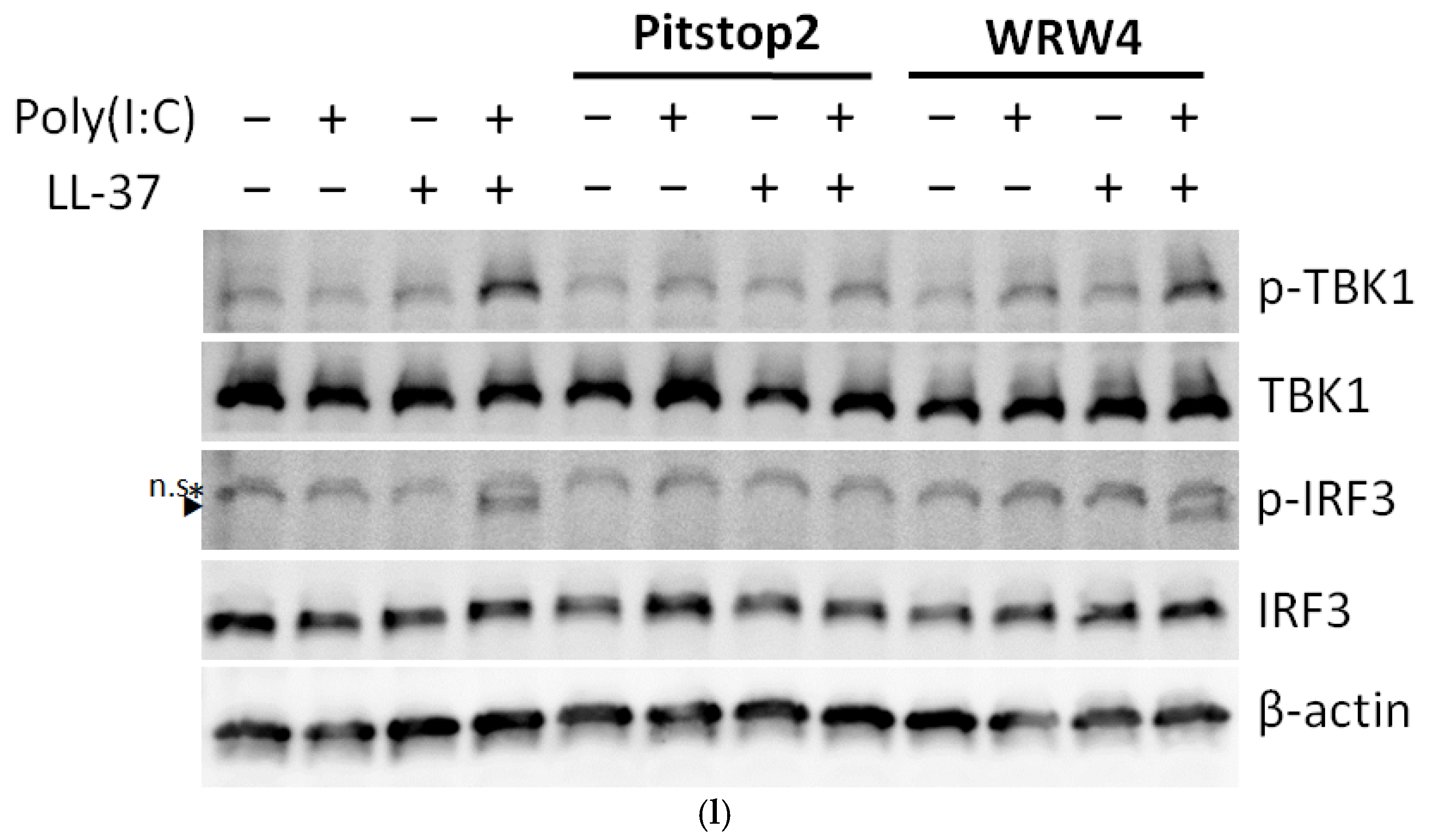
2.4. The Combination of LL-37 and poly(I:C) Was Recognized by TLRs on Endosomes and Cytosolic RNA Sensors, Followed by Clathrin-Dependent Endocytosis and Cytokine Induction
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Antibodies
4.3. Culture of Human Primary Keratinocytes
4.4. DNA Microarray
4.5. siRNA-Mediated Silencing of Gene Expression
4.6. Quantitative RT-PCR Analysis
4.7. Enzyme-Linked Immunosorbent Assay (ELISA)
4.8. Western Blotting
4.9. Proximity Ligation Assay (PLA)
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Gallo, R.L.; Ono, M.; Povsic, T.; Page, C.; Eriksson, E.; Klagsbrun, M.; Bernfield, M. Syndecans, cell surface heparan sulfate proteoglycans, are induced by a proline-rich antimicrobial peptide from wounds. Proc. Natl. Acad. Sci. USA 1994, 91, 11035–11039. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, K.; Schauber, J.; Coda, A.; Lin, H.; Dorschner, R.A.; Schechter, N.M.; Bonnart, C.; Descargues, P.; Hovnanian, A.; Gallo, R.L. Kallikrein-mediated proteolysis regulates the antimicrobial effects of cathelicidins in skin. FASEB J. 2006, 20, 2068–2080. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Gallo, R.L. AMPed up immunity: How antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009, 30, 131–141. [Google Scholar] [CrossRef]
- Méndez-Samperio, P. The human cathelicidin hCAP18/LL-37: A multifunctional peptide involved in mycobacterial infections. Peptides 2010, 31, 1791–1798. [Google Scholar] [CrossRef]
- Yamasaki, K.; Gallo, R.L. Antimicrobial peptides in human skin disease. Eur. J. Derm. 2008, 18, 11–21. [Google Scholar]
- Overhage, J.; Campisano, A.; Bains, M.; Torfs, E.C.W.; Rehm, B.H.A.; Hancock, R.E.W. Human Host Defense Peptide LL-37 Prevents Bacterial Biofilm Formation. Infect. Immun. 2008, 76, 4176–4182. [Google Scholar] [CrossRef]
- Murakami, M.; Lopez-Garcia, B.; Braff, M.; Dorschner, R.A.; Gallo, R.L. Postsecretory processing generates multiple cathelicidins for enhanced topical antimicrobial defense. J. Immunol. 2004, 172, 3070–3077. [Google Scholar] [CrossRef]
- Takahashi, T.; Gallo, R.L. The Critical and Multifunctional Roles of Antimicrobial Peptides in Dermatology. Derm. Clin 2017, 35, 39–50. [Google Scholar] [CrossRef]
- Scott, M.G.; Davidson, D.J.; Gold, M.R.; Bowdish, D.; Hancock, R.E.W. The Human Antimicrobial Peptide LL-37 Is a Multifunctional Modulator of Innate Immune Responses. J. Immunol. 2002, 169, 3883–3891. [Google Scholar] [CrossRef]
- Kurosaka, K.; Chen, Q.; Yarovinsky, F.; Oppenheim, J.J.; Yang, D. Mouse cathelin-related antimicrobial peptide chemoattracts leukocytes using formyl peptide receptor-like 1/mouse formyl peptide receptor-like 2 as the receptor and acts as an immune adjuvant. J. Immunol. 2005, 174, 6257–6265. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Chen, Q.; Schmidt, A.P.; Anderson, G.M.; Wang, J.M.; Wooters, J.; Oppenheim, J.J.; Chertov, O. Ll-37, the Neutrophil Granule–And Epithelial Cell–Derived Cathelicidin, Utilizes Formyl Peptide Receptor–Like 1 (Fprl1) as a Receptor to Chemoattract Human Peripheral Blood Neutrophils, Monocytes, and T Cells. J. Exp. Med. 2000, 192, 1069–1074. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, K.; Di Nardo, A.; Bardan, A.; Murakami, M.; Ohtake, T.; Coda, A.; Dorschner, R.A.; Bonnart, C.; Descargues, P.; Hovnanian, A.; et al. Increased serine protease activity and cathelicidin promotes skin inflammation in rosacea. Nat. Med. 2007, 13, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Koczulla, R.; von Degenfeld, G.; Kupatt, C.; Krotz, F.; Zahler, S.; Gloe, T.; Issbrucker, K.; Unterberger, P.; Zaiou, M.; Lebherz, C.; et al. An angiogenic role for the human peptide antibiotic LL-37/hCAP-18. J. Clin. Invest. 2003, 111, 1665–1672. [Google Scholar] [CrossRef] [PubMed]
- Geremia, A.; Biancheri, P.; Allan, P.; Corazza, G.R.; Di Sabatino, A. Innate and adaptive immunity in inflammatory bowel disease. Autoimmun. Rev. 2014, 13, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Beisswenger, C.; Herr, C.; Schmid, R.M.; Gallo, R.L.; Han, G.; Zakharkina, T.; Bals, R. Expression of the antimicrobial peptide cathelicidin in myeloid cells is required for lung tumor growth. Oncogene 2014, 33, 2709–2716. [Google Scholar] [CrossRef][Green Version]
- Hiemstra, P.S.; Amatngalim, G.D.; van der Does, A.M.; Taube, C. Antimicrobial peptides and innate lung defenses: Role in infectious and noninfectious lung diseases and therapeutic applications. Chest 2016, 149, 545–551. [Google Scholar] [CrossRef]
- Welling, M.M.; Nabuurs, R.J.A.; van der Weerd, L. Potential role of antimicrobial peptides in the early onset of Alzheimer’s disease. Alzheimer’s Dement. 2015, 11, 51–57. [Google Scholar] [CrossRef]
- Takahashi, T.; Asano, Y.; Nakamura, K.; Yamashita, T.; Saigusa, R.; Ichimura, Y.; Toyama, T.; Taniguchi, T.; Yoshizaki, A.; Tamaki, Z. A potential contribution of antimicrobial peptide LL-37 to tissue fibrosis and vasculopathy in systemic sclerosis. Br. J. Dermatol. 2016, 175, 1195–1203. [Google Scholar] [CrossRef]
- Kahlenberg, J.M.; Kaplan, M.J. Little peptide, big effects: The role of LL-37 in inflammation and autoimmune disease. J. Immunol. 2013, 191, 4895–4901. [Google Scholar] [CrossRef]
- Kulkarni, N.N.; Takahashi, T.; Sanford, J.A.; Tong, Y.; Gombart, A.F.; Hinds, B.; Cheng, J.Y.; Gallo, R.L. Innate Immune Dysfunction in Rosacea Promotes Photosensitivity and Vascular Adhesion Molecule Expression. J. Investig. Dermatol. 2020, 140, 645–655.e6. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, K.; Gallo, R.L. Rosacea as a disease of cathelicidins and skin innate immunity. J. Investig. Derm. Symp. Proc 2011, 15, 12–15. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Yamasaki, K. Psoriasis and Antimicrobial Peptides. Int. J. Mol. Sci. 2020, 21, 6791. [Google Scholar] [CrossRef] [PubMed]
- Niyonsaba, F.; Ushio, H.; Nagaoka, I.; Okumura, K.; Ogawa, H. The human beta-defensins (-1, -2, -3, -4) and cathelicidin LL-37 induce IL-18 secretion through p38 and ERK MAPK activation in primary human keratinocytes. J. Immunol. 2005, 175, 1776–1784. [Google Scholar] [CrossRef] [PubMed]
- Tjabringa, G.S.; Aarbiou, J.; Ninaber, D.K.; Drijfhout, J.W.; Sørensen, O.E.; Borregaard, N.; Rabe, K.F.; Hiemstra, P.S. The antimicrobial peptide LL-37 activates innate immunity at the airway epithelial surface by transactivation of the epidermal growth factor receptor. J. Immunol. 2003, 171, 6690–6696. [Google Scholar] [CrossRef] [PubMed]
- Chamorro, C.I.; Weber, G.; Grönberg, A.; Pivarcsi, A.; Ståhle, M. The Human Antimicrobial Peptide LL-37 Suppresses Apoptosis in Keratinocytes. J. Investig. Dermatol. 2009, 129, 937–944. [Google Scholar] [CrossRef]
- Roers, A.; Hiller, B.; Hornung, V. Recognition of Endogenous Nucleic Acids by the Innate Immune System. Immunity 2016, 44, 739–754. [Google Scholar] [CrossRef]
- Di Nardo, A.; Braff, M.H.; Taylor, K.R.; Na, C.; Granstein, R.D.; McInturff, J.E.; Krutzik, S.; Modlin, R.L.; Gallo, R.L. Cathelicidin antimicrobial peptides block dendritic cell TLR4 activation and allergic contact sensitization. J. Immunol. 2007, 178, 1829–1834. [Google Scholar] [CrossRef]
- Takahashi, T.; Kulkarni, N.N.; Lee, E.Y.; Zhang, L.-j.; Wong, G.C.L.; Gallo, R.L. Cathelicidin promotes inflammation by enabling binding of self-RNA to cell surface scavenger receptors. Sci. Rep. 2018, 8, 4032. [Google Scholar] [CrossRef]
- Zhang, L.-J.; Sen, G.L.; Ward, N.L.; Johnston, A.; Chun, K.; Chen, Y.; Adase, C.; Sanford, J.A.; Gao, N.; Chensee, M.; et al. Antimicrobial Peptide LL37 and MAVS Signaling Drive Interferon-β Production by Epidermal Keratinocytes during Skin Injury. Immunity 2016, 45, 119–130. [Google Scholar] [CrossRef]
- Ferrara, N.; Gerber, H.-P.; Lecouter, J. The biology of VEGF and its receptors. Nat. Med. 2003, 9, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.S.; Mann, M.; DuBois, R.N. The role of cyclooxygenases in inflammation, cancer, and development. Oncogene 1999, 18, 7908–7916. [Google Scholar] [CrossRef]
- Bradley, J. TNF-mediated inflammatory disease. J. Pathol. 2008, 214, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.Y.; Lee, Z.-H.; Song, Y.W. CXCL10 and autoimmune diseases. Autoimmun. Rev. 2009, 8, 379–383. [Google Scholar] [CrossRef] [PubMed]
- McNab, F.; Mayer-Barber, K.; Sher, A.; Wack, A.; O’Garra, A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015, 15, 87–103. [Google Scholar] [CrossRef]
- Loo, Y.-M.; Gale, M., Jr. Immune signaling by RIG-I-like receptors. Immunity 2011, 34, 680–692. [Google Scholar] [CrossRef]
- Nijnik, A.; Pistolic, J.; Filewod, N.C.J.; Hancock, R.E.W. Signaling Pathways Mediating Chemokine Induction in Keratinocytes by Cathelicidin LL-37 and Flagellin. J. Innate Immun. 2012, 4, 377–386. [Google Scholar] [CrossRef]
- Tomasinsig, L.; Pizzirani, C.; Skerlavaj, B.; Pellegatti, P.; Gulinelli, S.; Tossi, A.; Virgilio, F.D.; Zanetti, M. The Human Cathelicidin LL-37 Modulates the Activities of the P2X7 Receptor in a Structure-dependent Manner *. J. Biol. Chem. 2008, 283, 30471–30481. [Google Scholar]
- Wan, M.; Godson, C.; Guiry, P.J.; Agerberth, B.; Haeggström, J.Z. Leukotriene B4/antimicrobial peptide LL-37 proinflammatory circuits are mediated by BLT1 and FPR2/ALX and are counterregulated by lipoxin A4 and resolvin E1. FASEB J. 2011, 25, 1697–1705. [Google Scholar] [CrossRef]
- Kuznik, A.; Bencina, M.; Svajger, U.; Jeras, M.; Rozman, B.; Jerala, R. Mechanism of endosomal TLR inhibition by antimalarial drugs and imidazoquinolines. J. Immunol. 2011, 186, 4794–4804. [Google Scholar] [CrossRef]
- Yoshimori, T.; Yamamoto, A.; Moriyama, Y.; Futai, M.; Tashiro, Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J Biol Chem 1991, 266, 17707–17712. [Google Scholar] [CrossRef] [PubMed]
- Howell, M.D.; Jones, J.F.; Kisich, K.O.; Streib, J.E.; Gallo, R.L.; Leung, D.Y.M. Selective Killing of Vaccinia Virus by LL-37: Implications for Eczema Vaccinatum. J. Immunol. 2004, 172, 1763–1767. [Google Scholar] [CrossRef] [PubMed]
- Herster, F.; Bittner, Z.; Archer, N.K.; Dickhöfer, S.; Eisel, D.; Eigenbrod, T.; Knorpp, T.; Schneiderhan-Marra, N.; Löffler, M.W.; Kalbacher, H.; et al. Neutrophil extracellular trap-associated RNA and LL37 enable self-amplifying inflammation in psoriasis. Nat. Commun. 2020, 11, 105. [Google Scholar] [CrossRef] [PubMed]
- Canavese, M.; Altruda, F.; Ruzicka, T.; Schauber, J. Vascular endothelial growth factor (VEGF) in the pathogenesis of psoriasis--a possible target for novel therapies? J. Derm. Sci. 2010, 58, 171–176. [Google Scholar] [CrossRef]
- Xia, Y.P.; Li, B.; Hylton, D.; Detmar, M.; Yancopoulos, G.D.; Rudge, J.S. Transgenic delivery of VEGF to mouse skin leads to an inflammatory condition resembling human psoriasis. Blood 2003, 102, 161–168. [Google Scholar] [CrossRef]
- Benhadou, F.; Glitzner, E.; Brisebarre, A.; Swedlund, B.; Song, Y.; Dubois, C.; Rozzi, M.; Paulissen, C.; Marmol, V.D.; Sibilia, M.; et al. Epidermal autonomous VEGFA/Flt1/Nrp1 functions mediate psoriasis-like disease. Sci. Adv. 2020, 6, eaax5849. [Google Scholar] [CrossRef]
- Akman, A.; Yilmaz, E.; Mutlu, H.; Ozdogan, M. Complete remission of psoriasis following bevacizumab therapy for colon cancer. Clin. Exp. Dermatol. Clin. Dermatol. 2009, 34, e202–e204. [Google Scholar] [CrossRef]
- Bakry, O.A.; Samaka, R.M.; Shoeib, M.A.M.; Abdel Aal, S.M. Nuclear Factor Kappa B and Cyclo-Oxygenase-2: Two Concordant Players in Psoriasis Pathogenesis. Ultrastruct. Pathol. 2015, 39, 49–61. [Google Scholar] [CrossRef]
- Stetson, D.B.; Medzhitov, R. Type I Interferons in Host Defense. Immunity 2006, 25, 373–381. [Google Scholar] [CrossRef]
- Takiguchi, T.; Morizane, S.; Yamamoto, T.; Kajita, A.; Ikeda, K.; Iwatsuki, K. Cathelicidin antimicrobial peptide LL-37 augments interferon-β expression and antiviral activity induced by double-stranded RNA in keratinocytes. Br. J. Dermatol. 2014, 171, 492–498. [Google Scholar] [CrossRef]
- Fry, L.; Baker, B.S. Triggering psoriasis: The role of infections and medications. Clin. Dermatol. 2007, 25, 606–615. [Google Scholar] [CrossRef] [PubMed]
- Canton, J.; Neculai, D.; Grinstein, S. Scavenger receptors in homeostasis and immunity. Nat. Rev. Immunol. 2013, 13, 621–634. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, N.N.; O’Neill, A.M.; Dokoshi, T.; Luo, E.W.C.; Wong, G.C.L.; Gallo, R.L. Sequence determinants in the cathelicidin LL-37 that promote inflammation via presentation of RNA to scavenger receptors. J. Biol. Chem. 2021, 297, 100828. [Google Scholar] [PubMed]
- Nakagawa, Y.; Gallo, R.L. Endogenous intracellular cathelicidin enhances TLR9 activation in dendritic cells and macrophages. J. Immunol. 2015, 194, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Qi, R.; Jordan, J.L.; San Mateo, L.; Kao, C.C. The human antimicrobial peptide LL-37, but not the mouse ortholog, mCRAMP, can stimulate signaling by poly(I:C) through a FPRL1-dependent pathway. J. Biol. Chem. 2013, 288, 8258–8268. [Google Scholar] [CrossRef]
- Saito, R.; Hirakawa, S.; Ohara, H.; Yasuda, M.; Yamazaki, T.; Nishii, S.; Aiba, S. Nickel differentially regulates NFAT and NF-κB activation in T cell signaling. Toxicol. Appl. Pharmacol. 2011, 254, 245–255. [Google Scholar] [CrossRef]
- Zhang, L.J.; Vogel, W.K.; Liu, X.; Topark-Ngarm, A.; Arbogast, B.L.; Maier, C.S.; Filtz, T.M.; Leid, M. Coordinated regulation of transcription factor Bcl11b activity in thymocytes by the mitogen-activated protein kinase (MAPK) pathways and protein sumoylation. J. Biol. Chem. 2012, 287, 26971–26988. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amagai, R.; Takahashi, T.; Terui, H.; Fujimura, T.; Yamasaki, K.; Aiba, S.; Asano, Y. The Antimicrobial Peptide Cathelicidin Exerts Immunomodulatory Effects via Scavenger Receptors. Int. J. Mol. Sci. 2023, 24, 875. https://doi.org/10.3390/ijms24010875
Amagai R, Takahashi T, Terui H, Fujimura T, Yamasaki K, Aiba S, Asano Y. The Antimicrobial Peptide Cathelicidin Exerts Immunomodulatory Effects via Scavenger Receptors. International Journal of Molecular Sciences. 2023; 24(1):875. https://doi.org/10.3390/ijms24010875
Chicago/Turabian StyleAmagai, Ryo, Toshiya Takahashi, Hitoshi Terui, Taku Fujimura, Kenshi Yamasaki, Setsuya Aiba, and Yoshihide Asano. 2023. "The Antimicrobial Peptide Cathelicidin Exerts Immunomodulatory Effects via Scavenger Receptors" International Journal of Molecular Sciences 24, no. 1: 875. https://doi.org/10.3390/ijms24010875
APA StyleAmagai, R., Takahashi, T., Terui, H., Fujimura, T., Yamasaki, K., Aiba, S., & Asano, Y. (2023). The Antimicrobial Peptide Cathelicidin Exerts Immunomodulatory Effects via Scavenger Receptors. International Journal of Molecular Sciences, 24(1), 875. https://doi.org/10.3390/ijms24010875