Spectroscopic Characterization and Biological Activity of Hesperetin Schiff Bases and Their Cu(II) Complexes
Abstract
1. Introduction
2. Results and Discussion
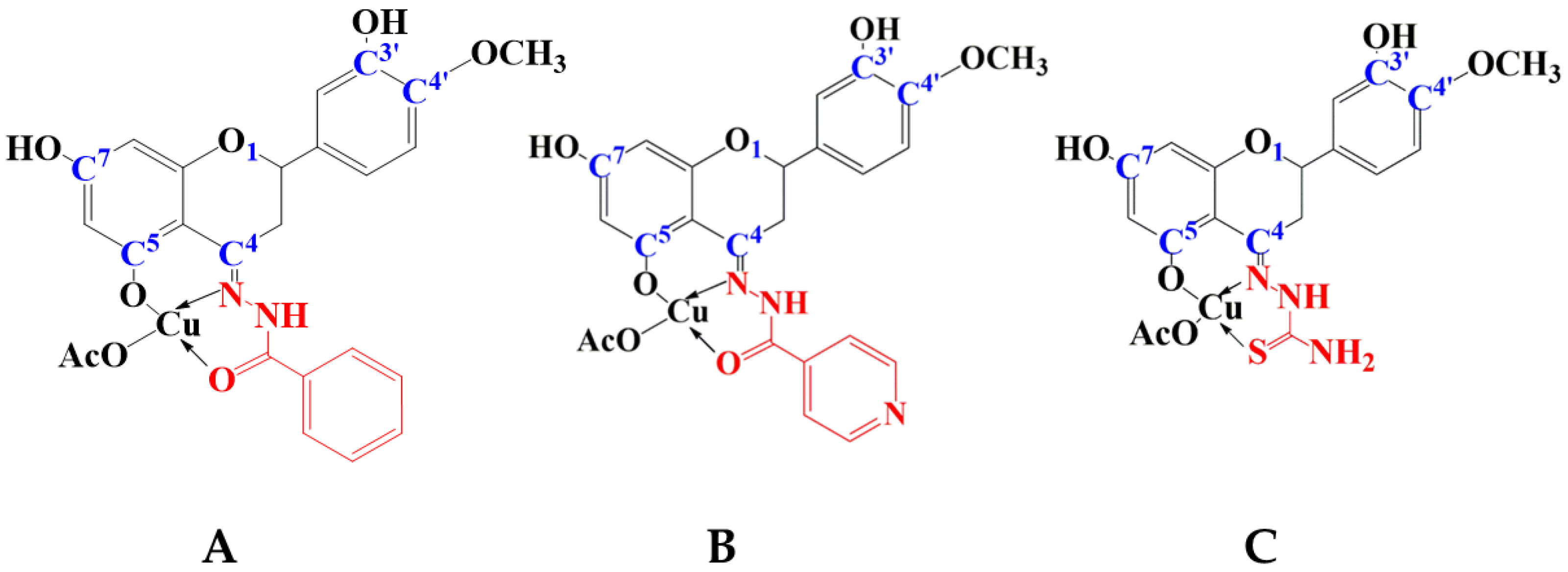
2.1. Characterization of the Complexes
2.1.1. IR Spectral Studies
2.1.2. UV-Vis Studies
2.1.3. PXRD Studies
2.1.4. TGA Studies
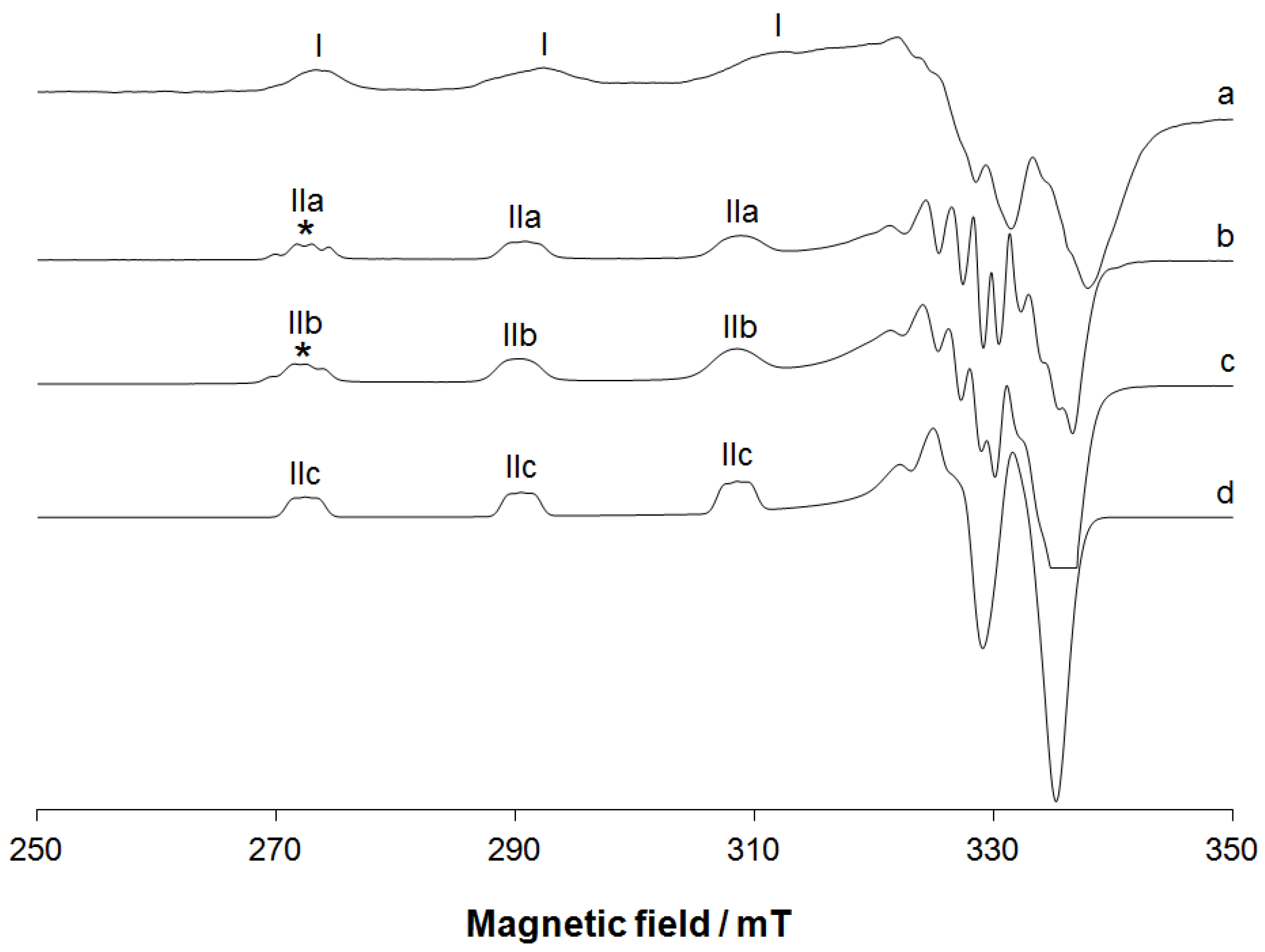
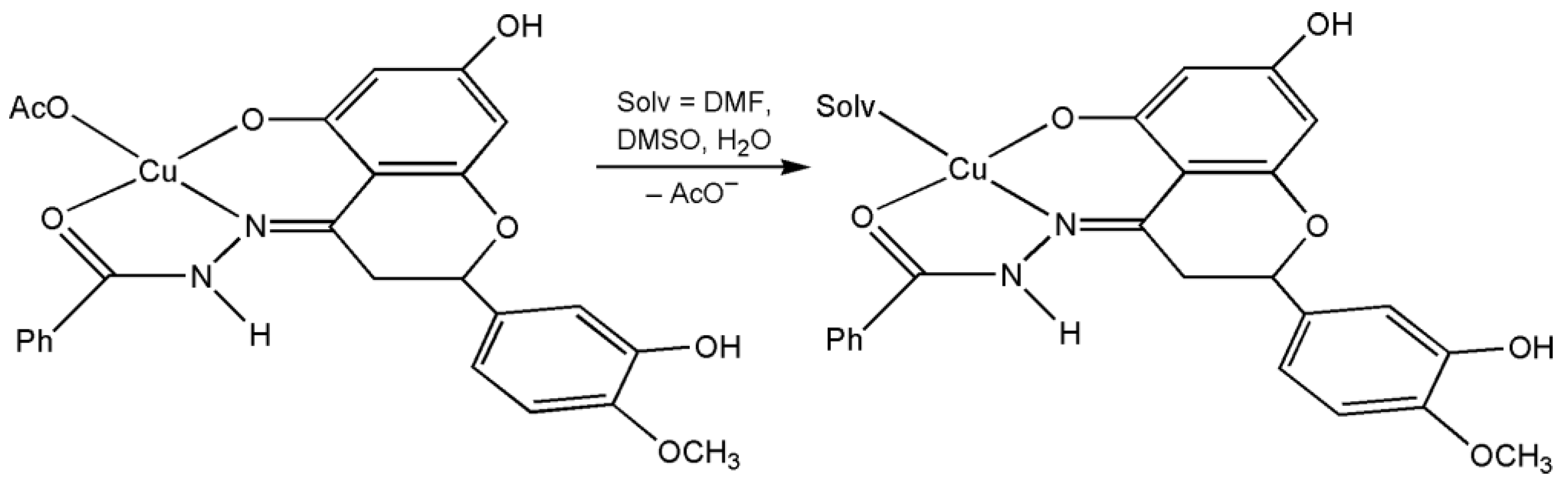
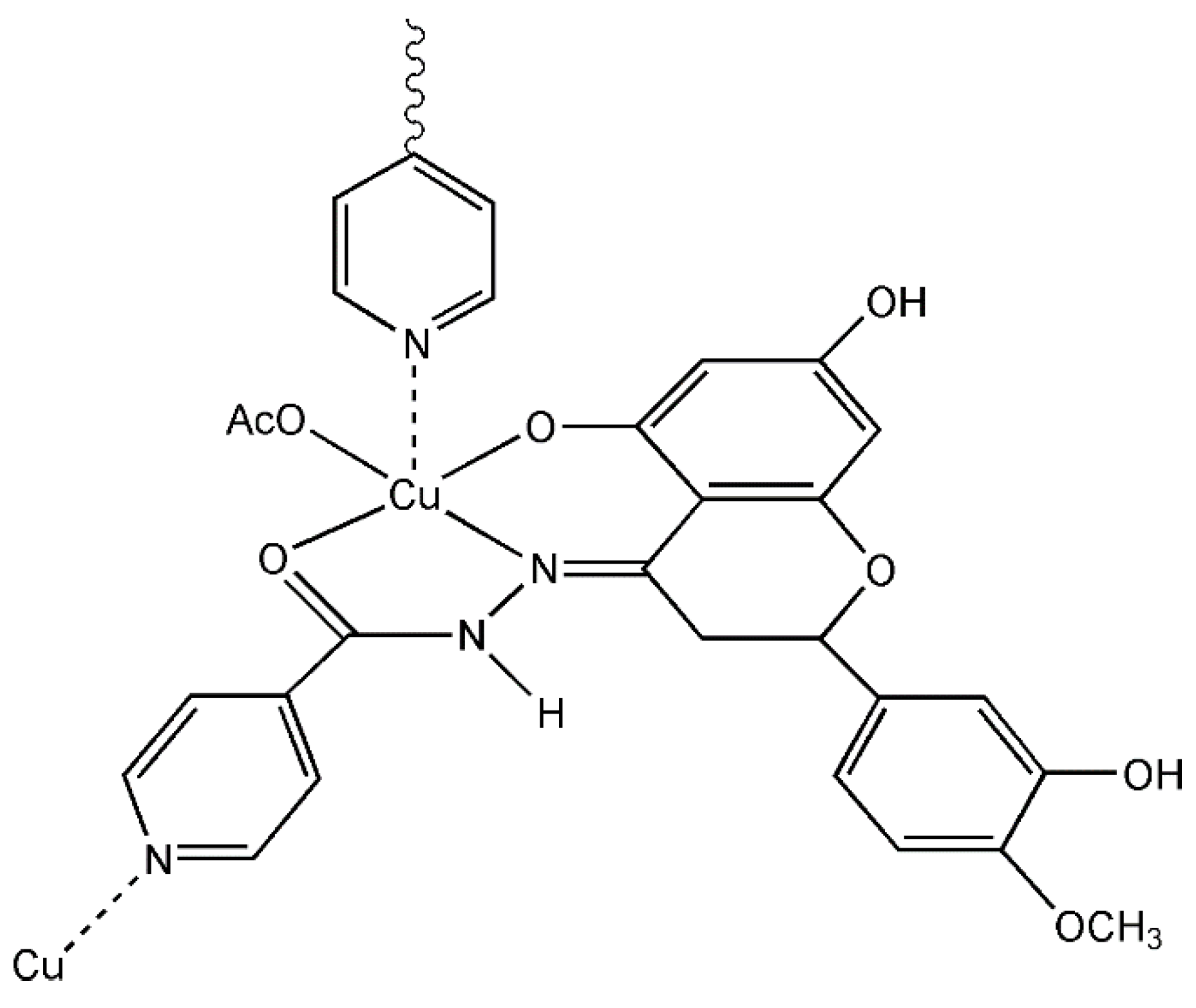
2.1.5. EPR Studies
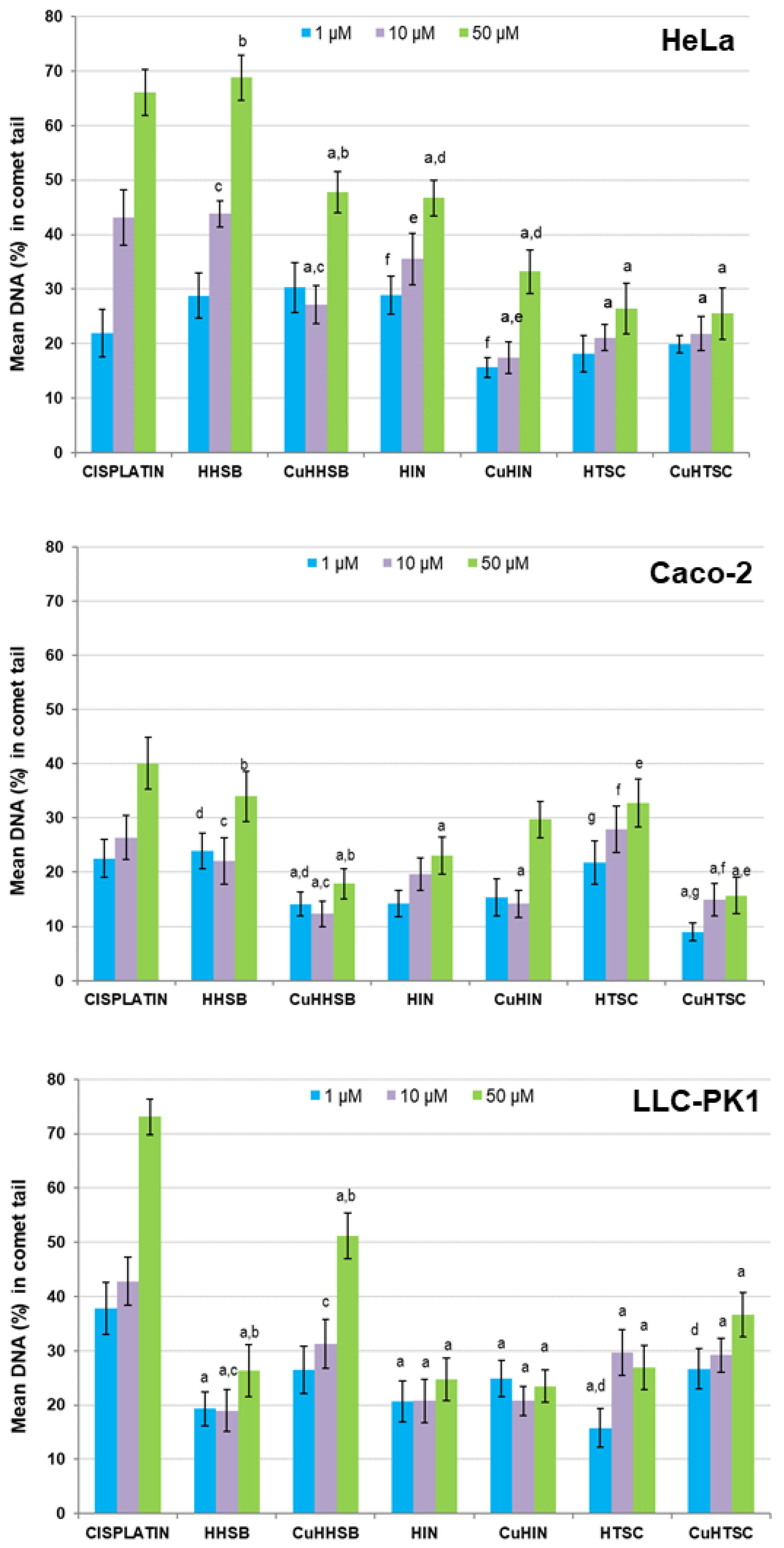
2.1.6. Biological Activity
- Basal endogenous DNA damage
- DSBs induction
- Endogenous oxidative DNA damage
3. Materials and Methods
3.1. Materials in the Synthesis of Copper Complexes with Hesperetin Derivatives
3.2. Apparatus
3.3. Characterization of Ligands
3.4. Characterization of Copper(II) Complexes in Solid State
3.4.1. Characterization of Copper(II) Complex with HHSB in Solid State
3.4.2. Characterization of Copper(II) Complex with HIN in Solid State
3.4.3. Characterization of Copper(II) Complex with HTSC in Solid State
3.5. EPR Experiments
3.6. Genotoxic Activity
3.6.1. Cell Culture
3.6.2. Single-Cell Gel Electrophoresis Assay (Comet Assay)
3.6.3. Double-Strain Breaks
3.6.4. Oxidative DNA Damage
3.6.5. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, J.; Bray, F. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2020; Available online: https://gco.iarc.fr/today/home (accessed on 25 November 2022).
- World Health Organization. Available online: https://www.who.int/news-room/fact-sheets/detail/cervical-cancer (accessed on 25 November 2022).
- López-Gastélum, K.A.; Velázquez-Contreras, E.F.; García, J.J.; Flores-Alamo, M.; Aguirre, G.; Chávez-Velasco, D.; Narayanan, J.; Rocha-Alonzo, F. Mononuclear and tetranuclear copper(Ii) complexes bearing amino acid schiff base ligands: Structural characterization and catalytic applications. Molecules 2021, 26, 7301. [Google Scholar] [CrossRef] [PubMed]
- Pitucha, M.; Korga-Plewko, A.; Czylkowska, A.; Rogalewicz, B.; Drozd, M.; Iwan, M.; Kubik, J.; Humeniuk, E.; Adamczuk, G.; Karczmarzyk, Z.; et al. Influence of complexation of thiosemicarbazone derivatives with Cu (II) ions on their antitumor activity against melanoma cells. Int. J. Mol. Sci. 2021, 22, 3104. [Google Scholar] [CrossRef] [PubMed]
- Al-Shboul, T.M.A.; El-khateeb, M.; Obeidat, Z.H.; Ababneh, T.S.; Al-Tarawneh, S.S.; Al Zoubi, M.S.; Alshaer, W.; Abu Seni, A.; Qasem, T.; Moriyama, H.; et al. Synthesis, Characterization, Computational and Biological Activity of Some Schiff Bases and Their Fe, Cu and Zn Complexes. Inorganics 2022, 10, 112. [Google Scholar] [CrossRef]
- Yusuf, T.L.; Oladipo, S.D.; Zamisa, S.; Kumalo, H.M.; Lawal, I.A.; Lawal, M.M.; Mabuba, N. Design of New Schiff-Base Copper(II) Complexes: Synthesis, Crystal Structures, DFT Study, and Binding Potency toward Cytochrome P450 3A4. ACS Omega 2021, 6, 13704–13718. [Google Scholar] [CrossRef] [PubMed]
- Subin Kumar, K.; Aravindakshan, K.K. Synthesis, cytotoxic, anticancer and antimicrobial activities of some metal complexes of a novel tetradentate Schiff base ligand, (E)-3-((2-((E)-(1-(2-hydroxyphenyl)ethylidene)amino)ethyl)imino)-N-phenylbutanamide. Results Chem. 2021, 3, 100129–100138. [Google Scholar] [CrossRef]
- More, M.S.; Joshi, P.G.; Mishra, Y.K.; Khanna, P.K. Metal complexes driven from Schiff bases and semicarbazones for biomedical and allied applications: A review. Mater. Today Chem. 2019, 14, 100195–100217. [Google Scholar] [CrossRef]
- Ebosie, N.P.; Ogwuegbu, M.O.C.; Onyedika, G.O.; Onwumere, F.C. Biological and analytical applications of Schiff base metal complexes derived from salicylidene-4-aminoantipyrine and its derivatives: A review. J. Iran. Chem. Soc. 2021, 18, 3145–3175. [Google Scholar] [CrossRef]
- Bilici, A.; Kaya, I.; Doǧan, F. Monomer/polymer schiff base copper(II) complexes for catalytic oxidative polymerization of 2,2′dihydroxybiphenyl. J. Polym. Sci. Part A Polym. Chem. 2009, 47, 2977–2984. [Google Scholar] [CrossRef]
- Jiang, S.; Ni, H.; Liu, F.; Gu, S.; Yu, P.; Gou, Y. Binuclear Schiff base copper(II) complexes: Syntheses, crystal structures, HSA interaction and anti-cancer properties. Inorg. Chim. Acta 2020, 499, 119186. [Google Scholar] [CrossRef]
- Roohbakhsh, A.; Parhiz, H.; Soltani, F.; Rezaee, R.; Iranshahi, M. Neuropharmacological properties and pharmacokinetics of the citrus flavonoids hesperidin and hesperetin—A mini-review. Life Sci. 2014, 113, 1–6. [Google Scholar] [CrossRef]
- Khan, A.; Ikram, M.; Hahm, J.R.; Kim, M.O. Antioxidant and Anti-Inflammatory Effects of Citrus Flavonoid Hesperetin: Special Focus on Neurological Disorders. Antioxidants 2020, 9, 609. [Google Scholar] [CrossRef]
- Yap, K.M.; Sekar, M.; Wu, Y.S.; Gan, S.H.; Rani, N.N.I.M.; Seow, L.J.; Subramaniyan, V.; Fuloria, N.K.; Fuloria, S.; Lum, P.T. Hesperidin and its aglycone hesperetin in breast cancer therapy: A review of recent developments and future prospects. Saudi J. Biol. Sci. 2021, 28, 6730–6747. [Google Scholar] [CrossRef] [PubMed]
- Sohel, M.; Sultana, H.; Sultana, T.; Al Amin, M.; Aktar, S.; Ali, M.C.; Bin Rahim, Z.; Hossain, M.A.; Al Mamun, A.; Amin, M.N.; et al. Chemotherapeutic potential of hesperetin for cancer treatment, with mechanistic insights: A comprehensive review. Heliyon 2022, 8, e08815. [Google Scholar] [CrossRef]
- Evans, J.A.; Mendonca, P.; Soliman, K.F.A. Neuroprotective Effects and Therapeutic Potential of the Citrus Flavonoid Hesperetin in Neurodegenerative Diseases. Nutrients 2022, 14, 2228. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Ding, Z.; Chen, J.; Chen, T.; Wang, T.; Huang, J. The AMPK-SIRT1-FoxO1-NF-κB signaling pathway participates in hesperetin-mediated neuroprotective effects against traumatic brain injury via the NLRP3 inflammasome. Immunopharmacol. Immunotoxicol. 2022, 44, 970–983. [Google Scholar] [CrossRef] [PubMed]
- Shirzad, M.; Heidarian, E.; Beshkar, P.; Gholami-Arjenaki, M. Biological effects of hesperetin on Interleukin-6/phosphorylated signal transducer and activator of transcription 3 pathway signaling in prostate cancer PC3 cells. Pharmacogn. Res. 2017, 9, 188–194. [Google Scholar] [CrossRef]
- Zalpoor, H.; Bakhtiyari, M.; Shapourian, H.; Rostampour, P.; Tavakol, C.; Nabi-Afjadi, M. Hesperetin as an anti-SARS-CoV-2 agent can inhibit COVID-19-associated cancer progression by suppressing intracellular signaling pathways. Inflammopharmacology 2022, 30, 1533–1539. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wolfram, J.; Shen, H.; Fang, X.; Ferrari, M. Hesperetin: An inhibitor of the transforming growth factor-β (TGF-β) signaling pathway. Eur. J. Med. Chem. 2012, 58, 390–395. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tan, M.; Zhu, J.; Pan, Y.; Chen, Z.; Liang, H.; Liu, H. Synthesis, cytotoxic activity, and DNA binding properties of copper (II) complexes with hesperetin, naringenin, and apigenin. Bioinorg. Chem. Appl. 2009, 2009, 347872. [Google Scholar] [CrossRef]
- Ferreira de Oliveira, J.M.P.; Santos, C.; Fernandes, E. Therapeutic potential of hesperidin and its aglycone hesperetin: Cell cycle regulation and apoptosis induction in cancer models. Phytomedicine 2020, 73, 152887. [Google Scholar] [CrossRef]
- Roohbakhsh, A.; Parhiz, H.; Soltani, F.; Rezaee, R.; Iranshahi, M. Molecular mechanisms behind the biological effects of hesperidin and hesperetin for the prevention of cancer and cardiovascular diseases. Life Sci. 2015, 124, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Iranshahi, M.; Rezaee, R.; Parhiz, H.; Roohbakhsh, A.; Soltani, F. Protective effects of flavonoids against microbes and toxins: The cases of hesperidin and hesperetin. Life Sci. 2015, 137, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Sykula, A.; Kowalska-Baron, A.; Dzeikala, A.; Bodzioch, A.; Lodyga-Chruscinska, E. An experimental and DFT study on free radical scavenging activity of hesperetin Schiff bases. Chem. Phys. 2019, 517, 91–103. [Google Scholar] [CrossRef]
- Lodyga-Chruscinska, E.; Symonowicz, M.; Sykula, A.; Bujacz, A.; Garribba, E.; Rowinska-Zyrek, M.; Oldziej, S.; Klewicka, E.; Janicka, M.; Krolewska, K.; et al. Chelating ability and biological activity of hesperetin Schiff base. J. Inorg. Biochem. 2015, 143, 34–47. [Google Scholar] [CrossRef]
- Qin, L.; Li, Y.; Liang, F.; Li, L.; Lan, Y.; Li, Z.; Lu, X.; Yang, M.; Ma, D. A microporous 2D cobalt-based MOF with pyridyl sites and open metal sites for selective adsorption of CO2. Microporous Mesoporous Mater. 2022, 341, 112098–112106. [Google Scholar] [CrossRef]
- Qin, L.; Liang, F.; Li, Y.; Wu, J.; Guan, S.; Wu, M.; Xie, S.; Luo, M.; Ma, D. A 2D Porous Zinc-Organic Framework Platform for Loading of 5-Fluorouracil. Inorganics 2022, 10, 202. [Google Scholar] [CrossRef]
- Dong, X.; Li, Y.; Li, D.; Liao, D.; Qin, T.; Prakash, O.; Kumar, A.; Liu, J. A new 3D 8-connected Cd(ii) MOF as a potent photocatalyst for oxytetracycline antibiotic degradation. CrystEngComm 2022, 24, 6933–6943. [Google Scholar] [CrossRef]
- Chandra, S.; Kumar, A. Spectral studies on Co(II), Ni(II) and Cu(II) complexes with thiosemicarbazone (L1) and semicarbazone (L2) derived from 2-acetyl furan. Spectrochim. Acta—Part A Mol. Biomol. Spectrosc. 2007, 66, 1347–1351. [Google Scholar] [CrossRef]
- Chandra, S.; Gupta, L.K. EPR, mass, IR, electronic, and magnetic studies on copper(II) complexes of semicarbazones and thiosemicarbazones. Spectrochim. Acta—Part A Mol. Biomol. Spectrosc. 2005, 61, 269–275. [Google Scholar] [CrossRef]
- Xie, M.X.; Xu, X.Y.; Wang, Y.D. Interaction between hesperetin and human serum albumin revealed by spectroscopic methods. Biochim. Biophys. Acta—Gen. Subj. 2005, 1724, 215–224. [Google Scholar] [CrossRef]
- Issa, R.M.; Khedr, A.M.; Rizk, H. 1H NMR, IR and UV/VIS Spectroscopic Studies of Some Schiff Bases Derived from 2-Aminobenzothiazole and 2-Amino-3-Hydroxypyridine. J. Chinese Chem. Soc. 2008, 55, 875–884. [Google Scholar] [CrossRef]
- Li, Y.; Yang, Z. yin DNA binding affinity and antioxidative activity of copper(II) and zinc(II) complexes with a novel hesperetin Schiff base ligand. Inorg. Chim. Acta 2009, 362, 4823–4831. [Google Scholar] [CrossRef]
- Levina, A.; Crans, D.C.; Lay, P.A. Speciation of metal drugs, supplements and toxins in media and bodily fluids controls in vitro activities. Coord. Chem. Rev. 2017, 352, 473–498. [Google Scholar] [CrossRef]
- Alves, W.A.; De Almeida Santos, R.H.; Paduan-Filho, A.; Becerra, C.C.; Borin, A.C.; Da Costa Ferreira, A.M. Molecular structure and intra- and intermolecular magnetic interactions in chloro-bridged copper(II) dimers. Inorg. Chim. Acta 2004, 357, 2269–2278. [Google Scholar] [CrossRef]
- Thakurta, S.; Roy, P.; Rosair, G.; Gómez-García, C.J.; Garribba, E.; Mitra, S. Ferromagnetic exchange coupling in a new bis(μ-chloro)-bridged copper(II) Schiff base complex: Synthesis, structure, magnetic properties and catalytic oxidation of cycloalkanes. Polyhedron 2009, 28, 695–702. [Google Scholar] [CrossRef]
- Ray, A.; Rizzoli, C.; Pilet, G.; Desplanches, C.; Garribba, E.; Rentschler, E.; Mitra, S. Two new supramolecular architectures of singly phenoxo-bridged copper(II) and doubly phenoxo-bridged manganese(ii) complexes derived from an unusual ONOO donor hydrazone ligand: Syntheses, structural variations, cryomagnetic, DFT, and EPR studies. Eur. J. Inorg. Chem. 2009, 2009, 2915–2928. [Google Scholar] [CrossRef]
- Datta, A.; Liu, P.H.; Huang, J.H.; Garribba, E.; Turnbull, M.; MacHura, B.; Hsu, C.L.; Chang, W.T.; Pevec, A. End-to-end thiocyanato-bridged zig-zag polymers of Cu II, Co II and Ni II with a hydrazone ligand: EPR, magnetic susceptibility and biological study. Polyhedron 2012, 44, 77–87. [Google Scholar] [CrossRef]
- Brodowska, K.; Correia, I.; Garribba, E.; Marques, F.; Klewicka, E.; Łodyga-Chruscińska, E.; Pessoa, J.C.; Dzeikala, A.; Chrusciński, L. Coordination ability and biological activity of a naringenin thiosemicarbazone. J. Inorg. Biochem. 2016, 165, 36–48. [Google Scholar] [CrossRef]
- Hathaway, B.J.; Billing, D.E. The electronic properties and stereochemistry of mono-nuclear complexes of the copper(II) ion. Coord. Chem. Rev. 1970, 5, 143–207. [Google Scholar] [CrossRef]
- Garribba, E.; Micera, G. The determination of the geometry of Cu(II) complexes. An EPR spectroscopy experiment. J. Chem. Educ. 2006, 83, 1229–1232. [Google Scholar] [CrossRef]
- Rizzi, A.C.; Neuman, N.I.; González, P.J.; Brondino, C.D. EPR as a tool for study of isolated and coupled paramagnetic centers in coordination compounds and macromolecules of biological interest. Eur. J. Inorg. Chem. 2016, 2016, 192–207. [Google Scholar] [CrossRef]
- Raj, D.; Kumar Padhi, S. The sporadic μ-pyridine bridge in transition metal complexes: A real bond or an interaction? Coord. Chem. Rev. 2022, 450, 214238. [Google Scholar] [CrossRef]
- Smith, T.D.; Pilbrow, J.R. The determination of structural properties of dimeric transition metal ion complexes from epr spectra. Coord. Chem. Rev. 1974, 13, 173–278. [Google Scholar] [CrossRef]
- Brodowska, K.; Sykuła, A.; Garribba, E.; Lodyga-Chruścińska, E.; Sójka, M. Naringenin Schiff base: Antioxidant activity, acid-base profile, and interactions with DNA. Transit. Met. Chem. 2016, 41, 179–189. [Google Scholar] [CrossRef]
- Garribba, E.; Micera, G.; Sanna, D.; Strinna-Erre, L. The Cu(II)-2,2′-bipyridine system revisited. Inorg. Chim. Acta 2000, 299, 253–261. [Google Scholar] [CrossRef]
- Collins, A.R.; Duthie, S.J.; Dobson, V.L. Direct enzymic detection of endogenous oxidative base damage in human lymphocyte DNA. Carcinogenesis 1993, 14, 1733–1735. [Google Scholar] [CrossRef]
- Sears, C.R.; Turchi, J.J. Complex cisplatin-double strand break (DSB) lesions directly impair cellular non-homologous end-joining (NHEJ) independent of downstream damage response (DDR) pathways. J. Biol. Chem. 2012, 287, 24263–24272. [Google Scholar] [CrossRef]
- Varga, T.; Aplan, P.D. Chromosomal aberrations induced by double strand DNA breaks. DNA Repair 2005, 4, 1038–1046. [Google Scholar] [CrossRef]
- Chen, J.; Cheng, F.; Luo, D.; Huang, J.; Ouyang, J.; Nezamzadeh-Ejhieh, A.; Khan, M.S.; Liu, J.; Peng, Y. Recent advances in Ti-based MOFs in biomedical applications. Dalt. Trans. 2022, 51, 14817–14832. [Google Scholar] [CrossRef]
- Zhang, W.; Ye, G.; Liao, D.; Chen, X.; Lu, C.; Nezamzadeh-Ejhieh, A.; Khan, M.S.; Liu, J.; Pan, Y.; Dai, Z. Recent Advances of Silver-Based Coordination Polymers on Antibacterial Applications. Molecules 2022, 27, 7166. [Google Scholar] [CrossRef]
- Zhang, T.; Sun, Y.; Cao, J.; Luo, J.; Wang, J.; Jiang, Z.; Huang, P. Intrinsic nucleus-targeted ultra-small metal–organic framework for the type I sonodynamic treatment of orthotopic pancreatic carcinoma. J. Nanobiotechnol. 2021, 19, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Guo, J.; Cai, X.; Bin, L.; Lu, C.; Singh, A.; Trivedi, M.; Kumar, A.; Liu, J. Manganese complexes and manganese-based metal-organic frameworks as contrast agents in MRI and chemotherapeutics agents: Applications and prospects. Colloids Surfaces B Biointerfaces 2022, 213, 112432–112454. [Google Scholar] [CrossRef] [PubMed]
- Han, I.; Choi, S.A.; Lee, D.N. Therapeutic Application of Metal–Organic Frameworks Composed of Copper, Cobalt, and Zinc: Their Anticancer Activity and Mechanism. Pharmaceutics 2022, 14, 378. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, S.M.; Hashemi, S.A.; Ghahramani, Y.; Azhdari, R.; Yousefi, K.; Gholami, A.; Fallahi Nezhad, F.; Vijayakameswara Rao, N.; Omidifar, N.; Chiang, W.H. Antiproliferative and Apoptotic Effects of Graphene Oxide @AlFu MOF Based Saponin Natural Product on OSCC Line. Pharmaceuticals 2022, 15, 1137. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Zhao, Y.; Sun, Y.; Cao, J. Recent Progress of Metal-Organic Framework-Based Photodynamic Therapy for Cancer Treatment. Int. J. Nanomed. 2022, 17, 2367–2395. [Google Scholar] [CrossRef]
- Khabour, O.F.; Saleh, N.; Alzoubi, K.H.; Hisaindee, S.; Al-Fyad, D.; Al-Kaabi, L.; Dodeen, A.; Esmadi, F.T. Genotoxicity of structurally related copper and zinc containing Schiff base complexes. Drug Chem. Toxicol. 2013, 36, 435–442. [Google Scholar] [CrossRef]
- Stolarczyk, M.; Wolska, A.; Mikołajczyk, A.; Bryndal, I.; Cieplik, J.; Lis, T.; Matera-Witkiewicz, A. A new pyrimidine schiff base with selective activities against enterococcus faecalis and gastric adenocarcinoma. Molecules 2021, 26, 2296. [Google Scholar] [CrossRef]
- Rao, N.N.; Kishan, E.; Gopichand, K.; Nagaraju, R.; Ganai, A.M.; Rao, P.V. Design, synthesis, spectral characterization, DNA binding, photo cleavage and antibacterial studies of transition metal complexes of benzothiazole Schiff base. Chem. Data Collect. 2020, 27, 100368–100381. [Google Scholar] [CrossRef]
- Bansal, A.; Saleh-E-In, M.M.; Kar, P.; Roy, A.; Sharma, N.R. Synthesis of Carvacrol Derivatives as Potential New Anticancer Agent against Lung Cancer. Molecules 2022, 27, 4597. [Google Scholar] [CrossRef]
- Li, Y.; Yang, Z.Y.; Wang, M.F. Synthesis, characterization, DNA binding properties, fluorescence studies and antioxidant activity of transition metal complexes with hesperetin-2-hydroxy benzoyl hydrazone. J. Fluoresc. 2010, 20, 891–905. [Google Scholar] [CrossRef]
- Singh, N.P.; McCoy, M.T.; Tice, R.R.; Schneider, E.L. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 1988, 175, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Blasiak, J.; Kowalik, J. A comparison of the in vitro genotoxicity of tri- and hexavalent chromium. Mutat. Res.—Genet. Toxicol. Environ. Mutagen. 2000, 469, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Klaude, M.; Eriksson, S.; Nygren, J.; Ahnström, G. The comet assay: Mechanisms and technical considerations. Mutat. Res.—DNA Repair 1996, 363, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Blasiak, J.; Synowiec, E.; Tarnawska, J.; Czarny, P.; Poplawski, T.; Reiter, R.J. Dental methacrylates may exert genotoxic effects via the oxidative induction of DNA double strand breaks and the inhibition of their repair. Mol. Biol. Rep. 2012, 39, 7487–7496. [Google Scholar] [CrossRef]
- Blasiak, J.; Arabski, M.; Krupa, R.; Wozniak, K.; Zadrozny, M.; Kasznicki, J.; Zurawska, M.; Drzewoski, J. DNA damage and repair in type 2 diabetes mellitus. Mutat. Res.—Fundam. Mol. Mech. Mutagen. 2004, 554, 297–304. [Google Scholar] [CrossRef]

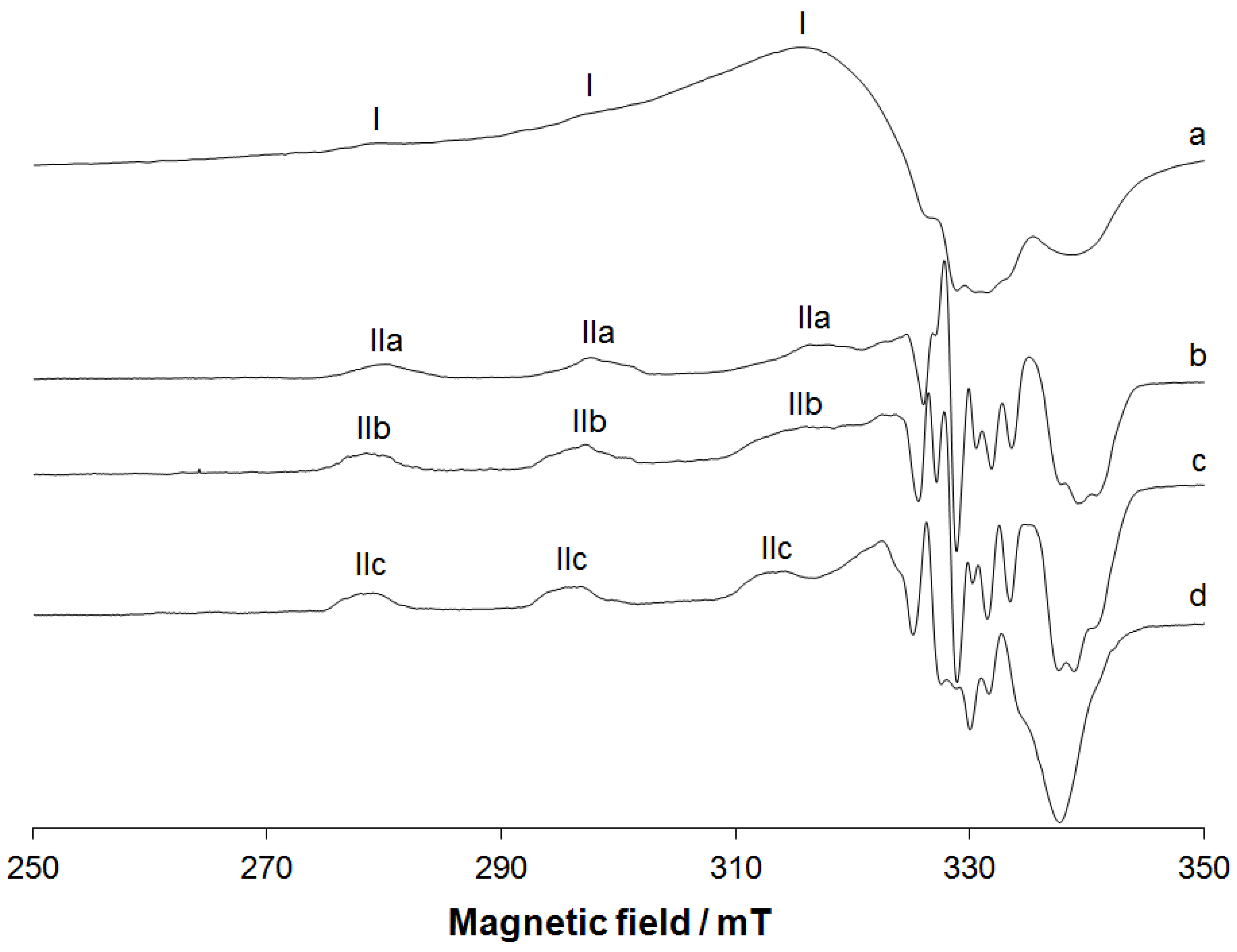


| Species | gz | Az/10−4 cm−1 | ∆H/mT a | Coordination Mode |
|---|---|---|---|---|
| [Cu(HHSB)(AcO)] | 2.230 | 198.6 | 3.6 | (O−, N, Oket); AcO– |
| [Cu(HHSB)(DMF)]+ | 2.243 | 189.8 | 3.8 | (O−, N, Oket); ODMF |
| [Cu(HHSB)(DMSO)]+ | 2.244 | 190.4 | 3.7 | (O−, N, Oket); ODMSO |
| [Cu(HHSB)(H2O)]+ | 2.243 | 188.8 | 3.8 | (O−, N, Oket); Owater b |
| [Cu(HTSC)(AcO)] | ~2.20 | ~185 | c | (O−, N, S); AcO– |
| [Cu(HTSC)(DMF)]+ | 2.181 | 192.7 | 3.9 | (O−, N, S); ODMF |
| [Cu(HTSC)(DMSO)]+ | 2.194 | 190.6 | 4.1 | (O−, N, S); ODMSO |
| [Cu(HTSC)(H2O)]+ | 2.200 | 185.0 | 3.7 | (O−, N, S); Owater d |
| Species | System | gz | gx, gy | Coordination Mode |
|---|---|---|---|---|
| {[Cu(HIN)](AcO)}n | Powder | ~2.19 | 2.063 | (O−, N, Oket); AcO–; Npyrax |
| {[Cu(HIN)](AcO)}n | DMF | ~2.20 | 2.064 | (O−, N, Oket); AcO–; Npyrax |
| {[Cu(HIN)](AcO)}n | DMSO/DMF | ~2.20 | 2.063 | (O−, N, Oket); AcO–; Npyrax |
| {[Cu(HIN)](AcO)}n | DMSO | 2.192 | 2.062 | (O−, N, Oket); AcO–; Npyrax |
| Compound | Mean DNA (%) in Comet Tail (±S.E.M.) of HeLa Cells | |
|---|---|---|
| DSBs | Endo III | |
| Cisplatin | 30.2 ± 3.4 | 26.5 ± 3.9 |
| HHSB | 23.3 ± 1.0 a | 7.5 ± 3.9 a |
| CuHHSB | 14.4 ± 0.7 a,b | 8.3 ± 2.7 a |
| HIN | 19.5 ± 0.7 a | 5.3 ± 3.5 a |
| CuHIN | 7.7 ± 3.6 a | 12.2 ± 4.2 a |
| HTSC | 13.5 ± 0.7 a,c | 10.2 ± 2.0 a |
| CuHTSC | 6.4 ± 2.3 a,c | 11.6 ± 1.9 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sykuła, A.; Nowak, A.; Garribba, E.; Dzeikala, A.; Rowińska-Żyrek, M.; Czerwińska, J.; Maniukiewicz, W.; Łodyga-Chruścińska, E. Spectroscopic Characterization and Biological Activity of Hesperetin Schiff Bases and Their Cu(II) Complexes. Int. J. Mol. Sci. 2023, 24, 761. https://doi.org/10.3390/ijms24010761
Sykuła A, Nowak A, Garribba E, Dzeikala A, Rowińska-Żyrek M, Czerwińska J, Maniukiewicz W, Łodyga-Chruścińska E. Spectroscopic Characterization and Biological Activity of Hesperetin Schiff Bases and Their Cu(II) Complexes. International Journal of Molecular Sciences. 2023; 24(1):761. https://doi.org/10.3390/ijms24010761
Chicago/Turabian StyleSykuła, Anna, Adriana Nowak, Eugenio Garribba, Aliaksandr Dzeikala, Magdalena Rowińska-Żyrek, Justyna Czerwińska, Waldemar Maniukiewicz, and Elżbieta Łodyga-Chruścińska. 2023. "Spectroscopic Characterization and Biological Activity of Hesperetin Schiff Bases and Their Cu(II) Complexes" International Journal of Molecular Sciences 24, no. 1: 761. https://doi.org/10.3390/ijms24010761
APA StyleSykuła, A., Nowak, A., Garribba, E., Dzeikala, A., Rowińska-Żyrek, M., Czerwińska, J., Maniukiewicz, W., & Łodyga-Chruścińska, E. (2023). Spectroscopic Characterization and Biological Activity of Hesperetin Schiff Bases and Their Cu(II) Complexes. International Journal of Molecular Sciences, 24(1), 761. https://doi.org/10.3390/ijms24010761






