Phosphodiesterase 5a Signalling in Skeletal Muscle Pathophysiology
Abstract
1. Introduction
2. Results
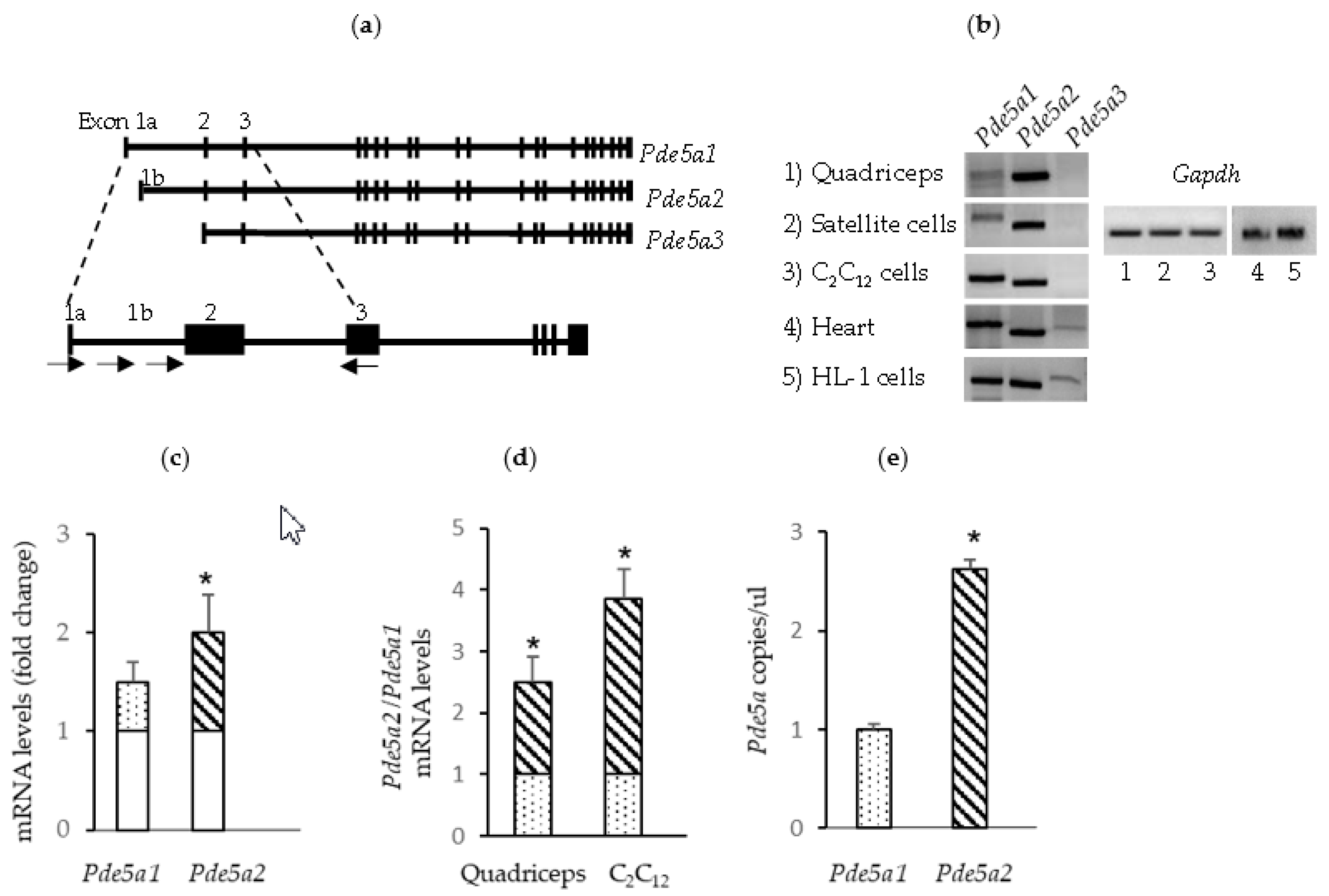
2.1. Pde5a Expression in Murine Skeletal Muscular Cells
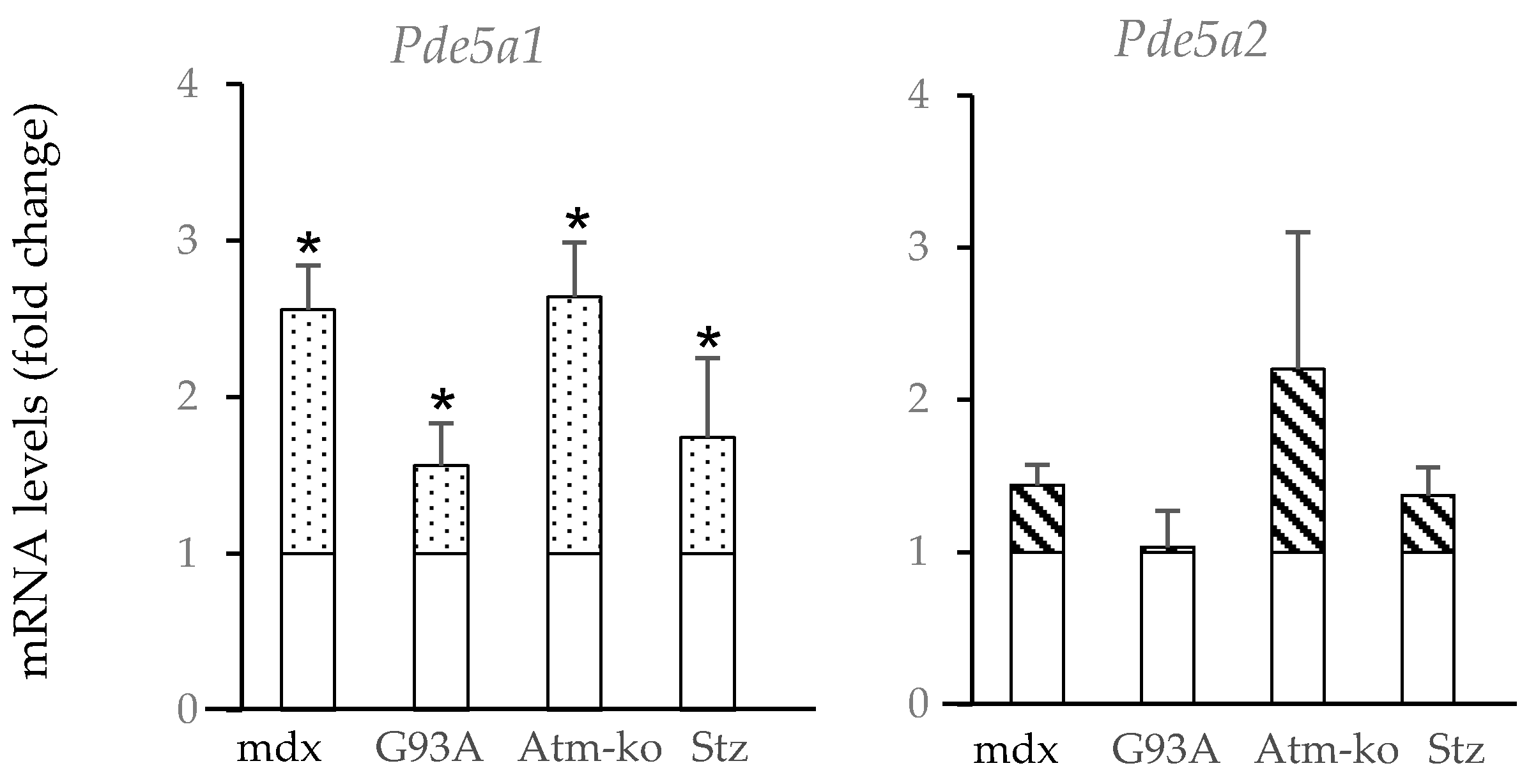
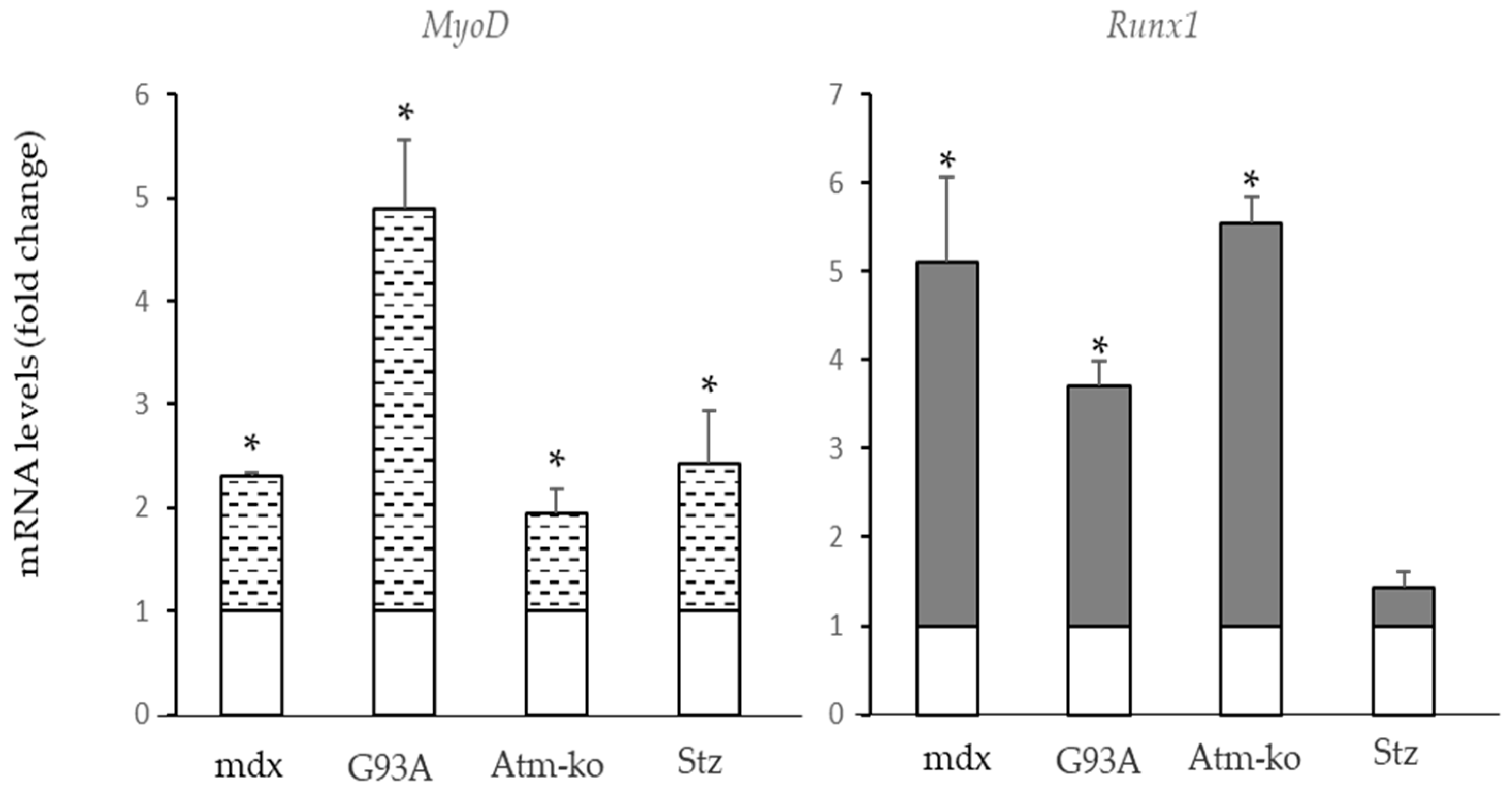
2.2. Pde5a1 Increases in Damaged Skeletal Muscles
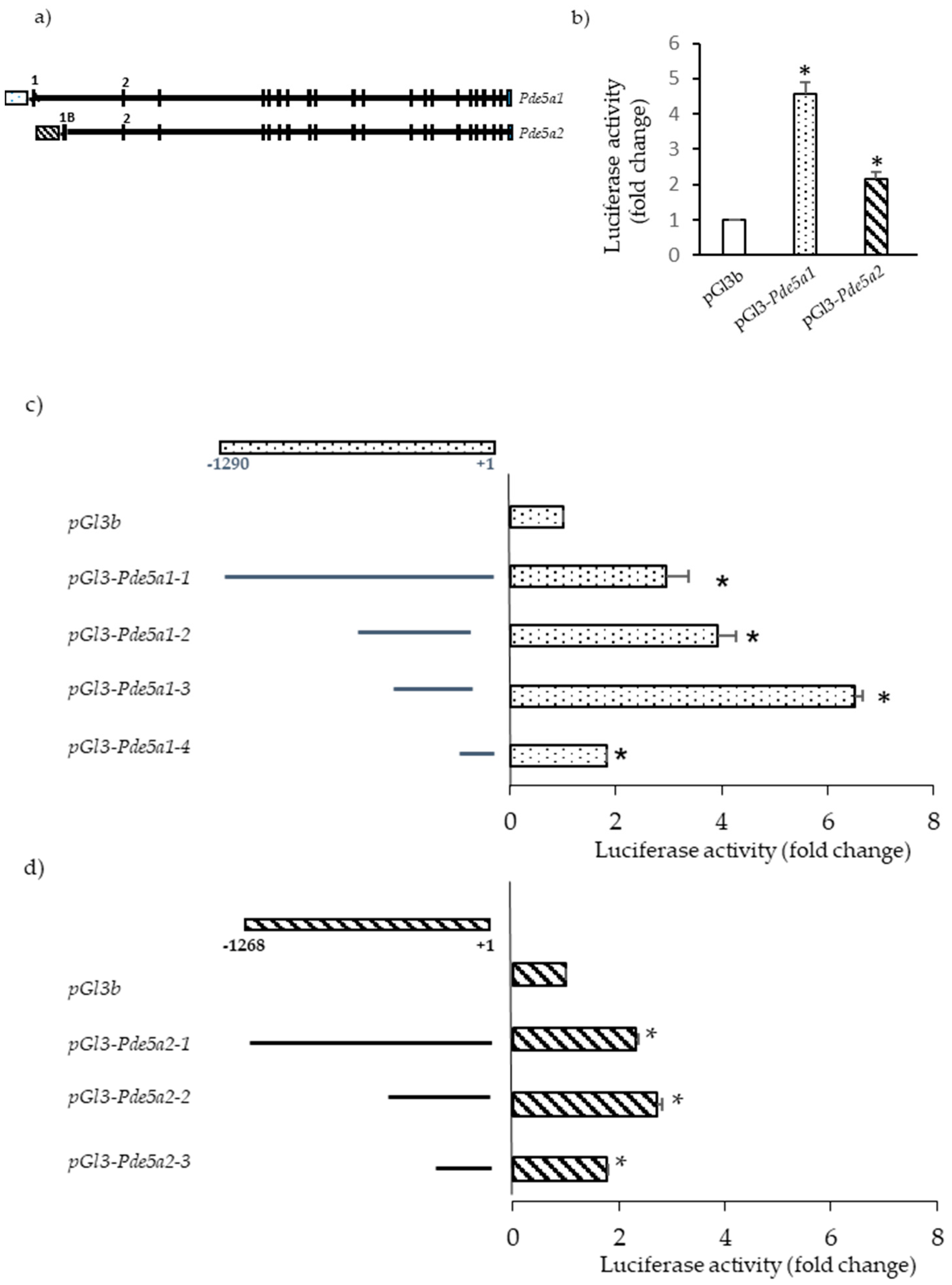
2.3. Promoter Analysis
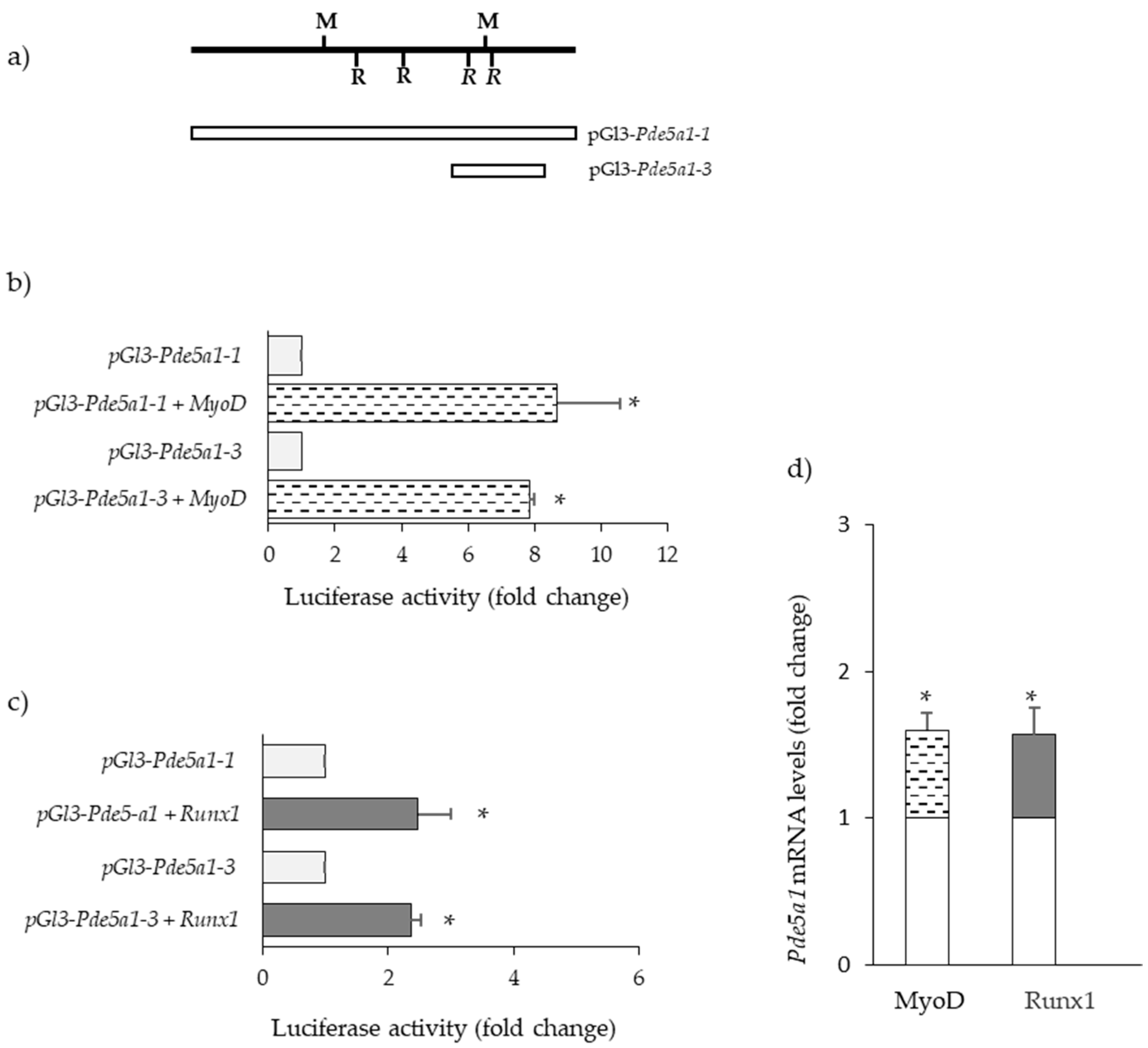
2.4. Transcription Factors Regulating Pde5a Promoters
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Cell Cultures
4.3. Cell Immunofluorescence
4.4. RNA Preparation and Analysis
4.5. Vector Constructs
4.6. Transfection and Luciferase Assays
4.7. Statistical Analysis
4.8. Bioinformatics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Piil, P.; Jørgensen, T.S.; Egelund, J.; Damsgaard, R.; Gliemann, L.; Hellsten, Y.; Nyberg, M. Exercise training improves blood flow to contracting skeletal muscle of older men via enhanced cGMP signalling. J. Appl. Physiol. 2018, 124, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M. Molecular Physiology of Membrane Guanylyl Cyclase Receptors. Physiol. Rev. 2016, 96, 751–804. [Google Scholar] [CrossRef] [PubMed]
- Blanton, R.M. cGMP Signaling and Modulation in Heart Failure. J. Cardiovasc. Pharmacol. 2020, 75, 385–398. [Google Scholar] [CrossRef]
- Jordan, E.; Balke, J.E.; Zhang, L.; Percival, J.M. Neuronal nitric oxide synthase (nNOS) splice variant function: Insights into nitric oxide signaling from skeletal muscle. Nitric Oxide 2019, 82, 35–47. [Google Scholar] [CrossRef]
- Conti, M.; Beavo, J. Biochemistry and Physiology of Cyclic Nucleotide Phosphodiesterases: Essential Components in Cyclic Nucleotide Signaling. Ann. Rev. Biochem. 2007, 76, 481–511. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, F.; Murata, T.; Simizu, K.; Degerman, E.; Maurice, D.; Manganiello, V. Cyclic Nucleotide Phosphodiesterases: Important signaling modulators and therapeutic targets. Oral Dis. 2015, 21, e25–e50. [Google Scholar] [CrossRef] [PubMed]
- Lugnier, C.; Meyer, A.; Talha, S.; Geny, B. Cyclic nucleotide phosphodiesterases: New targets in the metabolic syndrome? Pharmacol. Ther. 2020, 208, 107475. [Google Scholar] [CrossRef]
- Dunkerly, E.B.; Kass, D.A. Myocardial Phosphodiesterases and their Role in cGMP Regulation. J. Cardiovasc. Pharmacol. 2020, 75, 483–493. [Google Scholar] [CrossRef]
- Delhaye, S.; Bardoni, B. Role of phosphodiesterases in the pathophysiology of neurodevelopmental disorders. Mol. Psychiatry 2021, 26, 4570–4582. [Google Scholar] [CrossRef]
- Bisserier, M.; Pradhan, N.; Hadri, L. Current and emerging therapeutic approaches to pulmonary hypertension. Rev. Cardiovasc. Med. 2020, 21, 163–179. [Google Scholar] [CrossRef]
- Erro, R.; Mencacci, N.E.; Bhatia, K.P. The Emerging Role of Phosphodiesterases in Movement Disorders. Mov. Disord. 2021, 36, 2225–2243. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Goldminz, A.M.; Kim, N.; Gottlieb, A.B. Phosphodiesterase 4-targeted treatments for autoimmune diseases. BMC Med. 2013, 11, 96. [Google Scholar] [CrossRef] [PubMed]
- Schepers, M.; Tiane, A.; Paes, D.; Sanchez, S.; Rombaut, B.; Piccart, E.; Rutten, B.P.F.; Brone, B.; Hellings, N.; Prickaerts, J.; et al. Targeting Phosphodiesterases-Towards a Tailor-Made Approach in Multiple Sclerosis Treatment. Front. Immunol. 2019, 10, 1727. [Google Scholar] [CrossRef] [PubMed]
- Hanna, R.; Nour-Eldine, W.; Saliba, Y.; Dagher-Hamalian, C.; Hachem, P.; Abou-Khalil, P.; Mika, D.; Varin, A.; El Hayek, M.S.; Pereira, L.; et al. Cardiac Phosphodiesterases Are Differentially Increased in Diabetic Cardiomyopathy. Life Sci. 2021, 283, 119857. [Google Scholar] [CrossRef] [PubMed]
- Campolo, F.; Pofi, R.; Venneri, M.; Isidori, A. Priming metabolism with the type 5 phosphodiesterase: The role of cGMP-hydrolyzing enzymes. Curr. Opin. Pharmacol. 2021, 60, 298–305. [Google Scholar] [CrossRef]
- Cesarini, V.; Guida, E.; Campolo, F.; Crescioli, C.; Di Baldassaree, A.; Pisano, C.; Balistreri, C.M.; Ruvolo, G.; Jannini, E.A.; Dolci, S. Type 5 phosphodiesterase (PDE5) and the vascular tree: From embryogenesis to aging and disease. Mech. Ageing Dev. 2020, 190, 111311. [Google Scholar] [CrossRef]
- Loma, O.; Zaccolo, M. Phosphodiesterases Maintain Signaling Fidelity via Compartmentalization of Cyclic Nucleotides. Physiology 2014, 29, 141–149. [Google Scholar] [CrossRef]
- Calamera, G.; Roman-Moltzau, L.; Levy, F.O.; Andressen, K.W. Phosphodiesterases and Compartmentation of cAMP and cGMP Signaling in Regulation of Cardiac Contractility in Normal and Failing Hearts. Int. J. Mol. Sci. 2022, 23, 2145. [Google Scholar] [CrossRef]
- Barbagallo, F.; Xu, B.; Reddy, G.R.; West, T.; Wang, Q.; Fu, Q.; Li, M.; Shi, Q.; Ginsburg, K.S.; Ferrier, W.; et al. Genetically Encoded Biosensors Reveal PKA Hyperphosphorylation on the Myofilaments in Rabbit Heart Failure. Circ. Res. 2016, 119, 931–943. [Google Scholar] [CrossRef]
- Bloom, T.J. Cyclic nucleotide phosphodiesterase isozymes expressed in mouse skeletal muscle Can. J. Physiol. Pharmacol. 2002, 80, 1132–1135. [Google Scholar] [CrossRef]
- Genders, A.J.; Bradley, E.A.; Rattigan, S.; Richards, S.M. cGMP phosphodiesterase inhibition improves the vascular and metabolic actions of insulin in skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2011, 301, E342–E350. [Google Scholar] [CrossRef] [PubMed]
- Tetsi, L.; Charles, A.L.; Paradis, S.; Lejay, A.; Talha, S.; Geny, B.; Lugnier, C. Effects of cyclic nucleotide phosphodiesterases (PDEs) on mitochondrial skeletal muscle functions. Cell. Mol. Life Sci. 2017, 74, 1883–1893. [Google Scholar] [CrossRef] [PubMed]
- Bloom, T.J. Age-related alterations in cyclic nucleotide phosphodiesterase activity in dystrophic mouse leg muscle. Can. J. Physiol. Pharmacol. 2005, 83, 1055–1060. [Google Scholar] [CrossRef] [PubMed]
- Pokreisz, P.; Vandenwijngaert, S.; Bito, V.; Van den Bergh, A.; Lenaerts, I.; Busch, C.; Marsboom, G.; Gheysens, O.; Vermeersch, P.; Biesmans, L.; et al. Ventricular phosphodiesterase-5 expression is increased in patients with advanced heart failure and contributes to adverse ventricular remodeling after myocardial infarction in mice. Circulation 2009, 119, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Shan, X.; Quaile, M.P.; Monk, J.K.; French, B.; Cappola, T.P.; Margulies, K.B. Differential expression of PDE5 in failing and nonfailing human myocardium. Circ. Heart Fail. 2012, 5, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Isidori, A.M.; Cornacchione, M.; Barbagallo, F.; Di Grazia, A.; Barrios, F.; Fassina, L.; Monaco, L.; Giannetta, E.; Gianfrilli, D.; Garofalo, S.; et al. Inhibition of type 5 phosphodiesterase counteracts β2-adrenergic signalling in beating cardiomyocytes. Cardiovasc. Res. 2015, 106, 408–420. [Google Scholar] [CrossRef]
- Chen, S.; Yan, C. An update of cyclic nucleotide phosphodiesterase as a target for cardiac diseases. Expert Opin. Drug Discov. 2021, 16, 183–196. [Google Scholar] [CrossRef]
- Giannetta, E.; Isidori, A.M.; Galea, N.; Carbone, I.; Mandosi, E.; Vizza, C.D.; Naro, F.; Morano, S.; Fedele, F.; Lenzi, A. Chronic Inhibition of cGMP Phosphodiesterase 5A Improves Diabetic Cardiomyopathy A Randomized, Controlled Clinical Trial Using Magnetic Resonance Imaging With Myocardial Tagging. Circulation 2012, 125, 2323–2333. [Google Scholar] [CrossRef]
- Koka, S.; Das, A.; Zhu, S.G.; Durrant, D.; Xi, L.; Kukreja, R.C. Long-acting phosphodiesterase-5 inhibitor tadalafil attenuates doxorubicin-induced cardiomyopathy without interfering with chemotherapeutic effect. J. Pharmacol. Exp. Ther. 2010, 334, 1023–1030. [Google Scholar] [CrossRef]
- Tzoumas, N.; Tariq, E.; Farrah, T.E.; Dhaun, N.; Webb, D.J. Established and emerging therapeutic uses of PDE type 5 inhibitors in cardiovascular disease. Br. J. Pharmacol. 2020, 177, 5467–5488. [Google Scholar] [CrossRef]
- Percival, J.M.; Adamo, C.A.; Beavo, J.A.; Froehner, S.C. Evaluation of the Therapeutic Utility of Phosphodiesterase 5A Inhibition in the mdx Mouse Model of Duchenne Muscular Dystrophy. Handb. Exp. Pharmacol. 2011, 204, 323–344. [Google Scholar] [CrossRef]
- Percival, J.M.; Whitehead, N.P.; Adams, M.E.; Adamo, C.M.; Beavo, J.A.; Froehner, S.C. Sildenafil reduces respiratory muscle weakness and fibrosis in the mdx mouse model of Duchenne muscular dystrophy. J. Pathol. 2012, 228, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Asai, A.; Sahani, N.; Kaneki, M.; Ouchi, Y.; Martyn, J.A.J.; Yasuhara, S.E. Primary Role of Functional Ischemia, Quantitative Evidence for the Two-Hit Mechanism, and Phosphodiesterase-5 Inhibitor Therapy in Mouse Muscular Dystrophy. PLoS ONE 2007, 2, e806. [Google Scholar] [CrossRef] [PubMed]
- De Arcangelis, V.; Strimpakos, G.; Gabanella, F.; Corbi, N.; Luvisetto, S.; Magrelli, A.; Onori, A.; Passananti, C.; Pisani, C.; Rome, S.; et al. Pathways implicated in tadalafil amelioration of Duchenne Muscular Dystrophy. J. Cell Physiol. 2016, 231, 224–232. [Google Scholar] [CrossRef] [PubMed]
- Nyberg, M.; Piil, P.; Egelund, J.; Sprague, R.S.; Mortensen, S.P.; Hellsten, Y. Potentiation of cGMP signaling increases oxygen delivery and oxidative metabolism in contracting skeletal muscle of older but not young humans. Physiol. Rep. 2015, 3, e12508. [Google Scholar] [CrossRef] [PubMed]
- Duranti, G.; Ceci, R.; Sgrò, P.; Sabatini, S.; Di Luigi, L. Influence of the PDE5 inhibitor tadalafil on redox status and antioxidant defense system in C2C12 skeletal muscle cells. Cell Stress Chap. 2017, 22, 389–396. [Google Scholar] [CrossRef]
- Sheffield-Moore, M.; Wiktorowicz, J.E.; Soman, K.V.; Danesi, C.P.; Kinsky, M.P.; Dillon, E.L.; Randolph, K.M.; Casperson, S.L.; Gore, D.C.; Horstman, A.M.; et al. Sildenafil increases muscle protein synthesis and reduces muscle fatigue. Clin. Transl. Sci. 2013, 6, 463–468. [Google Scholar] [CrossRef]
- Campolo, F.; Zevini, A.; Cardarelli, S.; Monaco, L.; Barbagallo, F.; Pellegrini, M.; Cornacchione, M.; Di Grazia, A.; De Arcangelis, V.; Gianfrilli, D.; et al. Identification of Murine Phosphodiesterase 5A Isoforms and their Functional Characterization in HL-1 Cardiac Cell Line. J. Cell. Physiol. 2018, 233, 325–337. [Google Scholar] [CrossRef]
- Cardarelli, S.; Giorgi, M.; Naro, F.; Malatesta, F.; Biagioni, S.; Saliola, M. Use of the KlADH3 promoter for the quantitative production of the murine PDE5A isoforms in the yeast Kluyveromyces lactis. Microb. Cell Fact. 2017, 16, 159. [Google Scholar] [CrossRef]
- Cardarelli, S.; Miele, A.E.; Zamparelli, C.; Biagioni, S.; Naro, F.; Malatesta, F.; Giorgi, M.; Saliola, M. The oligomeric assembly of the phosphodiesterase-5 is a mixture of dimers and tetramers: A putative role in the regulation of function. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2183–2190. [Google Scholar] [CrossRef]
- Cardarelli, S.; Miele, A.E.; Campolo, F.; Massimi, M.; Mancini, P.; Biagioni, S.; Naro, F.; Giorgi, M.; Saliola, M. Cellular Redox Metabolism Is Modulated by the Distinct Localization of Cyclic Nucleotide Phosphodiesterase 5A Isoforms. Int. J. Mol. Sci. 2022, 23, 8587. [Google Scholar] [CrossRef] [PubMed]
- Shibata, N. Transgenic mouse model for familial amyotrophic lateral sclerosis with superoxide dismutase-1 mutation. Neuropathology 2001, 21, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Tassinari, V.; De Gennaro, V.; La Sala, G.; Marazziti, D.; Bolasco, G.; Aguanno, S.; De Angelis, L.; Naro, F.; Pellegrini, M. Atrophy, oxidative switching and ultrastructural defects in skeletal muscle of the ataxia telangiectasia mouse model. J. Cell Sci. 2019, 132, jcs223008. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Murakami, N.; Saito, Y.; ·Goto, Y.; Koishi, K.; Nonaka, I. Expression of MyoD and myogenin in dystrophic mice, mdx and dy, during regeneration. Acta Neuropathol. 2000, 99, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Monaco, L. 2022; Unpublished work.
- Jensen, L.; Jorgensen, L.H.; Bech, R.D.; Frandsen, U.; Schroder, H.D. Skeletal Muscle Remodelling as a Function of Disease Progression in Amyotrophic Lateral Sclerosis. BioMed Res. Int. 2016, 2016, 5930621. [Google Scholar] [CrossRef]
- Barns, M.; Gondro, C.; Tellam, R.L.; Radley-Crabb, H.G.; Grounds, M.D.; Shavlakadze, T. Molecular analyses provide insight into mechanisms underlying sarcopenia and myofibre denervation in old skeletal muscles of mice. Int. J. Biochem. Cell Biol. 2014, 53, 174–185. [Google Scholar] [CrossRef]
- Carvalho, T.M.D.C.S.; Cardarelli, S.; Giorgi, M.; Lenzi, A.; Isidori, A.M.; Naro, F. Phosphodiesterases Expression during Murine Cardiac Development. Int. J. Mol. Sci. 2021, 22, 2593. [Google Scholar] [CrossRef]
- Dehoux, M.; Van Beneden, R.; Pasko, N.; Lause, P.; Verniers, J.; Underwood, L.; Ketelslegers, J.M.; Thissen, J.P. Role of the Insulin-Like Growth Factor I Decline in the Induction of Atrogin-1/MAFbx during Fasting and Diabetes. Endocrinology 2004, 145, 4806–4812. [Google Scholar] [CrossRef][Green Version]
- O’Neill, B.T.; Bhardwaj, G.; Penniman, C.M.; Krumpoch, M.T.; Suarez Beltran, P.A.; Klaus, K.; Poro, K.; Li, M.; Pan, H.; Dreyfuss, J.M.; et al. FoxO Transcription Factors Are Critical Regulators of Diabetes-Related Muscle Atrophy. Diabetes 2019, 68, 556–570. [Google Scholar] [CrossRef]
- Zammit, P.S. Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin. Cell Dev. Biol. 2017, 72, 19–32. [Google Scholar] [CrossRef]
- Shirakawa, T.; Toyono, T.; Inoue, A.; Matsubara, T.; Kawamoto, T.; Shoichiro, K. Factors Regulating or Regulated by Myogenic Regulatory Factors in Skeletal Muscle Stem Cells. Cells 2022, 11, 1493. [Google Scholar] [CrossRef] [PubMed]
- Park, K.H.J.; Franciosi, S.; Leavitt, B.R. Postnatal muscle modification by myogenic factors modulates neuropathology and survival in an ALS mouse model. Nat. Commun. 2013, 4, 2906. [Google Scholar] [CrossRef] [PubMed]
- Chuang, L.S.H.; Ito, K.; Ito, Y. RUNX family: Regulation and diversification of roles through interacting proteins. Int. J. Cancer 2013, 132, 1260–1271. [Google Scholar] [CrossRef] [PubMed]
- Porter, J.D.; Merriam, A.P.; Leaky, P.; Gong, B.; Khanna, S. Dissection of temporal gene expression signatures of affected and spared muscle groups in dystrophin-deficient (mdx) mice. Hum. Mol. Genet. 2003, 12, 1813–1821. [Google Scholar] [CrossRef][Green Version]
- Jose-Luis Gonzalez de Aguilar, J.L.; Niederhauser-Wiederkehr, C.; Halter, B.; De Tapia, M.; Di Scala, F.; Demougin, P.; Dupuis, L.; Primig, M.; Meininger, V.; Loeffle, J.P. Gene profiling of skeletal muscle in an amyotrophic lateral sclerosis mouse model. Physiol. Genom. 2008, 32, 207–218. [Google Scholar] [CrossRef]
- Bao, M.; Liu, S.; Yu, X.Y.; Wu, C.; Chen, Q.; Ding, H.; Shen, C.; Wang, B.; Wang, S.; Song, Y.H.; et al. Runx1 promotes satellite cell proliferation during ischemia—Induced muscle regeneration. Biochem. Biophys. Res. Comm. 2018, 503, 2993–2997. [Google Scholar] [CrossRef]
- Riddell, A.; McBride, M.; Braun, T.; Stuart, A.; Nicklin, S.A.; Cameron, E.; Loughrey, C.; Martin, T.P. RUNX1: An emerging therapeutic target for cardiovascular disease. Cardiovas. Res. 2020, 116, 1410–1423. [Google Scholar] [CrossRef]
- Wang, X.; Blagen, C.; Fan, J.; Nowak, S.J.; Taniuchi, I.; Littman, D.R.; Burden, S.J. Runx1 prevents wasting, myofibrillar disorganization, and autophagy of skeletal muscle. Genes Dev. 2015, 19, 1715–1722. [Google Scholar] [CrossRef]
- Umansky, K.B.; Gruenbaum-Cohen, Y.; Tsoory, M.; Feldmesser, E.; Goldenberg, D.; Brenner, O.; Groner, Y. Runx1 Transcription Factor Is Required for Myoblasts Proliferation during Muscle Regeneration. PLoS Genet. 2015, 11, e1005457. [Google Scholar] [CrossRef]
- Gao, L.; Yang, M.; Wei, Z.; Gu, M.; Yang, L.; Bai, C.; Wu, Y.; Li, G. MSTN Mutant Promotes Myogenic Differentiation by Increasing Demethylase TET1 Expression via the SMAD2/SMAD3 Pathway. Int. J. Biol. Sci. 2020, 16, 1324–1334. [Google Scholar] [CrossRef]
- Gu, M.; Zou, X.; Zhu, L.; Gao, Y.; Bai, C.; Yang, L.; Li, G. Myostatin Mutation Promotes Glycolysis by Increasing Phosphorylation of Phosphofructokinase via Activation of PDE5A-cGMP-PKG in Cattle Heart. Front. Cell. Dev. Biol. 2022, 9, 774185. [Google Scholar] [CrossRef] [PubMed]
- Bolger, G.B. The PDE-Opathies: Diverse Phenotypes Produced by a Functionally Related Multigene Family. Trends Genet. 2021, 37, 669–681. [Google Scholar] [CrossRef]
- Dang, T.A.; Kessler, T.; Wobst, J.; Wierer, M.; Braenne, I.; Strom, T.M.; Tennstedt, S.; Sanger, H.B.; Meitinger, T.; Erdmann, J.; et al. Identification of a Functional PDE5A Variant at the Chromosome 4q27 Coronary Artery Disease Locus in an Extended Myocardial Infarction Family. Circulation 2021, 144, 662–665. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Yuno, M.; Suzuki, Y.; Sugano, S.; Fujii, H. Identification of physical interactions between genomic regions by enChIP-Seq. Genes Cells 2017, 22, 506–520. [Google Scholar] [CrossRef] [PubMed]
- Hamidian, A.; Vaapil, M.; von Stedingk, K.; Fujita, T.; Persson, C.U.; Eriksson, P.; Veerla, S.; De Preter, K.; Speleman, F.; Fujii, H.; et al. Promoter-associated proteins of EPAS1 identified by enChIP-MS—A putative role of HDX as a negative regulator. Biochem. Bhyophys. Res. Comm. 2018, 499, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Pofi, R.; Fiore, D.; De Gaetano, R.; Panio, G.; Gianfrilli, D.; Pozza, C.; Barbagallo, F.; Xiang, Y.K.; Giannakakis, K.; Morano, S.; et al. Phosphodiesterase-5 inhibition preserves renal hemodynamics and function in mice with diabetic kidney disease by modulating miR-22 and BMP7. Sci. Rep. 2017, 7, 44584. [Google Scholar] [CrossRef] [PubMed]
| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| Pde5a1 | ATGGAACGAGCGGGCCCCAACT | GGTCAACTTCTGCATTGAA |
| Pde5a2 | ATGTTGCCCTTTGGAGACAAAAC | GGTCAACTTCTGCATTGAA |
| Pde5a3 | GTTTTGTTCTACAGAGACATG | GGTCAACTTCTGCATTGAA |
| Pde5a1 (qPCR) | GGACTGGGTGGAAGCGTGGCTG | GCAAGAGCAGGACTCGGTATGGGC |
| Pde5a2 (qPCR) | ATGTTGCCCTTTGGAGACAAAACG | GCAAGAGCAGGACTCGGTATGGGC |
| Gapdh | TCGTCCCGTAGACAAAATGG | TTGAGGTCAATGAAGGGGTC |
| Pde5a1-1 | ggtaccAACCACTCAGCCAGAATGAAGT | aagcttCGTCCCTGCAGTGTCTCAGCG |
| Pde5a1-2 | ggtaccCCACCAAGAGTTGCTTA | aagcttGAGTTGGGGCCCGCTCGTTC |
| Pde5a1-3 | ggtaccCCACCAAGAGTTGCTTA | aagcttGAGTTGGGGCCCGCTCGTTC |
| Pde5a2-1 | agatctAGAAGAGGAGCCACAAAGGACACAG | aagcttCCAAAGGGCAACATAGCAAAG |
| Pde5a2-2 | agatctGGAGCTTTGCGGCGCGCACACAAA | aagcttCCAAAGGGCAACATAGCAAAG |
| MyoD | CCCCGGCGGCAGAATGGCTACG | GGTCTGGGTTCCCTGTTCTGTGT |
| Runx1 | CGGCCATGAAGAACCAGGTA | CAACTTGTGGCGGATTTGT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Arcangelis, V.; De Angelis, L.; Barbagallo, F.; Campolo, F.; de Oliveira do Rego, A.G.; Pellegrini, M.; Naro, F.; Giorgi, M.; Monaco, L. Phosphodiesterase 5a Signalling in Skeletal Muscle Pathophysiology. Int. J. Mol. Sci. 2023, 24, 703. https://doi.org/10.3390/ijms24010703
De Arcangelis V, De Angelis L, Barbagallo F, Campolo F, de Oliveira do Rego AG, Pellegrini M, Naro F, Giorgi M, Monaco L. Phosphodiesterase 5a Signalling in Skeletal Muscle Pathophysiology. International Journal of Molecular Sciences. 2023; 24(1):703. https://doi.org/10.3390/ijms24010703
Chicago/Turabian StyleDe Arcangelis, Valeria, Luciana De Angelis, Federica Barbagallo, Federica Campolo, Ana Gabriela de Oliveira do Rego, Manuela Pellegrini, Fabio Naro, Mauro Giorgi, and Lucia Monaco. 2023. "Phosphodiesterase 5a Signalling in Skeletal Muscle Pathophysiology" International Journal of Molecular Sciences 24, no. 1: 703. https://doi.org/10.3390/ijms24010703
APA StyleDe Arcangelis, V., De Angelis, L., Barbagallo, F., Campolo, F., de Oliveira do Rego, A. G., Pellegrini, M., Naro, F., Giorgi, M., & Monaco, L. (2023). Phosphodiesterase 5a Signalling in Skeletal Muscle Pathophysiology. International Journal of Molecular Sciences, 24(1), 703. https://doi.org/10.3390/ijms24010703








