CXCL4-RNA Complexes Circulate in Systemic Sclerosis and Amplify Inflammatory/Pro-Fibrotic Responses by Myeloid Dendritic Cells
Abstract
1. Introduction
2. Results
2.1. CXCL4 Binds Self-RNA in a Dose-Dependent Manner and Prevents Its Degradation
2.2. CXCL4-RNA Complexes Are Present in SSc Plasma and Correlate with IFN-I
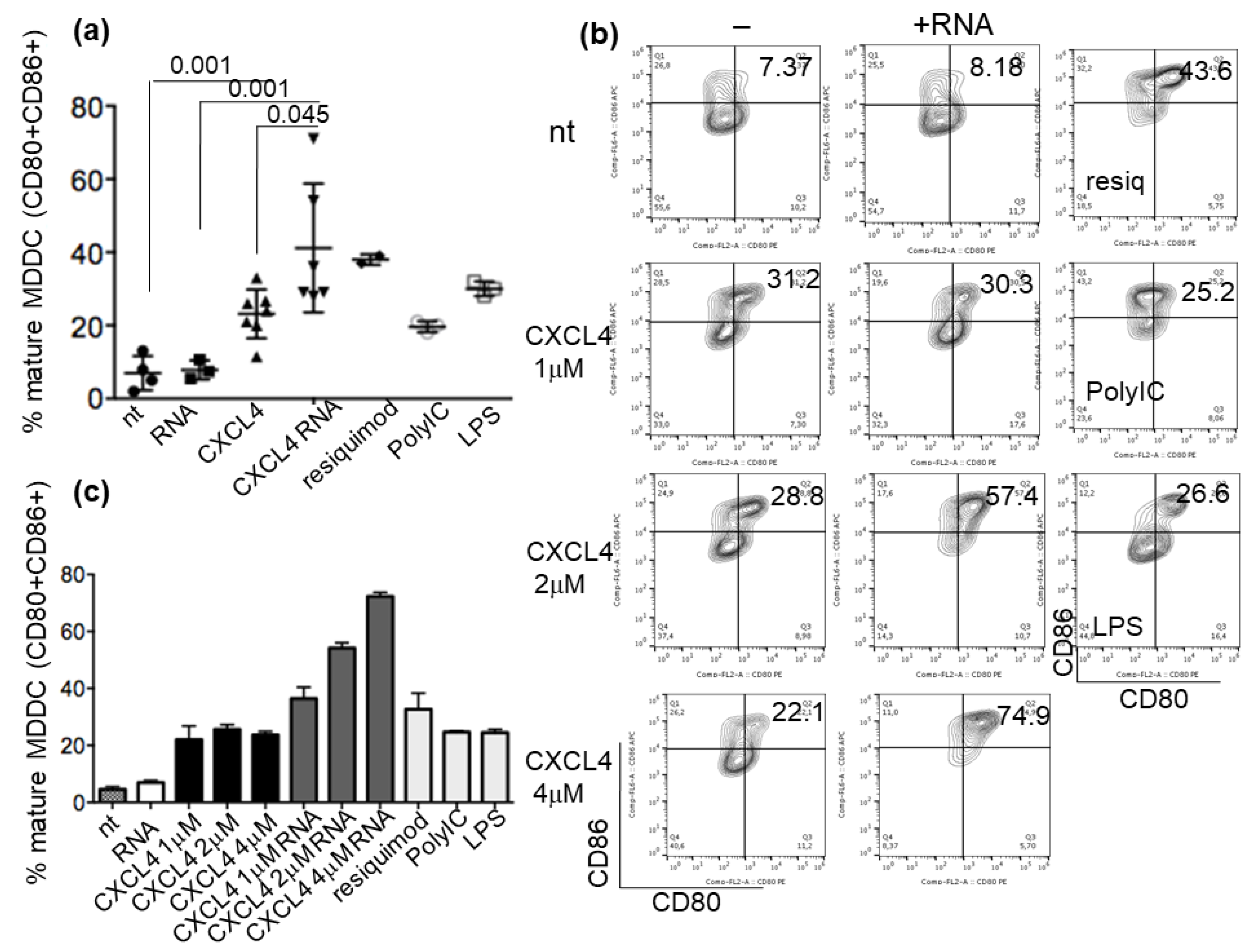
2.3. CXCL4-RNA Complexes Increase the Maturation of IFN-I-Primed Myeloid DCs
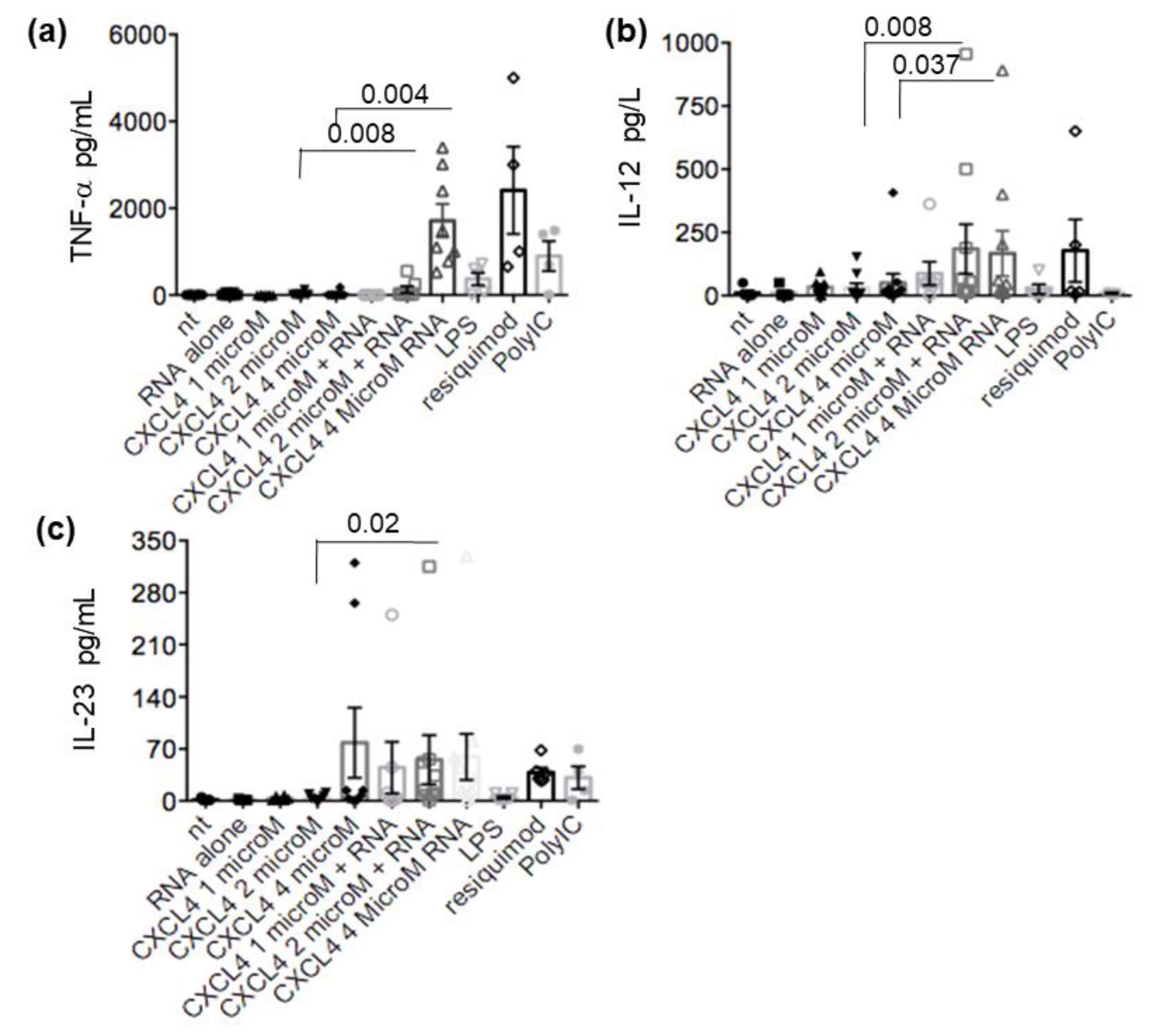
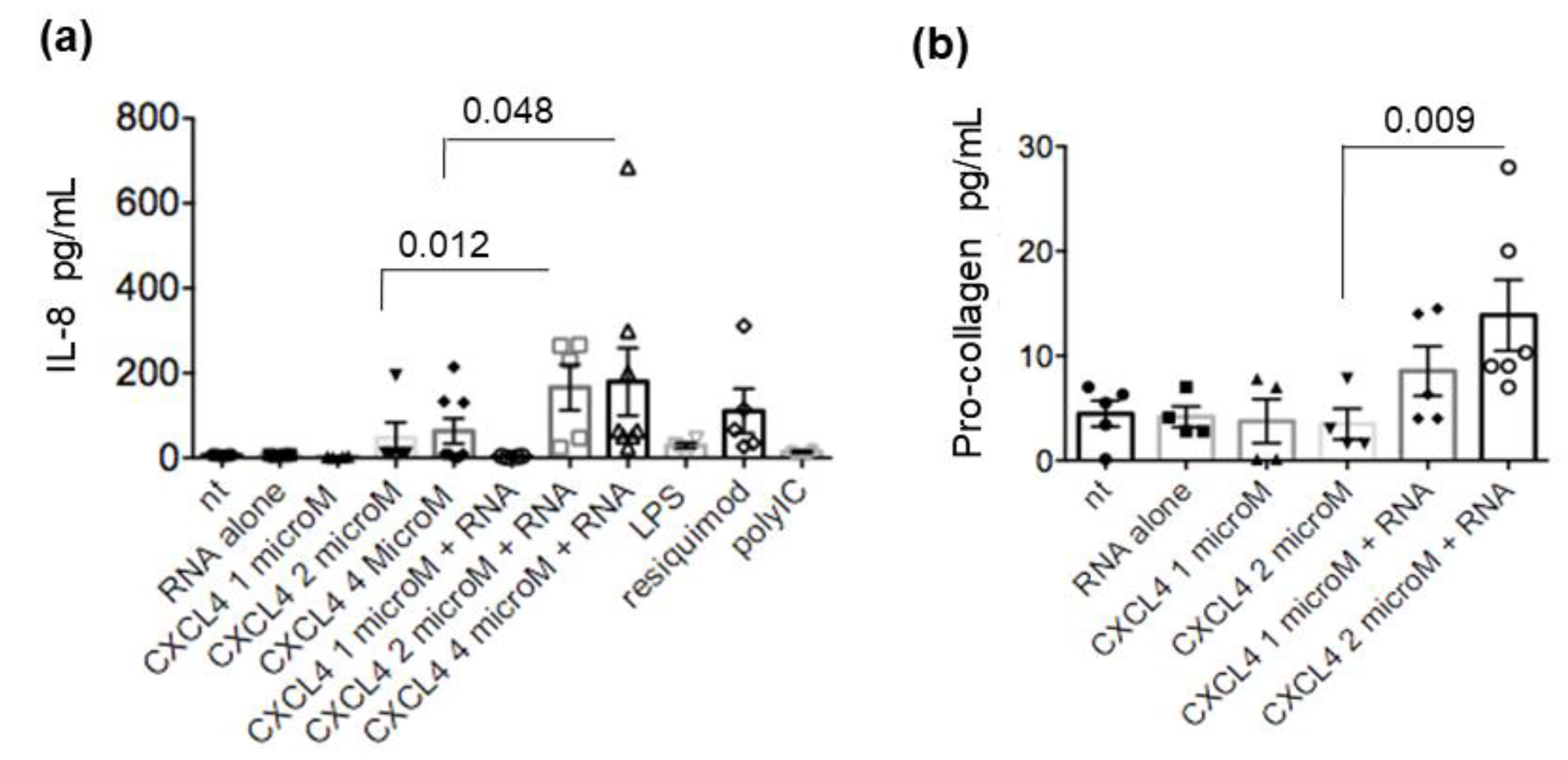
2.4. CXCL4-RNA Complexes Stimulate Pro-Inflammatory and Pro-Fibrotic Pathways
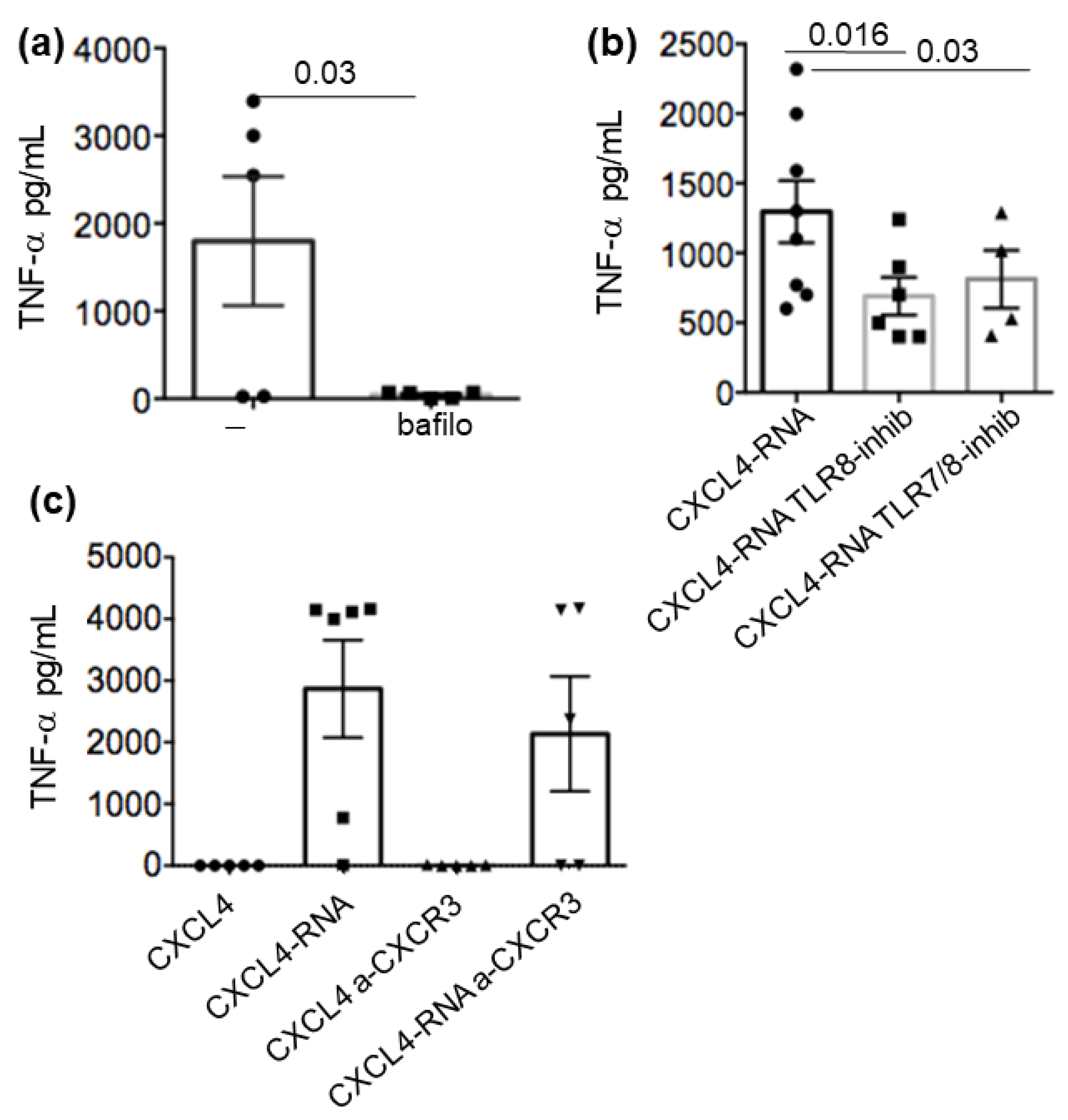
2.5. CXCL4-RNA Complex Stimulation Is RNA-Sensing–TLR-Dependent and Independent of CXCR3
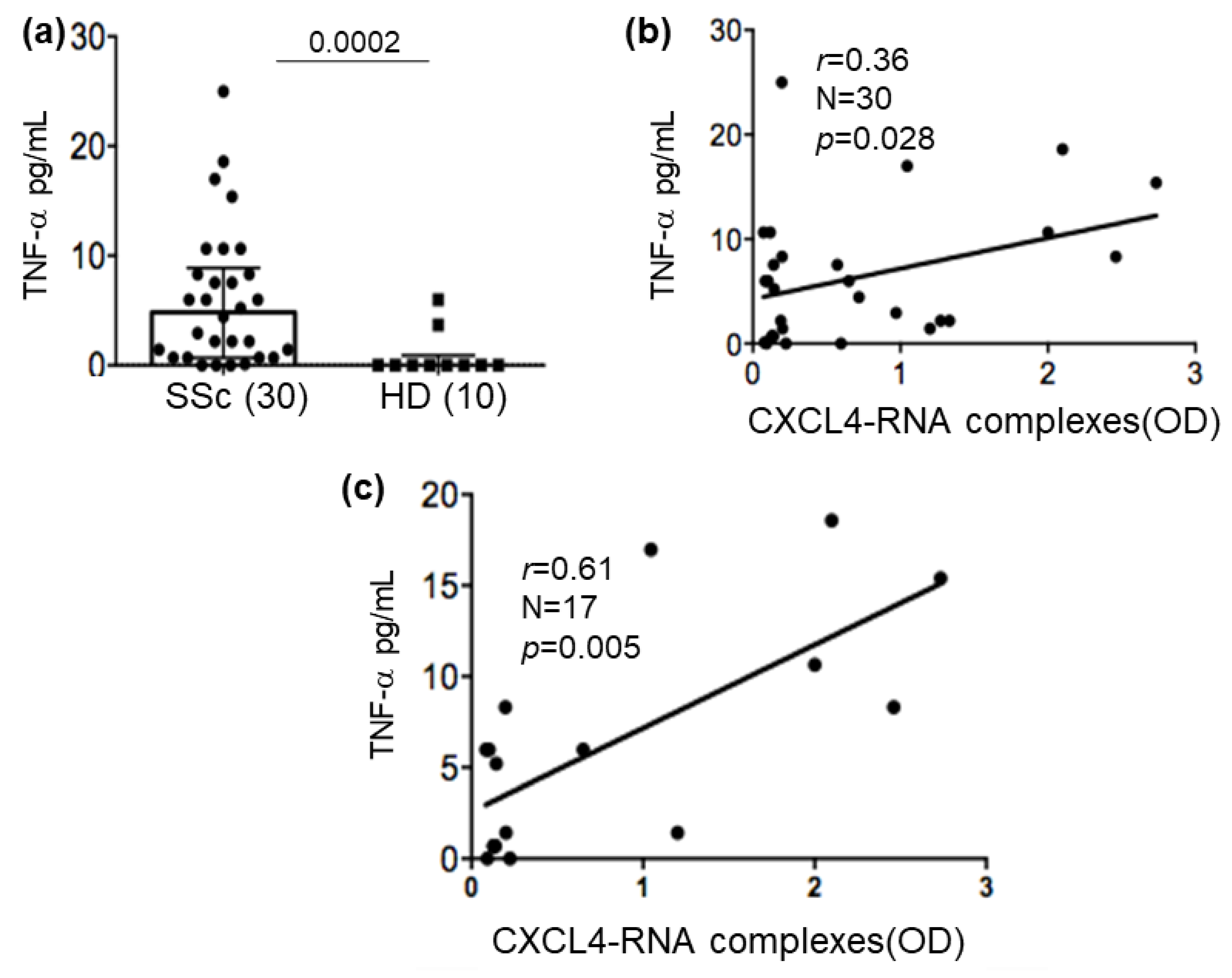
2.6. CXCL4-RNA Complexes Correlate with TNF-α in Plasma
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Reagents and Antibodies
4.3. Production of Human RNA
4.4. Binding of CXCL4 to RNA by Electrophoretic Mobility Shift (EMSA)
4.5. Ribonuclease Protection Assay of RNA Bound to CXCL4
4.6. Isolation/Stimulation of Monocytes and Production of MDDCs
4.7. Immunofluorescence and Flow Cytometry Analysis
4.8. ELISA for Cytokines/Chemokines in Cell Culture and Human Plasma
4.9. In-House ELISA for the Determination of CXCL4-RNA Complexes
4.10. Visualization of Protein-Bound RNA by Western Blot
4.11. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Ho, Y.Y.; Lagares, D.; Tager, A.M.; Kapoor, M. Fibrosis: A lethal component of systemic sclerosis. Nat. Rev. Rheumatol. 2014, 10, 390–402. [Google Scholar] [CrossRef] [PubMed]
- Denton, C.P.; Khanna, D. Systemic sclerosis. Lancet 2017, 390, 1685–1699. [Google Scholar] [CrossRef]
- Kim, D.; Peck, A.; Santer, D.; Patole, P.; Schwartz, S.M.; Molitor, J.A.; Arnett, F.C.; Elkon, K.B. Induction of interferon-α by scleroderma sera containing autoantibodies to topoisomerase I: Association of higher interferon-alpha activity with lung fibrosis. Arthritis Rheumatol. 2008, 58, 2163–2173. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Assassi, S. The role of type 1 interferon in systemic sclerosis. Front. Immunol. 2013, 4, 266. [Google Scholar] [CrossRef]
- Brkic, Z.; van Bon, L.; Cossu, M.; van Helden-Meeuwsen, C.G.; Vonk, M.C.; Knaapen, H.; van den Berg, W.; Dalm, V.A. The interferon type I signature is present in systemic sclerosis before overt fibrosis and might contribute to its pathogenesis through high BAFF gene expression and high collagen synthesis. Ann. Rheum. Dis. 2016, 75, 1567–1573. [Google Scholar] [CrossRef] [PubMed]
- Lande, R.; Lee, E.Y.; Palazzo, R.; Marinari, B.; Pietraforte, I.; Santos, G.S.; Mattenberger, Y.; Spadaro, F.; Stefanantoni, K.; Iannace, N.; et al. CXCL4 assembles DNA into liquid crystalline complexes to amplify TLR9-mediated interferon-α production in systemic sclerosis. Nat. Commun. 2019, 10, 1731. [Google Scholar] [CrossRef] [PubMed]
- Frasca, L.; Lande, R. Toll-like receptors in mediating pathogenesis in systemic sclerosis review. Clin. Exp. Immunol. 2020, 201, 14–24. [Google Scholar] [CrossRef]
- Lande, R.; Mennella, A.; Palazzo, R.; Pietraforte, I.; Stefanantoni, K.; Iannace, N.; Butera, A.; Boirivant, M.; Pica, R.; Conrad, C.; et al. Anti-CXCL4 antibody reactivity is present in Systemic Sclerosis (SSc) and correlates with the SSc Type I Interferon signature. Int. J. Mol. Sci. 2020, 21, 5102. [Google Scholar] [CrossRef]
- Romani, N.; Gruner, S.; Brang, D.; Kampgen, E.; Lenz, A.; Trockenbacher, B.; Konwalinka, G.; Fritsch, P.O.; Steinman, R.M.; Schuler, G. Proliferating dendritic cell progenitors in human blood. J. Exp. Med. 1994, 180, 83–93. [Google Scholar] [CrossRef]
- Sallusto, F.; Lanzavecchia, A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J. Exp. Med. 1994, 179, 1109–1118. [Google Scholar] [CrossRef]
- Randolph, G.J.; Beaulieu, S.; Lebecque, S.; Steinman, R.M.; Muller, W.A. Differentiation of monocytes into dendritic cells in a model of transendothelial trafficking. Science 1998, 282, 480–483. [Google Scholar] [CrossRef] [PubMed]
- Jarrossay, D.; Napolitani, G.; Colonna, M.; Sallusto, F.; Lanzavecchia, A. Specialization and complementarity in microbial molecule recognition by human myeloid and plasmacytoid dendritic cells. Eur. J. Immunol. 2001, 31, 3388–3393. [Google Scholar] [CrossRef] [PubMed]
- Kadowaki, N.; Ho, S.; Antonenko, S.; Malefyt, R.W.; Kastelein, R.A.; Bazan, F.; Liu, Y.J. Subsets of human dendritic cell precursors express different toll-like receptors and respond to different microbial antigens. J. Exp. Med. 2001, 194, 863–869. [Google Scholar] [CrossRef] [PubMed]
- Hornung, V.; Rothenfusser, S.; Britsch, S.; Krug, A.; Jahrsdörfer, B.; Giese, T.; Endres, S.; Hartmann, G. Quantitative expression of toll-like receptor 1–10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J. Immunol. 2002, 168, 4531–4537. [Google Scholar] [CrossRef]
- Severa, M.; Remoli, M.E.; Giacomini, E.; Annibali, V.; Gafa, V.; Lande, R.; Tomai, M.; Salvetti, M.; Coccia, E.M. Sensitization to TLR7 agonist in IFN-β-preactivated dendritic cells. J. Immunol. 2007, 178, 6208–6216. [Google Scholar] [CrossRef]
- Collin, M.; Bigley, V. Human dendritic cell subsets: An update. Immunology 2018, 154, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Silva-Cardoso, S.C.; Affandi, A.J.; Spel, L.; Cossu, M.; van Roon, J.A.G.; Boes, M.; Radstake, T.R.D.J. CXCL4 exposure potentiates TLR-driven polarization of human monocyte-derived dendritic cells and increases stimulation of T-cells. J. Immunol. 2017, 199, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Silva-Cardoso, S.C.; Tao, W.; Angiolilli, C.; Lopes, A.P.; Bekker, C.P.J.; Devaprasad, A.; Giovannone, B.; van Laar, J.; Cossu, M.; Marut, W.; et al. CXCL4 links inflammation and fibrosis by reprogramming monocyte-derived dendritic cells in vitro. Front. Immunol. 2020, 11, 2149. [Google Scholar] [CrossRef]
- Van Bon, L.; Affandi, A.J.; Broen, J.; Christmann, R.B.; Marijnissen, R.J.; Stawski, L.; Farina, G.A.; Stifano, G.; Mathes, A.L.; Cossu, M.; et al. Proteome-wide analysis and CXCL4 as a biomarker in systemic sclerosis. N. Engl. J. Med. 2014, 370, 433–443. [Google Scholar] [CrossRef]
- Banchereau, J.; Steinman, R.M. Dendritic cells and the control of immunity. Nature 1998, 392, 245–252. [Google Scholar] [CrossRef]
- Raja, J.; Denton, C.P. Cytokines in the immunopathology of systemic sclerosis. Semin. Immunopathol. 2015, 37, 543–557. [Google Scholar] [CrossRef] [PubMed]
- Murdaca, G.; Spano, F.; Contatore, M.; Guastalla, A.; Puppo, F. Potential use of TNF-α inhibitors in systemic sclerosis. Immunotherapy 2014, 6, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Mukaida, N. Pathophysiological roles of interleukin-8/CXCL8 in pulmonary diseases. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2003, 284, L566–L577. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Herrera, J.; Gilbertsen, A.; Xia, H.; Smith, K.; Benyumov, A.; Bitterman, P.B.; Henke, C.A. IL-8 mediates idiopathic pulmonary fibrosis mesenchymal progenitor cellfibrogenicity. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2018, 314, L127–L136. [Google Scholar] [CrossRef]
- Becker, M.O.; Radic, M.; Schmidt, K.; Huscher, D.; Riedlinger, A.; Michelfelder, M.; Meisel, C.; Ewert, R.; Burmester, G.R.; Riemekasten, G. Serum cytokines and their predictive value in pulmonary involvement of systemic sclerosis. Sarcoidosis Vasc. Diffuse Lung Dis. 2019, 36, 274–284. [Google Scholar] [CrossRef]
- Naka, T.; Nishimoto, N.; Kishimoto, T. The paradigm of IL-6: From basic science to medicine. Arthritis Res. Ther. 2002, 4 (Suppl. 3), S233–S242. [Google Scholar] [CrossRef]
- Wei, J.; Bhattacharyya, S.; Tourtellotte, W.G.; Varga, J. Fibrosis in systemic sclerosis: Emerging concepts and implications for targeted therapy. Autoimmun. Rev. 2011, 10, 267–275. [Google Scholar] [CrossRef]
- Karsdal, M.A.; Nielsen, S.H.; Leeming, D.J.; Langholm, L.L.; Nielsen, M.J.; Manon-Jensen, T.; Siebuhr, A.; Gudmann, N.S.; Rønnow, S.; Sand, J.M.; et al. The good and the bad collagens of fibrosis—Their role in signaling and organ function. Adv. Drug Deliv. Rev. 2017, 121, 43–56. [Google Scholar] [CrossRef]
- Kania, G.; Rudnik, M.; Distler, O. Involvement of the Myeloid Cell Compartment in Fibrogenesis and Systemic Sclerosis. Nat. Rev. Rheumatol. 2019, 15, 288–302. [Google Scholar] [CrossRef]
- Ahmad-Nejad, P.; Hacker, H.; Rutz, M.; Bauer, S.; Vabulas, R.M.; Wagner, H. Bacterial CpG-DNA and lipopolysaccharides activate Toll-like receptorsat distinct cellular compartments. Eur. J. Immunol. 2002, 32, 1958–1968. [Google Scholar] [CrossRef]
- Tatematstu, M.; Funami, K.; Seya, T.; Matsumoto, M. Extracellular RNA Sensing by Pattern Recognition Receptors. J. Innate Immun. 2018, 10, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Vandercappellen, J.; Van Damme, J.; Struyf, S. The role of the CXC chemokines platelet factor-4 (CXCL4/PF-4) and its variant (CXCL4L1/PF-4var) in inflammation, angiogenesis and cancer. Cytokine Growth Factor Rev. 2011, 22, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Bradley, J.R. TNF-Mediated inflammatory disease. J. Pathol. 2008, 214, 149–160. [Google Scholar] [CrossRef]
- Komura, K.; Fujimoto, M.; Hasegawa, M.; Ogawa, F.; Hara, T.; Muroi, E.; Takehara, K.; Sato, S. Increased serum interleukin 23 in patients with systemic sclerosis. J. Rheumatol. 2008, 35, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Rolla, G.; Fusaro, E.; Nicola, S.; Bucca, C.; Peroni, C.; Parisi, S.; Cassinis, M.C.; Ferraris, A.; Angelino, F.; Heffler, E.; et al. Th-17 cytokines and interstitial lung involvement in systemic sclerosis. J. Breath Res. 2016, 10, 046013. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, R.S.G.; Pereira, M.C.; Dantas, A.T.; Almeida, A.R.; Marques, C.D.L.; Rego, M.J.B.M.; Pitta, I.R.; Duarte, A.L.B.P.; Pitta, M.G.R. IL-17 and related cytokines involved in systemic sclerosis: Perspectives. Autoimmunity 2018, 51, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Ma, T.; Cao, H.; Chen, Y.; Wang, C.; Chen, X.; Xiang, Z.; Han, X. TNF-α-induced NF-κB activation promotes myofibroblast differentiation of LR-MSCs and exacerbates bleomycin-induced pulmonary fibrosis. J. Cell. Physiol. 2018, 233, 2409–2419. [Google Scholar] [CrossRef]
- Bonhomme, O.; André, B.; Gester, F.; de Seny, D.; Moermans, C.; Struman, I.; Louis, R.; Malaise, M.; Guiot, J. Biomarkers in systemic sclerosis-associated interstitial lung disease: Review of the literature. Rheumatology 2019, 58, 1534–1546. [Google Scholar] [CrossRef]
- Xing, X.; Li, A.; Tan, H.; Zhou, Y. IFN-γ+ IL-17+ Th17 cells regulate fibrosis through secreting IL-21 in systemic scleroderma. J. Cell. Mol. Med. 2020, 24, 13600–13608. [Google Scholar] [CrossRef]
- Sato, S.S.; Hanakawa, H.; Hasegawa, M.; Nagaoka, T.; Hamaguchi, Y.; Nishijima, C.; Komatsu, K.; Hirata, A.; Takehara, K. Levels of interleukin 12, a cytokine of type 1 helper T cells, are elevated in sera from patients with systemic sclerosis. J. Rheumatol. 2000, 27, 2838–2842. [Google Scholar]
- López-Isac, E.; Campillo-Davo, D.; Bossini-Castillo, L.; Guerra, S.G.; Assassi, S.; Simeón, C.P.; Carreira, P.; Ortego-Centeno, N.; de la Peña, P.G.; Spanish Scleroderma Group; et al. Influence of TYK2 in systemic sclerosis susceptibility: A new locus in the IL-12 pathway. Ann. Rheum. Dis. 2016, 75, 1521–1526. [Google Scholar] [CrossRef] [PubMed]
- Van den Hoogen, F.; Khanna, D.; Fransen, J.; Johnson, S.R.; Baron, M.; Tyndall, A.; Matucci-Cerinic, M.; Naden, R.P.; Medsger, T.A., Jr.; Carreira, P.A.; et al. 2013 Classification criteria for systemic sclerosis: An American college of rheumatology/European league against rheumatism collaborative initiative. Ann. Rheum. Dis. 2013, 72, 1747–1755. [Google Scholar] [CrossRef] [PubMed]
- Melsens, K.F.; De Keyser, S.; Decuman, Y.; Piette, E.; Vandecasteele, E.; Smith, V. Disease activity indices in systemic sclerosis: A systematic literature review. Clin. Exp. Rheumatol. 2016, 34 (Suppl. 100), 186–192. [Google Scholar] [PubMed]


| Characteristics 1 | SSc (34) | HD (24) |
|---|---|---|
| Sex (M/F) | 1/33 | 13/11 |
| Age | 52 (32–71) | 50 (31–55) |
| Early | 14 | - |
| Long-lasting | 20 | - |
| RNA pol | 6 | - |
| ANA positivity (ATA/ACA) | 29/2 | - |
| PAH | 20/14 | - |
| Lung fibrosis (yes/no) | 20/13 | - |
| Prednisolone use | 59% | - |
| Diff/lim | 33/1 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pietraforte, I.; Butera, A.; Gaddini, L.; Mennella, A.; Palazzo, R.; Campanile, D.; Stefanantoni, K.; Riccieri, V.; Lande, R.; Frasca, L. CXCL4-RNA Complexes Circulate in Systemic Sclerosis and Amplify Inflammatory/Pro-Fibrotic Responses by Myeloid Dendritic Cells. Int. J. Mol. Sci. 2023, 24, 653. https://doi.org/10.3390/ijms24010653
Pietraforte I, Butera A, Gaddini L, Mennella A, Palazzo R, Campanile D, Stefanantoni K, Riccieri V, Lande R, Frasca L. CXCL4-RNA Complexes Circulate in Systemic Sclerosis and Amplify Inflammatory/Pro-Fibrotic Responses by Myeloid Dendritic Cells. International Journal of Molecular Sciences. 2023; 24(1):653. https://doi.org/10.3390/ijms24010653
Chicago/Turabian StylePietraforte, Immacolata, Alessia Butera, Lucia Gaddini, Anna Mennella, Raffaella Palazzo, Doriana Campanile, Katia Stefanantoni, Valeria Riccieri, Roberto Lande, and Loredana Frasca. 2023. "CXCL4-RNA Complexes Circulate in Systemic Sclerosis and Amplify Inflammatory/Pro-Fibrotic Responses by Myeloid Dendritic Cells" International Journal of Molecular Sciences 24, no. 1: 653. https://doi.org/10.3390/ijms24010653
APA StylePietraforte, I., Butera, A., Gaddini, L., Mennella, A., Palazzo, R., Campanile, D., Stefanantoni, K., Riccieri, V., Lande, R., & Frasca, L. (2023). CXCL4-RNA Complexes Circulate in Systemic Sclerosis and Amplify Inflammatory/Pro-Fibrotic Responses by Myeloid Dendritic Cells. International Journal of Molecular Sciences, 24(1), 653. https://doi.org/10.3390/ijms24010653








