Semi-Biosynthetic Production of Surface-Binding Adhesive Antimicrobial Peptides Using Intein-Mediated Protein Ligation
Abstract
:1. Introduction
2. Results and Discussion
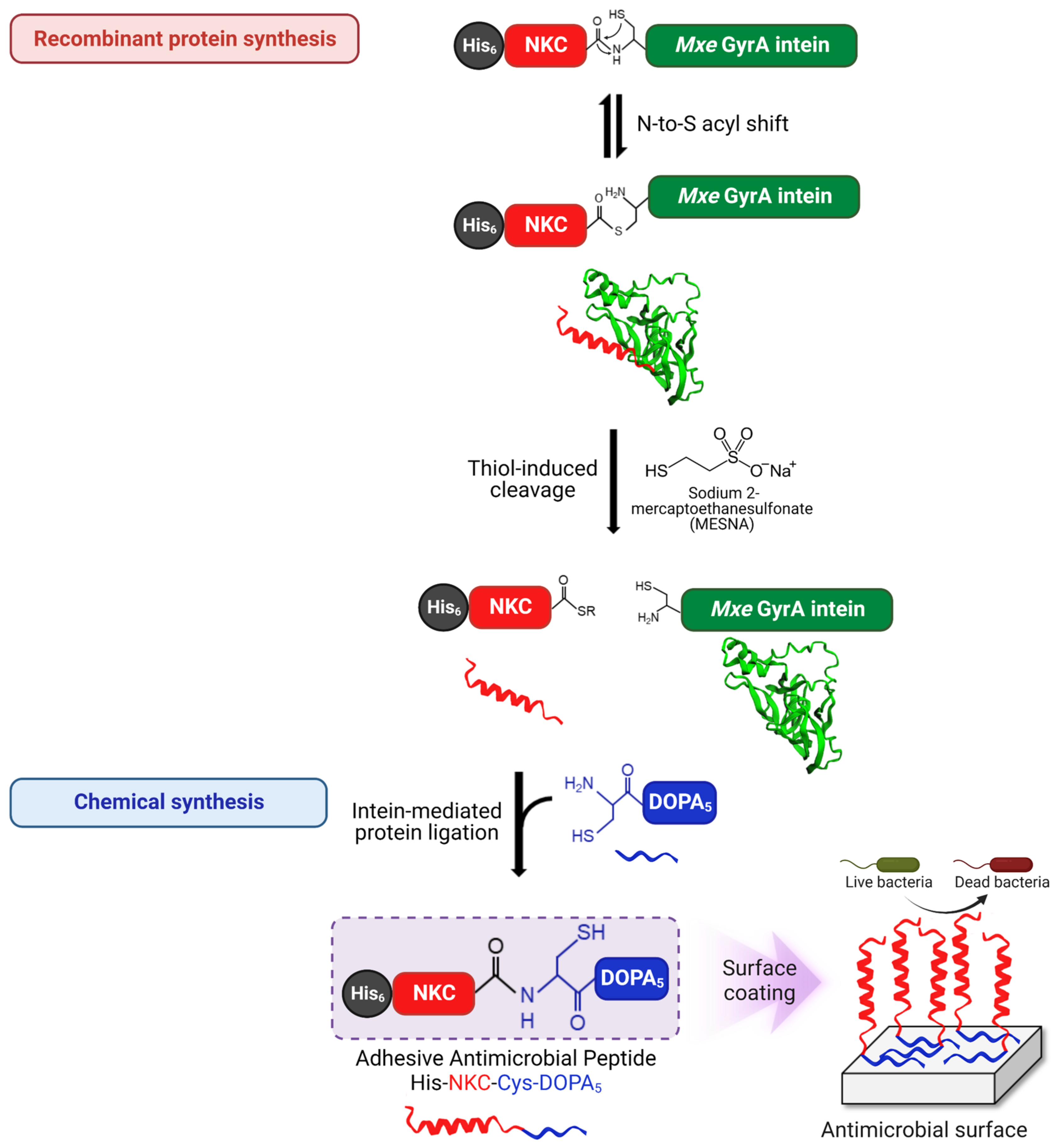
2.1. Semi-Synthesis of Adhesive AMP Containing DOPA
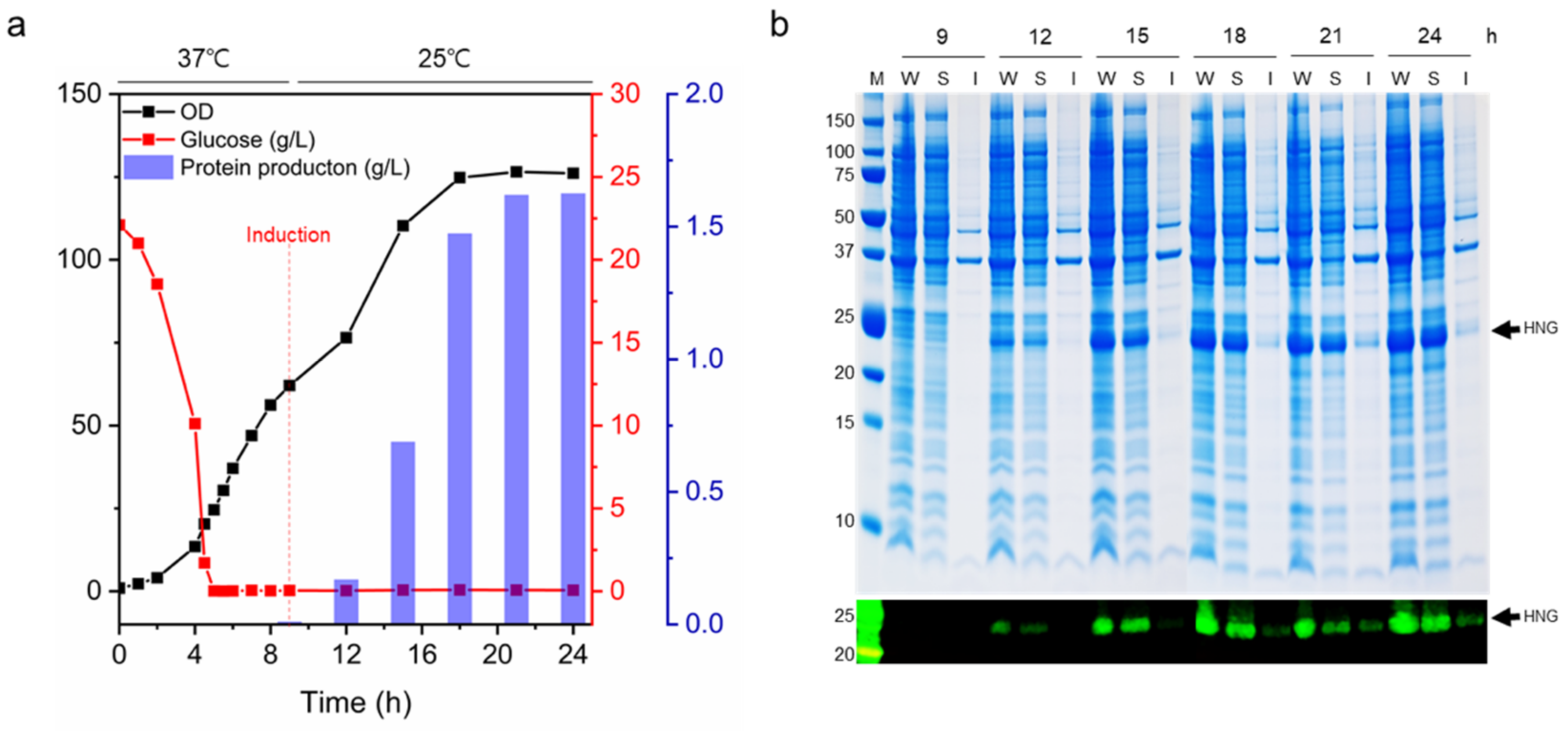
2.2. Expression and Purification of HNG Fusion Proteins
2.3. Fed-Batch Fermentation
2.4. Intein-Mediated Cleavage and Protein Ligation
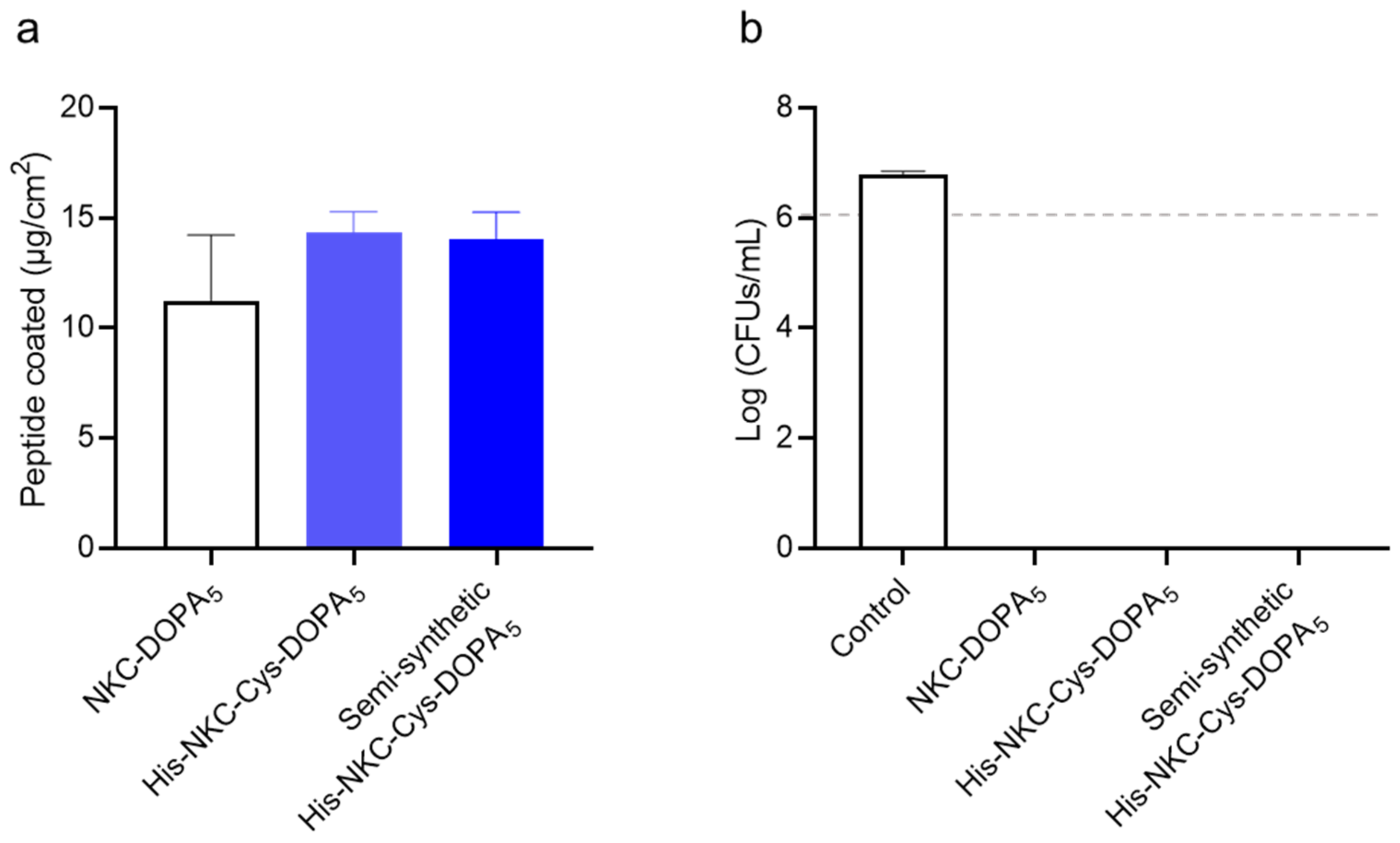
2.5. Purification of the Ligated Adhesive Peptide
2.6. Characterization of His-NKC-Cys-DOPA5
3. Materials and Methods
3.1. Peptide Synthesis
3.2. Bacterial Strains, Vectors, and Reagents
3.3. Construction of Expression Plasmids
3.4. Expression and Purification of HNG Fusion Protein
3.5. Mass Production of HNG Protein Using Fed-Batch Fermentation
3.6. Intein-Mediated Cleavage and Ligation
3.7. SDS-PAGE, Western Blotting, and NBT Staining
3.8. Protein Quantification
3.9. RP-HPLC
3.10. Antibacterial Activity Assay
3.11. Surface-Binding and Antimicrobial Activity Assays
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef]
- Zhang, L.J.; Gallo, R.L. Antimicrobial peptides. Curr. Biol. 2016, 26, R14–R19. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.V.R.; Yedery, R.D.; Aranha, C. Antimicrobial peptides: Premises and promises. Int. J. Antimicrob. Agents 2004, 24, 536–547. [Google Scholar] [CrossRef] [PubMed]
- Kardani, K.; Bolhassani, A. Antimicrobial/anticancer peptides: Bioactive molecules and therapeutic agents. Immunotherapy 2021, 13, 669–684. [Google Scholar] [CrossRef]
- Zhang, R.; Jiang, X.H.; Qiao, J.X.; Wang, Z.; Tong, A.P.; Yang, J.L.; Yang, S.Y.; Yang, L. Antimicrobial peptide DP7 with potential activity against SARS coronavirus infections. Signal Transduct. Target. Ther. 2021, 6, 140. [Google Scholar] [CrossRef]
- Zhao, J.; Zhao, C.; Liang, G.; Zhang, M.; Zheng, J. Engineering antimicrobial peptides with improved antimicrobial and hemolytic activities. J. Chem. Inf. Model. 2013, 53, 3280–3296. [Google Scholar] [CrossRef]
- Chen, X.; Han, J.; Cai, X.; Wang, S. Antimicrobial peptides: Sustainable application informed by evolutionary constraints. Biotechnol. Adv. 2022, 60, 108012. [Google Scholar] [CrossRef]
- Monteiro, C.; Costa, F.; Pirttilä, A.M.; Tejesvi, M.V.; Martins, M.C.L. Prevention of urinary catheter-associated infections by coating antimicrobial peptides from crowberry endophytes. Sci. Rep. 2019, 9, 10753. [Google Scholar] [CrossRef] [Green Version]
- Yasir, M.; Dutta, D.; Hossain, K.R.; Chen, R.; Ho, K.K.K.; Kuppusamy, R.; Clarke, R.J.; Kumar, N.; Willcox, M.D.P. Mechanism of action of surface immobilized antimicrobial peptides against Pseudomonas aeruginosa. Front. Microbiol. 2019, 10, 3053. [Google Scholar] [CrossRef]
- Akhavan, B.; Michl, T.D.; Giles, C.; Ho, K.; Martin, L.; Sharifahmadian, O.; Wise, S.G.; Coad, B.R.; Kumar, N.; Griesser, H.J.; et al. Plasma activated coatings with dual action against fungi and bacteria. Appl. Mater. Today 2018, 12, 72–84. [Google Scholar] [CrossRef]
- Xiao, M.; Jasensky, J.; Gerszberg, J.; Chen, J.; Tian, J.; Lin, T.; Lu, T.; Lahann, J.; Chen, Z. Chemically immobilized antimicrobial peptide on polymer and self-assembled monolayer substrates. Langmuir 2018, 34, 12889–12896. [Google Scholar] [CrossRef]
- Yazici, H.; Habib, G.; Boone, K.; Urgen, M.; Utku, F.S.; Tamerler, C. Self-assembling antimicrobial peptides on nanotubular titanium surfaces coated with calcium phosphate for local therapy. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 94, 333–343. [Google Scholar] [CrossRef]
- Yucesoy, D.T.; Hnilova, M.; Boone, K.; Arnold, P.M.; Snead, M.L.; Tamerler, C. Chimeric peptides as implant functionalization agents for titanium alloy implants with antimicrobial properties. JOM 2015, 67, 754–766. [Google Scholar] [CrossRef] [Green Version]
- Yazici, H.; O’Neill, M.B.; Kacar, T.; Wilson, B.R.; Oren, E.E.; Sarikaya, M.; Tamerler, C. Engineered chimeric peptides as antimicrobial surface coating agents toward infection-free implants. ACS Appl. Mater. Interfaces 2016, 8, 5070–5081. [Google Scholar] [CrossRef] [Green Version]
- Geng, H.; Yuan, Y.; Adayi, A.; Zhang, X.; Song, X.; Gong, L.; Zhang, X.; Gao, P. Engineered chimeric peptides with antimicrobial and titanium-binding functions to inhibit biofilm formation on Ti implants. Mater. Sci. Eng. C Mater. Biol. Appl. 2018, 82, 141–154. [Google Scholar] [CrossRef]
- Meyers, S.R.; Khoo, X.; Huang, X.; Walsh, E.B.; Grinstaff, M.W.; Kenan, D.J. The development of peptide-based interfacial biomaterials for generating biological functionality on the surface of bioinert materials. Biomaterials 2009, 30, 277–286. [Google Scholar] [CrossRef] [Green Version]
- Lim, K.; Chua, R.R.; Bow, H.; Tambyah, P.A.; Hadinoto, K.; Leong, S.S. Development of a catheter functionalized by a polydopamine peptide coating with antimicrobial and antibiofilm properties. Acta Biomater. 2015, 15, 127–138. [Google Scholar] [CrossRef]
- Cheng, H.; Yue, K.; Kazemzadeh-Narbat, M.; Liu, Y.; Khalilpour, A.; Li, B.; Zhang, Y.S.; Annabi, N.; Khademhosseini, A. Mussel-inspired multifunctional hydrogel coating for prevention of infections and enhanced osteogenesis. ACS Appl. Mater. Interfaces 2017, 9, 11428–11439. [Google Scholar] [CrossRef] [Green Version]
- Alves, D.; Vaz, A.T.; Grainha, T.; Rodrigues, C.F.; Pereira, M.O. Design of an antifungal surface embedding liposomal amphotericin B through a mussel adhesive-inspired coating strategy. Front. Chem. 2019, 7, 431. [Google Scholar] [CrossRef]
- Dhand, C.; Ong, C.Y.; Dwivedi, N.; Varadarajan, J.; Halleluyah Periayah, M.; Jianyang Lim, E.; Mayandi, V.; Goh, E.T.L.; Najjar, R.P.; Chan, L.W.; et al. Mussel-inspired durable antimicrobial contact lenses: The role of covalent and noncovalent attachment of antimicrobials. ACS Biomater. Sci. Eng. 2020, 6, 3162–3173. [Google Scholar] [CrossRef]
- Singha, P.; Locklin, J.; Handa, H. A review of the recent advances in antimicrobial coatings for urinary catheters. Acta Biomater. 2017, 50, 20–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakala, G.P.; Reches, M. Peptide-based approaches to fight biofouling. Adv. Mater. Interfaces 2018, 5, 180073. [Google Scholar] [CrossRef]
- Stillger, L.; Müller, D. Peptide-coating combating antimicrobial contaminations: A review of covalent immobilization strategies for industrial applications. J. Mater. Sci. 2022, 57, 10863–10885. [Google Scholar] [CrossRef]
- Vargas-Rodriguez, O.; Sevostyanova, A.; Söll, D.; Crnković, A. Upgrading aminoacyl-tRNA synthetases for genetic code expansion. Curr. Opin. Chem. Biol. 2018, 46, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Sung, B.H.; Kim, S.C.; Lee, H.S. Genetic incorporation of l-dihydroxyphenylalanine (DOPA) biosynthesized by a tyrosine phenol-lyase. Chem. Commun. 2018, 54, 3002–3005. [Google Scholar] [CrossRef] [PubMed]
- Alfonta, L.; Zhang, Z.W.; Uryu, S.; Loo, J.A.; Schultz, P.G. Site-specific incorporation of a redox-active amino acid into proteins. J. Am. Chem. Soc. 2003, 125, 14662–14663. [Google Scholar] [CrossRef]
- Kim, S.; Bae, G.; Shin, M.; Kang, E.; Park, T.Y.; Choi, Y.S.; Cha, H.J. Oriented in situ immobilization of a functional tyrosinase on microcrystalline cellulose effectively incorporates DOPA residues in bioengineered mussel adhesive protein. Biotechnol. J. 2021, 16, e2100216. [Google Scholar] [CrossRef]
- Tay, D.K.; Rajagopalan, G.; Li, X.; Chen, Y.; Lua, L.H.; Leong, S.S. A new bioproduction route for a novel antimicrobial peptide. Biotechnol. Bioeng. 2011, 108, 572–581. [Google Scholar] [CrossRef]
- Kim, D.S.; Kim, S.W.; Song, J.M.; Kim, S.Y.; Kwon, K.C. A new prokaryotic expression vector for the expression of antimicrobial peptide abaecin using SUMO fusion tag. BMC Biotechnol. 2019, 19, 13. [Google Scholar] [CrossRef]
- Tanhaiean, A.; Azghandi, M.; Razmyar, J.; Mohammadi, E.; Sekhavati, M.H. Recombinant production of a chimeric antimicrobial peptide in E. coli and assessment of its activity against some avian clinically isolated pathogens. Microb. Pathog. 2018, 122, 73–78. [Google Scholar] [CrossRef]
- Zhou, L.X.; Liu, Z.Y.; Xu, G.Y.; Li, L.H.; Xuan, K.; Xu, Y.; Zhang, R.Z. Expression of melittin in fusion with GST in Escherichia coli and its purification as a pure peptide with good bacteriostatic efficacy. ACS Omega 2020, 5, 9251–9258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dawson, P.E.; Muir, T.W.; Clark-Lewis, I.; Kent, S.B. Synthesis of proteins by native chemical ligation. Science 1994, 266, 776–779. [Google Scholar] [CrossRef] [PubMed]
- Conibear, A.C. Deciphering protein post-translational modifications using chemical biology tools. Nat. Rev. Chem. 2020, 4, 674–695. [Google Scholar] [CrossRef]
- Thompson, R.E.; Stevens, A.J.; Muir, T.W. Protein engineering through tandem transamidation. Nat. Chem. 2019, 11, 737–743. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.E.; Im, S.; Kim, H.; Sohn, J.H.; Cho, B.K.; Cho, J.H.; Sung, B.H.; Kim, S.C. Adhesive antimicrobial peptides containing 3,4-dihydroxy-L-phenylalanine residues for direct one-step surface coating. Int. J. Mol. Sci. 2021, 22, 11915. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.S.; Sung, B.H.; Park, M.K.; Lee, J.H.; Lim, K.J.; Park, S.C.; Kim, S.J.; Kim, H.K.; Sohn, J.H.; Kim, H.M.; et al. Recombinant lipase engineered with amphipathic and coiled-coil peptides. ACS Catal. 2015, 5, 5016–5025. [Google Scholar] [CrossRef]
- Kim, D.J.; Lee, Y.W.; Park, M.K.; Shin, J.R.; Lim, K.J.; Cho, J.H.; Kim, S.C. Efficacy of the designer antimicrobial peptide SHAP1 in wound healing and wound infection. Amino Acids 2014, 46, 2333–2343. [Google Scholar] [CrossRef] [Green Version]
- Freitas, E.D.; Bataglioli, R.A.; Oshodi, J.; Beppu, M.M. Antimicrobial peptides and their potential application in antiviral coating agents. Colloids Surf. B Biointerfaces 2022, 217, 112693. [Google Scholar] [CrossRef]
- Raducanu, V.S.; Raducanu, D.V.; Ouyang, Y.; Tehseen, M.; Takahashi, M.; Hamdan, S.M. TSGIT: An N- and C-terminal tandem tag system for purification of native and intein-mediated ligation-ready proteins. Protein Sci. 2021, 30, 497–512. [Google Scholar] [CrossRef]
- Harvey, J.A.; Itzhaki, L.S.; Main, E.R.G. Programmed protein self-assembly driven by genetically encoded intein-mediated native chemical ligation. ACS Synth. Biol. 2018, 7, 1067–1074. [Google Scholar] [CrossRef]
- Handley, T.N.G.; Kleffmann, T.; Butler, M.I. Development of ULYSSIS, a tool for the biosynthesis of cyclotides and cyclic knottins. Int. J. Pept. Res. Ther. 2022, 28, 21. [Google Scholar] [CrossRef]
- Priemel, T.; Palia, R.; Babych, M.; Thibodeaux, C.J.; Bourgault, S.; Harrington, M.J. Compartmentalized processing of catechols during mussel byssus fabrication determines the destiny of DOPA. Proc. Natl. Acad. Sci. USA 2020, 117, 7613–7621. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Mishra, B.; Dey, S.; Leong, S.S.J. Intein based bioprocess for production of a synthetic antimicrobial peptide: An alternative route to solid phase peptide synthesis. RSC Adv. 2014, 4, 31564–31572. [Google Scholar] [CrossRef]
- Lee, Y.J.; Jeong, K.J. Enhanced production of antibody fragment via SRP pathway engineering in Escherichia coli. Biotechnol. Bioprocess Eng. 2013, 18, 751–758. [Google Scholar] [CrossRef]
- Paz, M.A.; Flückiger, R.; Boak, A.; Kagan, H.M.; Gallop, P.M. Specific detection of quinoproteins by redox-cycling staining. J. Biol. Chem. 1991, 266, 689–692. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, C.; Zieger, M.; Rode, C.; Schroter, K.; Krahmer, A.; Wyrwa, R.; Raithel, H.; Hipler, U.C. JIS L 1902 and ISO 22196 for determination of antifungal properties of textiles and ceramic surfaces. Mycoses 2011, 54, 416–417. [Google Scholar]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]


| Name | Amino Acid Sequence * | MW ** (kDa) | MIC *** (µM) | Reference |
|---|---|---|---|---|
| His-NKC-GyrA (HNG) | HHHHHHAPKAMKLLKKLLKLQKKGIGGGGSGCITGDALVALPEGESVRIADIVPGARPNSDNAIDLKVLDRHGNPVLADRLFHSGEHPVYTVRTVEGLRVTGTANHPLLCLVDVAGVPTLLWKLIDEIKPGDYAVIQRSAFSVDCAGFARGKPEFAPTTYTVGVPGLVRFLEAHHRDPDAQAIADELTDGRFYYAKVASVTDAGVQPVYSLRVDTADHAFITNGFVSHA | 24.597 | - | This study |
| His-NKC-L | HHHHHHAPKAMKLLKKLLKLQKKGIGGGGSG | 3.345 | 16 | This study |
| Cys-DOPA5 | CYYYYY | 1.017 | - | This study |
| His-NKC-Cys-DOPA5 | HHHHHHAPKAMKLLKKLLKLQKKGIGGGGSGCYYYYY | 4.344 | 16 | This study |
| NKC-DOPA5 | APKAMKLLKKLLKLQKKGIGGGGSGGGGSYYYYY | 3676.3 | 16 | [35] |
| Procedure | Total Protein | HNG Protein | His-NKC-Cys-DOPA5 Peptide |
|---|---|---|---|
| High-pressure homogenization | 15.6 g/L | 985.4 mg/L (40.1 µM) | |
| His-tag purification and ultrafiltration | 0.77 g/L | 483.6 mg/L (19.7 µM) | |
| Ligation | 66.4 mg/L (15.4 µM) | ||
| Preparative purification | 45.2 mg/L (10.5 µM) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hwang, Y.E.; Im, S.; Cho, J.H.; Lee, W.; Cho, B.-K.; Sung, B.H.; Kim, S.C. Semi-Biosynthetic Production of Surface-Binding Adhesive Antimicrobial Peptides Using Intein-Mediated Protein Ligation. Int. J. Mol. Sci. 2022, 23, 15202. https://doi.org/10.3390/ijms232315202
Hwang YE, Im S, Cho JH, Lee W, Cho B-K, Sung BH, Kim SC. Semi-Biosynthetic Production of Surface-Binding Adhesive Antimicrobial Peptides Using Intein-Mediated Protein Ligation. International Journal of Molecular Sciences. 2022; 23(23):15202. https://doi.org/10.3390/ijms232315202
Chicago/Turabian StyleHwang, Young Eun, Seonghun Im, Ju Hyun Cho, Wonsik Lee, Byung-Kwan Cho, Bong Hyun Sung, and Sun Chang Kim. 2022. "Semi-Biosynthetic Production of Surface-Binding Adhesive Antimicrobial Peptides Using Intein-Mediated Protein Ligation" International Journal of Molecular Sciences 23, no. 23: 15202. https://doi.org/10.3390/ijms232315202
APA StyleHwang, Y. E., Im, S., Cho, J. H., Lee, W., Cho, B.-K., Sung, B. H., & Kim, S. C. (2022). Semi-Biosynthetic Production of Surface-Binding Adhesive Antimicrobial Peptides Using Intein-Mediated Protein Ligation. International Journal of Molecular Sciences, 23(23), 15202. https://doi.org/10.3390/ijms232315202






