Proteomic Approaches to Uncover Salt Stress Response Mechanisms in Crops
Abstract
1. Introduction
2. Morphological and Physiological Effects of Salt Stress on Crops
2.1. Rice
2.2. Wheat
2.3. Soybean
2.4. Other Crops
3. Proteomic Techniques to Analyze the Effect of Salt Stress on Crops
3.1. Rice
3.2. Wheat
3.3. Soybean
3.4. Other Crops
4. Roles of Proteomic Techniques for Improvement of Salt Tolerance of Crops
4.1. Chemical Application
4.2. Biotechnological Tool Genome Editing
4.3. Other Techniques
5. Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic strategies for improving crop yields. Nature 2019, 575, 109–118. [Google Scholar] [CrossRef]
- Li, X.; Wang, A.; Wan, W.; Luo, X.; Zheng, L.; He, G.; Huang, D.; Chen, W.; Huang, Q. High salinity inhibits soil bacterial community mediating nitrogen cycling. Appl. Environ. Microbiol. 2021, 87, e0136621. [Google Scholar] [CrossRef]
- Sarri, E.; Termentzi, A.; Abraham, E.M.; Papadopoulos, G.K.; Baira, K.; Machera, K.; Loukas, V.; Komaitis, F.; Tani, E. Salinity stress alters the secondary metabolic profile of M. sativa, M. arborea and their hybrid (Alborea). Int. J. Mol. Sci. 2021, 22, 4882. [Google Scholar] [CrossRef]
- Singh, A. Soil salinization management for sustainable development: A review. J. Environ. Manag. 2021, 277, 111383. [Google Scholar] [CrossRef]
- Yuvaraj, M.; Bose, K.S.C.; Elavarasi, P.; Tawfik, E. Soil salinity and its management. In Soil Moisture Importance; Meena, R.S., Datta, R., Eds.; IntechOpen: London, UK, 2020. [Google Scholar] [CrossRef]
- McDonald, G.K.; Tavakkoli, E.; Rengasamy, P. Commentary: Bread wheat with high salinity and sodicity tolerance. Front. Plant Sci. 2020, 11, 1194. [Google Scholar] [CrossRef]
- Munns, R.; Day, D.A.; Fricke, W.; Watt, M.; Arsova, B.; Barkla, B.J.; Bose, J.; Byrt, C.S.; Chen, Z.-H.; Foster, K.J. Energy costs of salt tolerance in crop plants. New Phytol. 2020, 225, 1072–1090. [Google Scholar] [CrossRef]
- Tarchoun, N.; Saadaoui, W.; Mezghani, N.; Pavli, O.I.; Falleh, H.; Petropoulos, S.A. The effects of salt stress on germination, seedling growth and biochemical responses of tunisian squash (Cucurbita maxima Duchesne) germplasm. Plants 2022, 11, 800. [Google Scholar] [CrossRef]
- Zulfiqar, F.; Ashraf, M. Nanoparticles potentially mediate salt stress tolerance in plants. Plant Physiol. Biochem. 2021, 160, 257–268. [Google Scholar] [CrossRef]
- Yung, W.S.; Li, M.W.; Sze, C.C.; Wang, Q.; Lam, H.M. Histone modifications and chromatin remodelling in plants in response to salt stress. Physiol. Plant 2021, 173, 1495–1513. [Google Scholar] [CrossRef]
- Seif El-Yazal, S.A.; Seif El-Yazal, M.A.; Dwidar, E.F.; Rady, M.M. Phytohormone crosstalk research: Cytokinin and its crosstalk with other phytohormones. Curr. Protein Pept. Sci. 2015, 16, 395–405. [Google Scholar] [CrossRef]
- van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How plant hormones mediate salt stress responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef]
- Hessini, K.; Issaoui, K.; Ferchichi, S.; Saif, T.; Abdelly, C.; Siddique, K.H.M.; Cruz, C. Interactive effects of salinity and nitrogen forms on plant growth, photosynthesis and osmotic adjustment in maize. Plant Physiol. Biochem. 2019, 139, 171–178. [Google Scholar] [CrossRef]
- Azzam, C.R.; Zaki, S.-N.S.; Bamagoos, A.A.; Rady, M.M.; Alharby, H.F. Soaking maize seeds in zeatin-type cytokinin biostimulators improves salt tolerance by enhancing the antioxidant system and photosynthetic efficiency. Plants 2022, 11, 1004. [Google Scholar] [CrossRef]
- Khalifa, S.A.M.; Elshafiey, E.H.; Shetaia, A.A.; El-Wahed, A.A.; Algethami, A.F.; AlAjmi, M.F.; Musharaf, S.G.; Zhao, C.; Masry, S.H.; El-Seedi, H.R. Overview of bee pollination and its economic value for crop production. Insects 2021, 12, 688. [Google Scholar] [CrossRef]
- Arif, Y.; Singh, P.; Siddiqui, H.; Bajguz, A.; Hayat, S. Salinity induced physiological and biochemical changes in plants: An omic approach towards salt stress tolerance. Plant Physiol. Biochem. 2020, 156, 64–77. [Google Scholar] [CrossRef]
- Mansour, M.M.F.; Hassan, F.A.S. How salt stress-responsive proteins regulate plant adaptation to saline conditions. Plant Mol. Biol. 2022, 108, 175–224. [Google Scholar] [CrossRef]
- Wang, K.; Li, F.; Gao, M.; Huang, Y.; Song, Z. Mechanisms of trehalose-mediated mitigation of Cd toxicity in rice seedlings. J. Clean. Prod. 2020, 267, 121982. [Google Scholar] [CrossRef]
- Chen, F.; Fang, P.; Peng, Y.; Zeng, W.; Zhao, X.; Ding, Y.; Zhuang, Z.; Gao, Q.; Ren, B. Comparative proteomics of salt-tolerant and salt-sensitive maize inbred lines to reveal the molecular mechanism of salt tolerance. Int. J. Mol. Sci. 2019, 20, 4725. [Google Scholar] [CrossRef]
- Szypulska, E.; Jankowski, K.; Weidner, S. ABA pretreatment can limit salinity-induced proteome changes in growing barley sprouts. Acta Physiol. Plant 2017, 39, 190. [Google Scholar] [CrossRef]
- Soltabayeva, A.; Ongaltay, A.; Omondi, J.O.; Srivastava, S. Morphological, physiological and molecular markers for salt-stressed plants. Plants 2021, 10, 243. [Google Scholar] [CrossRef]
- Sangwongchai, W.; Krusong, K.; Thitisaksakul, M. Salt tolerance at vegetative stage is partially associated with changes in grain quality and starch physicochemical properties of rice exposed to salinity stress at reproductive stage. J. Sci. Food Agric. 2022, 102, 370–382. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, B.; Li, J.; Song, Z.; Lu, B.; Chi, M.; Yang, B.; Qin, D.; Lam, Y.-W.; Li, J.; et al. Salt response analysis in two rice cultivars at seedling stage. Acta Physiol. Plant. 2017, 39, 215. [Google Scholar] [CrossRef]
- Sheteiwy, M.S.; Shao, H.; Qi, W.; Hamoud, Y.; Shaghaleh, H.; Khan, N.; Yang, R.; Tang, B. GABA-alleviated oxidative injury induced by salinity, osmotic stress and their combination by regulating cellular and molecular signals in rice. Int. J. Mol. Sci. 2019, 20, 5709. [Google Scholar] [CrossRef]
- Chang, J.; Cheong, B.E.; Natera, S.; Roessner, U. Morphological and metabolic responses to salt stress of rice (Oryza sativa L.) cultivars which differ in salinity tolerance. Plant Physiol. Biochem. 2019, 144, 427–435. [Google Scholar] [CrossRef]
- Ninmanont, P.; Wongchai, C.; Pfeiffer, W.; Chaidee, A. Salt stress of two rice varieties: Root border cell response and multi-logistic quantification. Protoplasma 2021, 258, 1119–1131. [Google Scholar] [CrossRef]
- Huang, Y.; Zhou, J.; Li, Y.; Quan, R.; Wang, J.; Huang, R.; Qin, H. Salt stress promotes abscisic acid accumulation to affect cell proliferation and expansion of primary roots in rice. Int. J. Mol. Sci. 2021, 22, 10892. [Google Scholar] [CrossRef]
- Bhanbhro, N.; Xiao, B.; Han, L.; Lu, H.; Wang, H.; Yang, C. Adaptive strategy of allohexaploid wheat to long-term salinity stress. BMC Plant Biol. 2020, 20, 210. [Google Scholar] [CrossRef]
- Wang, J.; Lv, P.; Yan, D.; Tian, X.; Wang, J.; Palta, J.A.; Xu, S.; Fang, Y.; Wang, Z. Exogenous melatonin improves seed germination of wheat (Triticum aestivum L.) under salt stress. Int. J. Mol. Sci. 2022, 23, 8436. [Google Scholar] [CrossRef]
- Chaurasia, S.; Singh, A.K.; Songachan, L.S.; Sharma, A.D.; Bhardwaj, R.; Singh, K. Multi-locus genome-wide association studies reveal novel genomic regions associated with vegetative stage salt tolerance in bread wheat (Triticum aestivum L.). Genomics 2020, 112, 4608–4621. [Google Scholar] [CrossRef]
- Liu, L.; Huang, L.; Lin, X.; Sun, C. Hydrogen peroxide alleviates salinity-induced damage through enhancing proline accumulation in wheat seedlings. Plant Cell Rep. 2020, 39, 567–575. [Google Scholar] [CrossRef] [PubMed]
- Mbarki, S.; Sytar, O.; Zivcak, M.; Abdelly, C.; Cerda, A.; Brestic, M. Anthocyanins of coloured wheat genotypes in species response to salt stress. Molecules 2018, 23, 1518. [Google Scholar] [CrossRef]
- Mehmood, S.; Ahmed, W.; Ikram, M.; Imtiaz, M.; Mahmood, S.; Tu, S.; Chen, D. Chitosan modified biochar increases soybean (Glycine max L.) resistance to salt stress by augmenting root morphology, antioxidant defense mechanisms and the expression of stress-responsive genes. Plants 2020, 9, 1173. [Google Scholar] [CrossRef] [PubMed]
- Silva, B.R.S.; Batista, B.L.; Lobato, A.K.S. Anatomical changes in stem and root of soybean plants submitted to salt stress. Plant Biol. 2020, 23, 57–65. [Google Scholar] [CrossRef] [PubMed]
- El-Esawi, M.A.; Alaraidh, I.A.; Alsahli, A.A.; Alamri, S.A.; Ali, H.M.; Alayafi, A.A. Bacillus firmus (SW5) augments salt tolerance in soybean (Glycine max L.) by modulating root system architecture, antioxidant defense systems and stress-responsive genes expression. Plant Physiol. Biochem. 2018, 132, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Wang, J.; Li, K. Integrated physiological, transcriptomic, and metabolomic analyses revealed molecular mechanism for salt resistance in soybean roots. Int. J. Mol. Sci. 2021, 22, 12848. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.M.; Mota, T.R.; Salatta, F.V.; Sinzker, R.C.; Gomez, L.D.; dos Santos, W.D.; Ferrarese-Filho, O. Cell wall remodeling under salt stress: Insights into changes in polysaccharides, feruloylation, lignification, and phenolic metabolism in maize. Plant Cell Environ. 2020, 43, 2172–2191. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, A.G.; de Oliveira, P.-M.S.; de Paiva, P.-S.K.; de Castro, M.E.; de Sousa, L.L.; Camelo, M.E.; de Carvalho, H.H.; Gomes-Filho, E. H2O2 priming promotes salt tolerance in maize by protecting chloroplasts ultrastructure and primary metabolites modulation. Plant Sci. 2021, 303, 110774. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Cao, B.; Chen, Z.; Xu, K. Root morphology ion absorption and antioxidative defense system of two Chinese cabbage cultivars (Brassica rapa L.) reveal the different adaptation mechanisms to salt and alkali stress. Protoplasma 2022, 259, 385–398. [Google Scholar] [CrossRef]
- Ho, W.W.H.; Hill, C.B.; Doblin, M.S. Integrative multi-omics analyses of barley rootzones under salinity stress reveal two distinctive salt tolerance mechanisms. Plant Commun. 2020, 1, 100031. [Google Scholar] [CrossRef]
- Temme, A.A.; Kerr, K.L.; Masalia, R.R.; Burke, J.M.; Donovan, L.A. Key traits and genes associate with salinity tolerance independent from vigor in cultivated sunflower. Plant Physiol. 2020, 184, 865–880. [Google Scholar] [CrossRef] [PubMed]
- Yichie, Y.; Brien, C.; Berger, B.; Roberts, T.H.; Atwell, B.J. Salinity tolerance in Australian wild Oryza species varies widely and matches that observed in O. sativa. Rice 2018, 11, 66. [Google Scholar] [CrossRef] [PubMed]
- Isayenkov, S.V.; Maathuis, F.J.M. Plant salinity stress: Many unanswered questions remain. Front. Plant Sci. 2019, 10, 80. [Google Scholar] [CrossRef] [PubMed]
- Ijaz, B.; Formentin, E.; Ronci, B.; Locato, V.; Berizza, E.; Hyder, M.Z.; Schiova, F.L.; Yasmin, T. Salt tolerance in indica rice cell cultures depends on a fine tuning of ROS signalling and homeostasis. PLoS ONE 2019, 14, e0213986. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Wang, K.; Chang, Y.; Zhang, B.; Li, F.; Meng, Y.; Li, M.; Zhao, Q.; An, S. OsHyPRP06/R3L1 regulates root system development and salt tolerance via apoplastic ROS homeostasis in rice (Oryza sativa L.). Plant Cell Environ. 2022, 45, 900–914. [Google Scholar] [CrossRef]
- Iqbal, M.; Irshad, S.; Nadeem, M.; Fatima, T.; Itrat, A.B. Salinity effects on wheat (Triticum aestivum L.) characteristics: A review article. Int. J. Agric. Biol. 2018, 12, 1–15. [Google Scholar]
- Dadshani, S.; Sharma, R.C.; Baum, M.; Ogbonnaya, F.C.; Léon, J.; Ballvora, A. Multi-dimensional evaluation of response to salt stress in wheat. PLoS ONE 2019, 14, e0222659. [Google Scholar] [CrossRef]
- Hu, C.-H.; Wei, X.-Y.; Yuan, B.; Yao, L.-B.; Ma, T.-T.; Zhang, P.-P.; Wang, X.; Wang, P.-Q.; Liu, W.-T.; Li, W.-Q.; et al. Genome-wide identification and functional analysis of NADPH oxidase family genes in wheat during development and environmental stress responses. Front. Plant Sci. 2018, 9, 906. [Google Scholar] [CrossRef]
- Yan, K.; He, W.; Bian, L.; Zhang, Z.; Tang, X.; An, M.; Li, L.; Han, G. Salt adaptability in a halophytic soybean (Glycine soja) involves photosystems coordination. BMC Plant Biol. 2020, 20, 155. [Google Scholar] [CrossRef]
- Hameed, A.; Ahmed, M.Z.; Hussain, T.; Aziz, I.; Ahmad, N.; Gul, B.; Nielsen, B.L. Effects of salinity stress on chloroplast structure and function. Cells 2021, 10, 2023. [Google Scholar] [CrossRef]
- Sarker, U.; Islam, M.T.; Oba, S. Salinity stress accelerates nutrients, dietary fiber, minerals, phytochemicals and antioxidant activity in Amaranthus tricolor leaves. PLoS ONE 2018, 13, e0206388. [Google Scholar] [CrossRef] [PubMed]
- Zahra, N.; Wahid, A.; Hafeez, M.B.; Lalarukh, I.; Batool, A.; Uzair, M.; El-Sheikh, M.A.; Alansi, S.; Kaushik, P. Effect of salinity and plant growth promoters on secondary metabolism and growth of milk thistle ecotypes. Life 2022, 12, 1530. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Zhang, Y.; Testerink, C. Root dynamic growth strategies in response to salinity. Plant Cell Environ. 2022, 45, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Zhu, C.; Bai, Z.; Huang, J.; Zhu, L.; Cao, X.; Nanda, S.; Hussain, S.; Riaz, A.; Liang, Q.; et al. iTRAQ-based protein profiling and biochemical analysis of two contrasting rice genotypes revealed their differential responses to salt stress. Int. J. Mol. Sci. 2019, 20, 547. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Iqbal, S.; Zhang, Y.; Zhang, G.; Ali, U.; Lu, S.; Yao, X.; Guo, L. Proteome-wide identification of S-sulphenylated cysteines in Brassica napus. Plant Cell Environ. 2021, 44, 3571–3582. [Google Scholar] [CrossRef]
- Jain, N.; Farhat, S.; Kumar, R.; Singh, N.; Singh, S.; Sreevathsa, R.; Kalia, S.; Singh, K.; Teruhiro, T.; Rai, V. Alteration of proteome in germinating seedlings of piegonpea (Cajanus cajan) after salt stress. Physiol. Mol. Biol. Plants 2021, 27, 2833–2848. [Google Scholar] [CrossRef]
- Yichie, Y.; Hasan, M.T.; Tobias, P.A.; Pascovici, D.; Goold, H.D.; Sluyter, S.C.V.; Roberts, T.H.; Atwell, B.J. Salt-treated roots of Oryza australiensis seedlings are enriched with proteins involved in energetics and transport. Proteomics 2019, 19, e1900175. [Google Scholar] [CrossRef]
- Nguyen, V.Q.; Sreewongchai, T.; Siangliw, M.; Roytrakul, S.; Yokthongwattana, C. Comparative proteomic analysis of chromosome segment substitution lines of Thai jasmine rice KDML105 under short-term salinity stress. Planta 2022, 256, 12. [Google Scholar] [CrossRef]
- Lakra, N.; Kaur, C.; Singla-Pareek, S.L.; Pareek, A. Mapping the ‘early salinity response’ triggered proteome adaptation in contrasting rice genotypes using iTRAQ approach. Rice 2019, 12, 3. [Google Scholar] [CrossRef]
- Peng, P.; Gao, Y.D.; Li, Z.; Yu, Y.W.; Qin, H.; Guo, Y.; Huang, R.F.; Wang, J. Proteomic analysis of a rice mutant sd58 possessing a novel d1 allele of heterotrimeric G protein alpha subunit (RGA1) in salt stress with a focus on ROS scavenging. Int. J. Mol. Sci. 2019, 20, 167. [Google Scholar] [CrossRef]
- Xiong, E.; Zhang, C.; Ye, C.; Jiang, Y.; Zhang, Y.; Chen, F.; Dong, G.; Zeng, D.; Yu, Y.; Wu, L. iTRAQ-based proteomic analysis provides insights into the molecular mechanisms of rice formyl tetrahydrofolate deformylase in salt response. Planta 2021, 254, 76. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Xiao, C.; Xiao, B.; Wang, M.; Liu, J.; Bhanbhro, N.; Khan, A.; Wang, H.; Wang, H.; Yang, C. Proteomic profiling sheds light on alkali tolerance of common wheat (Triticum aestivum L.). Plant Physiol. Biochem. 2019, 138, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Li, X.; Niu, F.; Sun, X.; Hu, Z.; Zhang, H. iTRAQ-based quantitative proteomic analysis of wheat roots in response to salt stress. Proteomics 2017, 17, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, D.; Zhu, D.; Liu, N.; Yan, Y. Endoplasmic reticulum subproteome analysis reveals underlying defense mechanisms of wheat seedling leaves under salt stress. Int. J. Mol. Sci. 2021, 22, 4840. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Luo, F.; Zou, R.; Liu, J.; Yan, Y. Integrated physiological and chloroplast proteome analysis of wheat seedling leaves under salt and osmotic stresses. J. Proteomics 2021, 234, 104097. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Ding, L.L.; Yao, Y.; Cao, Y.; Pan, Z.H.; Kong, D.H. Extracellular proteome analysis and flavor formation during soy sauce fermentation. Front. Microbiol. 2018, 9, 1872. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, M.; Cheng, H.; Sun, N.; Liu, S.; Li, S.; Wang, Y.; Zheng, Y.; Uversky, V.N. The effect of phosphorylation on the salt-tolerance-related functions of the soybean protein PM18, a member of the group-3 LEA protein family. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 1291–1301. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Qi, F.; Gao, L.; Rao, S.; Yang, Z.; Fang, W. iTRAQ-based quantitative proteomic analysis of dark-germinated soybeans in response to salt stress. RSC Adv. 2018, 8, 17905. [Google Scholar] [CrossRef]
- Pi, E.; Zhu, C.; Fan, W.; Huang, Y.; Qu, L.; Li, Y.; Zhao, Q.; Ding, F.; Qiu, L.; Wang, H.; et al. Quantitative phosphoproteomic and metabolomic analyses reveal GmMYB173 optimizes flavonoid metabolism in soybean under salt stress. Mol. Cell. Proteom. 2018, 17, 1209–1224. [Google Scholar] [CrossRef]
- Ji, W.; Cong, R.; Li, S.; Li, R.; Qin, Z.; Li, Y.; Zhou, X.; Chen, S.; Li, J. Comparative proteomic analysis of soybean leaves and roots by iTRAQ provides insights into response mechanisms to short-term salt stress. Front. Plant Sci. 2016, 7, 573. [Google Scholar] [CrossRef]
- Liu, A.; Xiao, Z.; Wang, Z.; Lam, H.M.; Chye, M.L. Galactolipid and phospholipid profile and proteome alterations in soybean leaves at the onset of salt stress. Front. Plant Sci. 2021, 12, 644408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Yan, Y.; Zeng, X.; Wang, Y.; Zhang, Y. Quantitative proteomics analysis reveals proteins associated with high melatonin content in barley seeds under NaCl-induced salt stress. J. Agric. Food Chem. 2022, 70, 8492–8510. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Liu, S.; Dong, T.; Xu, D.; Pan, S.; Li, Z.; Zhu, M. Comparative transcriptome and proteome analysis of salt-tolerant and salt-sensitive sweet potato and overexpression of IbNAC7 confers salt tolerance in Arabidopsis. Front. Plant Sci. 2020, 11, 572540. [Google Scholar] [CrossRef] [PubMed]
- Chiconato, D.A.; de Santana, M.G.; Balbuena, T.S.; Munns, R.; Dos Santos, D.M.M. Proteomic analysis of young sugarcane plants with contrasting salt tolerance. Funct. Plant Biol. 2021, 48, 588–596. [Google Scholar] [CrossRef]
- Arefian, M.; Vessal, S.; Malekzadeh-Shafaroudi, S.; Siddique, K.H.M.; Bagheri, A. Comparative proteomics and gene expression analyses revealed responsive proteins and mechanisms for salt tolerance in chickpea genotypes. BMC Plant Biol. 2019, 19, 300. [Google Scholar] [CrossRef]
- Ganie, S.A.; Molla, K.A.; Henry, R.J.; Bhat, K.V.; Mondal, T.K. Advances in understanding salt tolerance in rice. Theor. Appl. Genet. 2019, 132, 851–870. [Google Scholar] [CrossRef]
- Zhang, K.; Tang, J.; Wang, Y.; Kang, H.; Zeng, J. The tolerance to saline–alkaline stress was dependent on the roots in wheat. Physiol. Mol. Biol. Plants 2020, 26, 947–954. [Google Scholar] [CrossRef]
- Amirbakhtiar, N.; Ismaili, A.; Ghaffari, M.R.; Nazarian, F.F.; Shobbar, Z.S. Transcriptome response of roots to salt stress in a salinity-tolerant bread wheat cultivar. PLoS ONE 2019, 14, e0213305. [Google Scholar] [CrossRef]
- Dai, L.; Li, P.; Li, Q.; Leng, Y.; Zeng, D.; Qian, Q. Integrated multi-omics perspective to strengthen the understanding of salt tolerance in rice. Int. J. Mol. Sci. 2022, 23, 5236. [Google Scholar] [CrossRef]
- Saini, S.; Kaur, N.; Marothia, D.; Singh, B.; Singh, V.; Gantet, P.; Pati, P.K. Morphological analysis, protein profiling and expression analysis of auxin homeostasis genes of roots of two contrasting cultivars of rice provide inputs on mechanisms involved in rice adaptation towards salinity stress. Plants 2021, 10, 1544. [Google Scholar] [CrossRef]
- Roy Choudhury, A.; Roy, S.K.; Trivedi, P.; Choi, J.; Cho, K.; Yun, S.H.; Walitang, D.I.; Park, J.-H.; Kim, K.; Sa, T. Label-free proteomics approach reveals candidate proteins in rice (Oryza sativa L.) important for ACC deaminase producing bacteria-mediated tolerance against salt stress. Environ. Microbiol. 2022, 24, 3612–3624. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Long, R.; Kang, J.; Wang, Z.; Zhang, T.; Sun, H.; Li, X.; Yang, Q. Comparative proteomic analysis reveals that antioxidant system and soluble sugar metabolism contribute to salt tolerance in alfalfa (Medicago sativa L.) leaves. J. Proteome Res. 2019, 18, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, D.; Kaundal, A. Dynamics of salt tolerance: Molecular perspectives. In Biotechnologies of Crop Improvement; Gosal, S.S., Wani, S.H., Eds.; Springer International Publishing AG: Cham, Switzerland, 2018; Volume 3, pp. 25–40. [Google Scholar]
- Moin, M.; Saha, A.; Bakshi, A.; Madhav, M.S.; Kirti, P.B. Constitutive expression of ribosomal protein L6 modulates salt tolerance in rice transgenic plants. Gene 2021, 789, 145670. [Google Scholar] [CrossRef] [PubMed]
- Lv, D.W.; Zhu, G.R.; Zhu, D.; Bian, Y.-W.; Liang, X.-N.; Cheng, Z.-W.; Deng, X.; Yan, Y.-M. Proteomic and phosphoproteomic analysis reveals the response and defense mechanism in leaves of diploid wheat T. monococcum under salt stress and recovery. J. Proteom. 2016, 143, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Du, X.; Zhang, J.; Li, J.; Chen, S.; Duanmu, H.; Li, H. Quantitative redox proteomics revealed molecular mechanisms of salt tolerance in the roots of sugar beet monomeric addition line M14. Bot. Stud. 2022, 63, 5. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Shi, C.; Su, C.; Liu, Y. Complementary analyses of the transcriptome and iTRAQ proteome revealed mechanism of ethylene dependent salt response in bread wheat (Triticum aestivum L.). Food Chem. 2020, 325, 126866. [Google Scholar] [CrossRef]
- Ahmed, I.M.; Nadira, U.A.; Qiu, C.W.; Cao, F.; Chen, Z.-H.; Vincze, E.; Wu, F. The barley S-adenosylmethionine synthetase 3 gene HvSAMS3 positively regulates the tolerance to combined drought and salinity stress in Tibetan wild barley. Cells 2020, 9, 1530. [Google Scholar] [CrossRef]
- Zulfiqar, F.; Nafees, M.; Chen, J.; Darras, A.; Ferrante, A.; Hancock, J.T.; Ashraf, M.; Zaid, A.; Latif, N.; Corpas, F.J.; et al. Chemical priming enhances plant tolerance to salt stress. Front. Plant Sci. 2022, 13, 946922. [Google Scholar] [CrossRef]
- Zhu, M.; Liu, Y.; Cai, P.; Duan, X.; Sang, S.; Qiu, Z. Jasmonic acid pretreatment improves salt tolerance of wheat by regulating hormones biosynthesis and antioxidant capacity. Front Plant Sci. 2022, 13, 968477. [Google Scholar] [CrossRef]
- Tiwari, J.K.; Buckseth, T.; Zinta, R.; Bhatia, N.; Dalamu, D.; Naik, S.; Poonia, A.K.; Kardile, H.B.; Challam, C.; Singh, R.K.; et al. Germplasm, breeding, and genomics in potato improvement of biotic and abiotic stresses tolerance. Front. Plant Sci. 2022, 13, 805671. [Google Scholar] [CrossRef]
- Farhat, S.; Jain, N.; Singh, N.; Sreevathsa, R.; Dash, P.K.; Rai, R.; Yadav, S.; Kumar, P.; Sarkar, A.K.; Jain, A.; et al. CRISPR-Cas9 directed genome engineering for enhancing salt stress tolerance in rice. Semin. Cell Dev. Biol. 2019, 96, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.B.; Li, J.; Qin, R.Y.; Xu, R.F.; Li, H.; Yang, Y.C.; Ma, H.; Li, L.; Wei, P.C.; Yang, J.B. Identification of a regulatory element responsible for salt induction of rice OsRAV2 through ex situ and in situ promoter analysis. Plant Mol. Biol. 2016, 90, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Hu, J.; Dong, L.; Zeng, D.; Guo, L.; Zhang, G.; Zhu, L.; Qian, Q. The tolerance of salinity in rice requires the presence of a functional copy of FLN2. Biomolecules 2020, 10, 17. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.D.; Yang, C.C.; Qin, R.; Alamin, M.; Yue, E.K.; Jin, X.L.; Shi, C.H. A guanine insert in OsBBS1 leads to early leaf senescence and salt stress sensitivity in rice (Oryza sativa L.). Plant Cell Rep. 2018, 37, 933–946. [Google Scholar] [CrossRef]
- Zhang, A.; Liu, Y.; Wang, F.; Li, T.; Chen, Z.; Kong, D.; Bi, J.; Zhang, F.; Luo, X.; Wang, J. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. 2019, 39, 47. [Google Scholar] [CrossRef]
- Lan, T.; Zheng, Y.; Su, Z.; Yu, S.; Song, H.; Zheng, X.; Lin, G.; Wu, W. OsSPL10, a SBP-Box gene, plays a dual role in salt tolerance and trichome formation in rice (Oryza sativa L.). G3 2019, 9, 4107–4114. [Google Scholar] [CrossRef]
- Thudi, M.; Palakurthi, R.; Schnable, J.C.; Chitikineni, A.; Dreisigacker, S.; Mace, E.; Srivastava, R.K.; Satyavathi, C.T.; Odeny, D.; Tiwari, V.K. Genomic resources in plant breeding for sustainable agriculture. J. Plant Physiol. 2021, 257, 153351. [Google Scholar] [CrossRef]
- Singh, V.K.; Singh, B.D.; Kumar, A.; Maurya, S.; Krishnan, S.G.; Vinod, K.K.; Singh, M.P.; Ellur, R.K.; Bhowmick, P.K.; Singh, A.K. Marker-assisted introgression of Saltol QTL enhances seedling stage salt tolerance in the rice variety “Pusa Basmati 1”. Int. J. Genom. 2018, 2018, 8319879. [Google Scholar] [CrossRef]
- Kong, W.; Zhang, C.; Zhang, S. Uncovering the novel QTLs and candidate genes of salt tolerance in rice with linkage mapping, RTM-GWAS, and RNA-seq. Rice 2021, 14, 93. [Google Scholar] [CrossRef]
- Bohra, A.; Chand-Jha, U.; Godwin, I.D.; Varshney, K.R. Genomic interventions for sustainable agriculture. Plant Biotechnol. J. 2020, 18, 2388–2405. [Google Scholar] [CrossRef]
- Hoyos-Villegas, V.; Song, Q.; Kelly, J.D. Genome-wide association analysis for drought tolerance and associated traits in common bean. Plant Genome 2017, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Soren, K.R.; Madugula, P.; Kumar, N.; Barmukh, R.; Sengar, M.S.; Bharadwaj, C.; Sharma, P.C.; Singh, S.; Bhandari, A.; Singh, J. Genetic dissection and identification of candidate genes for salinity tolerance using Axiom®CicerSNP array in chickpea. Int. J. Mol. Sci. 2020, 21, 5058. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.R.; Naveed, S.A.; Zhang, Y.; Li, Z.; Zhao, X.; Fiaz, S.; Zhang, F.; Wu, Z.; Hu, Z.; Fu, B.; et al. Identification of candidate genes for salinity and anaerobic tolerance at the germination stage in rice by genome wide association analyses. Front. Genet. 2022, 13, 822516. [Google Scholar] [CrossRef] [PubMed]
- Ondrasek, G.; Rathod, S.; Manohara, K.K.; Gireesh, C.; Anantha, M.S.; Sakhare, A.S.; Parmar, B.; Yadav, B.K.; Bandumula, N.; Raihan, F. Salt stress in plants and mitigation approaches. Plants 2022, 11, 717. [Google Scholar] [CrossRef] [PubMed]
- Abiala, A.; Abdelrahman, M.; Burritt, D.J.; Tran, L.-S.P. Salt stress tolerance mechanisms and potential applications of legumes for sustainable reclamation of salt-degraded soils. Land Degrad. Dev. 2018, 29, 3812–3822. [Google Scholar] [CrossRef]
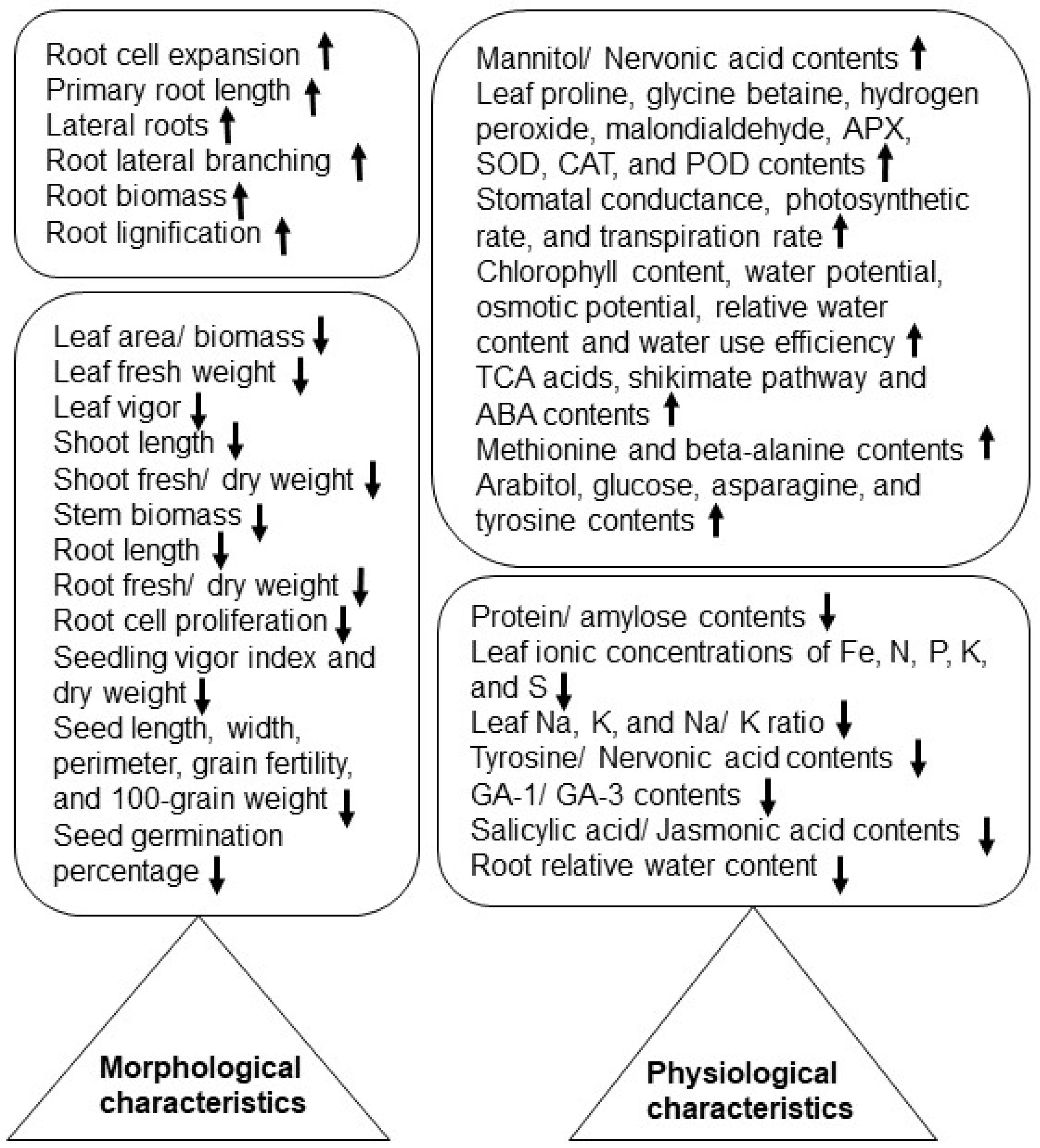
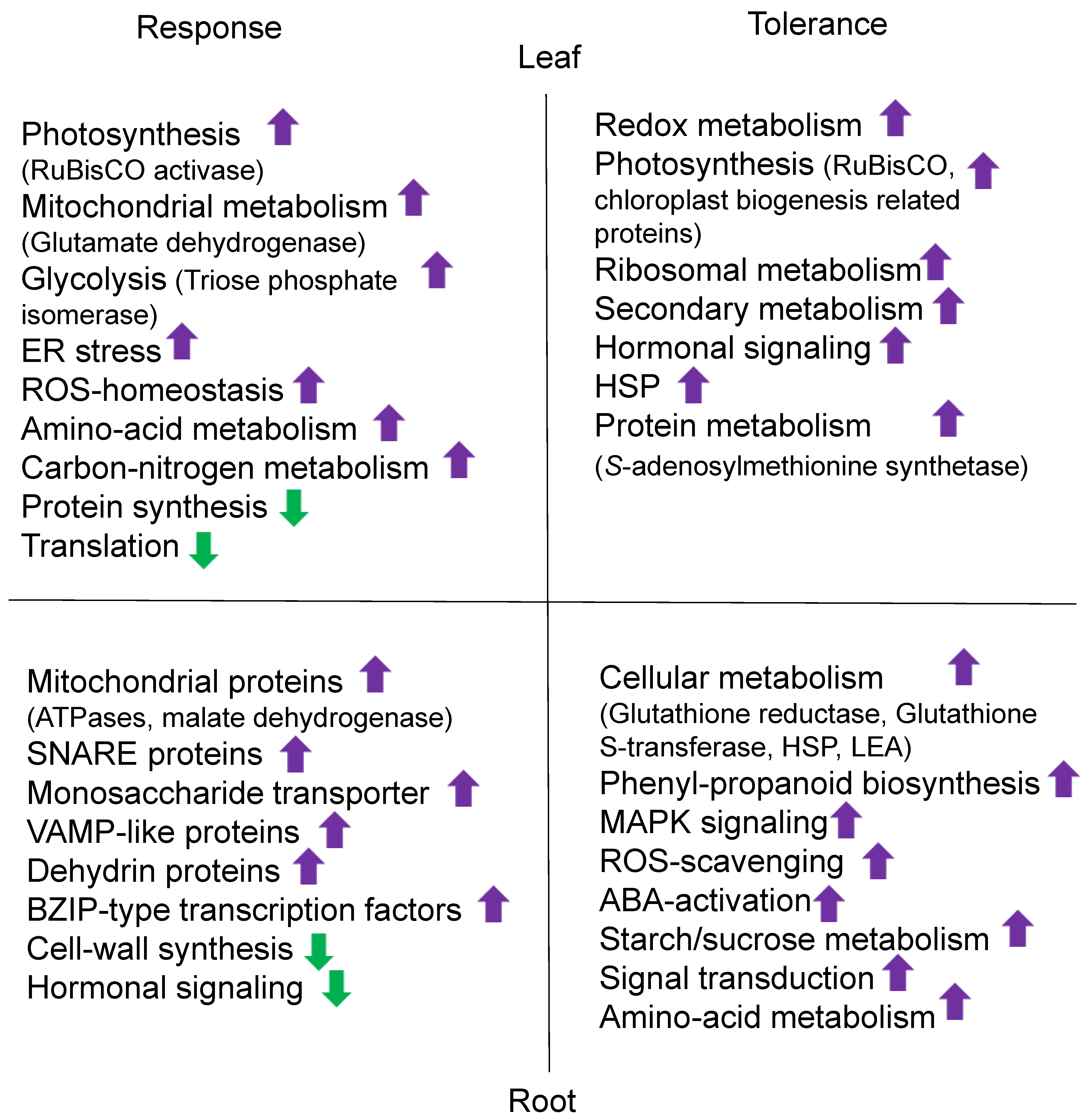
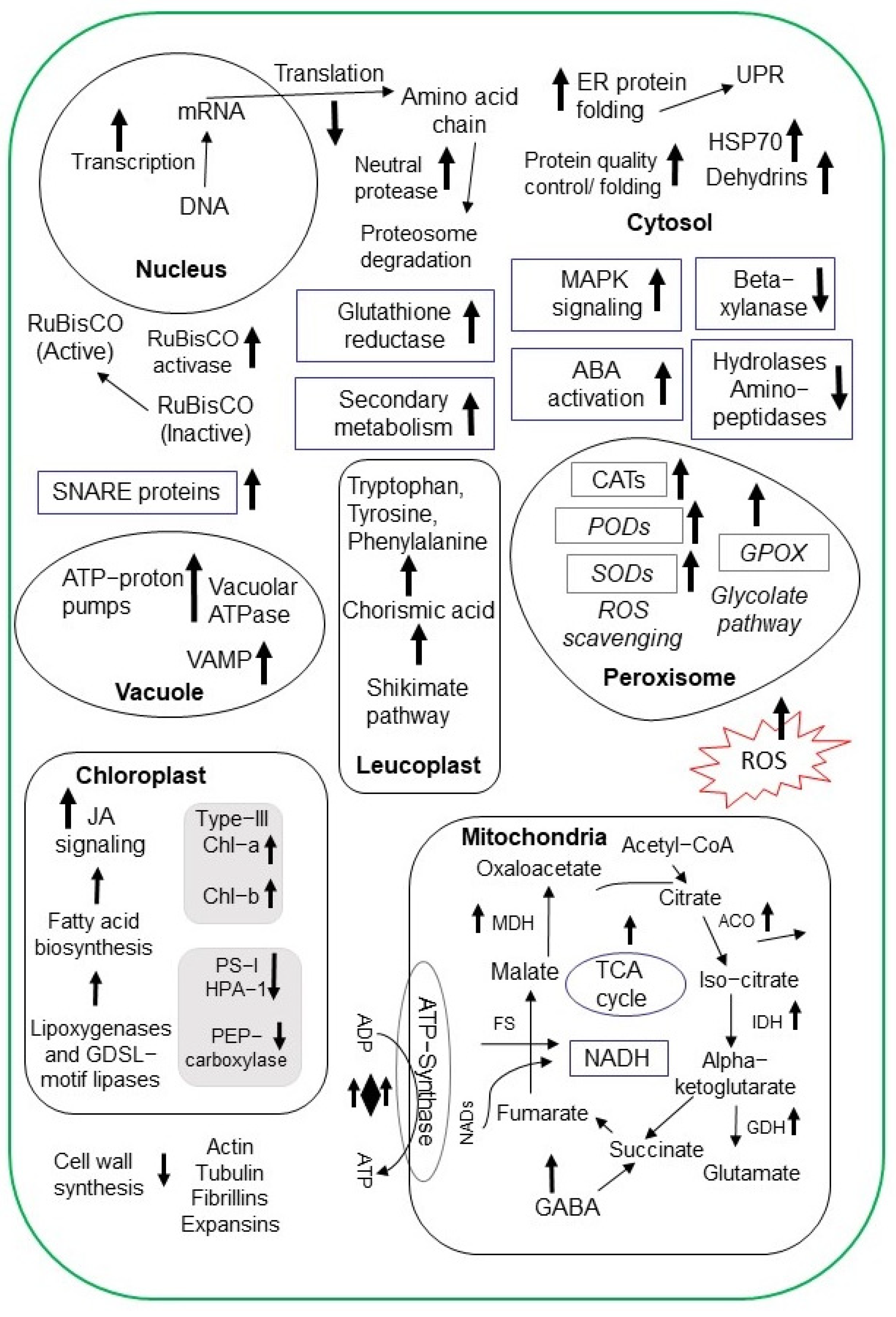
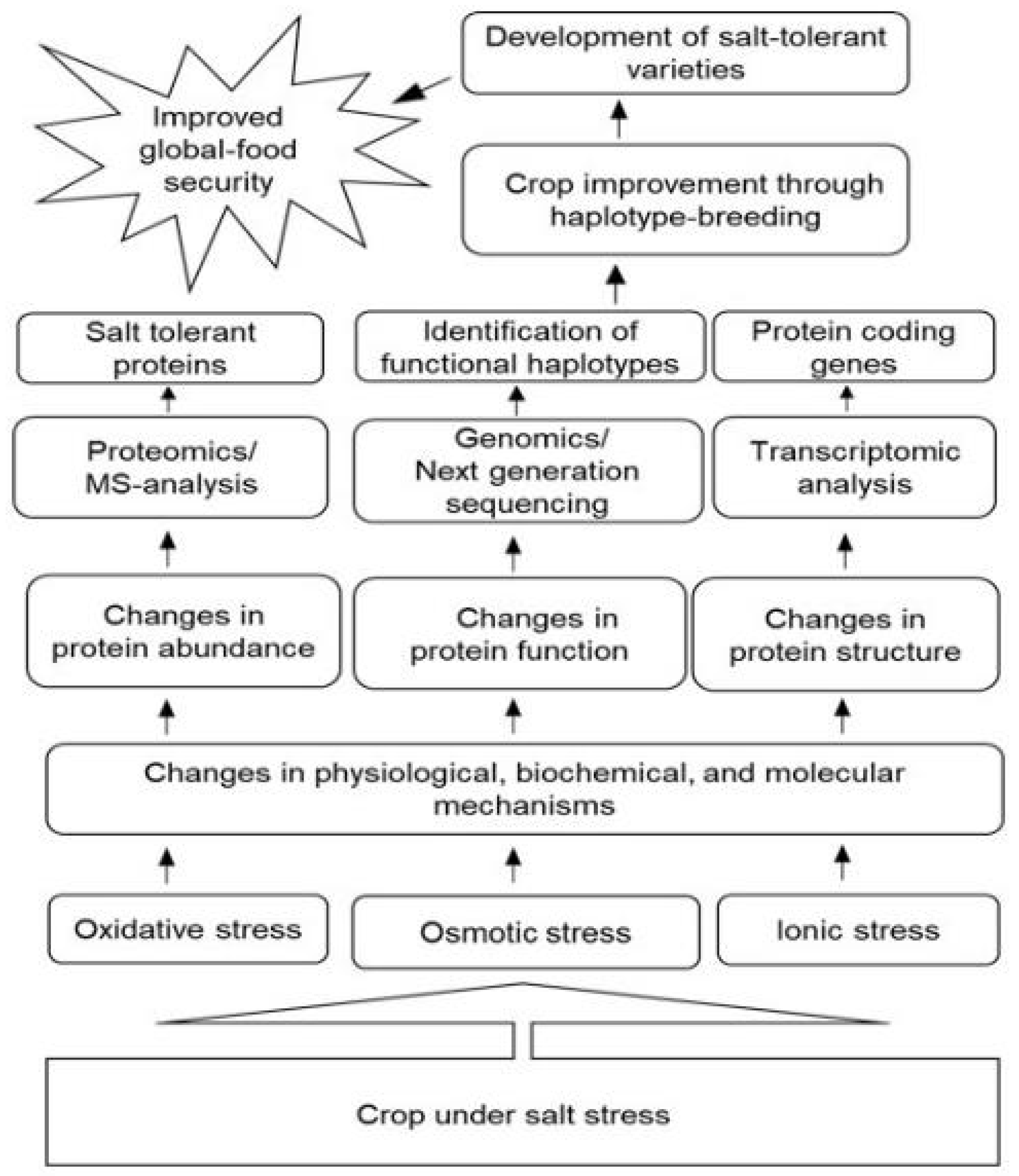
| Crop | Organ | NaCl Conc. * | Morphological Effects | Physiological Effects | Metabolites | Ref. ** |
|---|---|---|---|---|---|---|
| Rice | Shoot, leaf, root | 60 mM | The shoot length, leaf area, shoot fresh weight, and root fresh weight decreased in the sensitive variety. | The Na/K ratio decreased in tolerant and sensitive varieties. | (-) | [24] |
| Shoot, leaf, root | 150 mM | The germination percentage, root length, and seedling dry weight reduced. | The stomatal conductance, photosynthetic rate, transpiration rate, chlorophyll content, water potential, osmotic potential, relative water content, and water use efficiency decreased. | Proline, malonaldehyde, and glutathione reductase increased. | [25] | |
| Leaf, root | 50 mM | The shoot length, root weight. and root lateral branching decreased in the sensitive cultivar only. | In the leaf, most organic acids involved in TCA and the shikimate pathway decreased in sensitive and tolerant cultivars. In the root, mannitol increased in the tolerant cultivar. | Raffinose, fructose, ribose, sucrose, shikimate, and quinate decreased. | [26] | |
| Shoot, root | 60 mM | The shoot fresh/dry weight and root dry weight decreased in tolerant and sensitive varieties. Root border cells increased in the tolerant variety. | The relative water content of the shoot and root decreased in the sensitive variety but was maintained in the tolerant one. | (-) | [27] | |
| Root | 100 mM | In the root apical meristem, cell proliferation was inhibited and cell expansion was enhanced. | The ABA content accumulated in the primary root. | Ethylene increased. | [28] | |
| Grain | 40 mM | The length, width, perimeter, and grain fertility/100-grain weight decreased in salt-sensitive varieties but was maintained in the salt-tolerant one. | Endosperm-starch accumulation was enhanced, and the protein/amylose content decreased in salt-susceptible varieties. | (-) | [23] | |
| Wheat | Leaf, root | 100 mM | The aerenchyma and packing density of thylakoids increased. | ABA and GA3/JA increased and decreased, respectively, in the leaf, but ABA/JA and GA1/SA increased and decreased, respectively, in the root. | Flavonoids increased. | [29] |
| Seed | 200 mM | The germination rate/index, mean germination time, and vigor index decreased. | Malondialdehyde and soluble sugars increased, and superoxide dismutase decreased. | Zeatin, anthocyanin, flavone, and flavonol increased. | [30] | |
| Seedling | 100 mM | The total fresh weight, shoot dry weight, leaf chlorophyll, root dry weight, and root length decreased. | Ionic concentrations of Na, K, and Na/K decreased in the leaf. | (-) | [31] | |
| Root | 100 mM | Proline/malondialdehyde contents increased along with the maximum electrolyte leakage. | NADPH oxidase increased. | [32] | ||
| Shoot, leaf | 200 mM | The shoot dry weight decreased in yellow/blue and increased in purple/dark-purple-colored grain cultivars. | The simple fluorescence ratio, flavonoid content, modified flavonoid index, proline content, and anthocyanins content increased. | The malondialdehyde content increased. | [33] | |
| Soybean | Root | 40/80 mM | The root length decreased. | Proline and glycine betaine increased. | Soluble sugars decreased. | [34] |
| Stem, root | 100 mM | The stem and root biomass decreased. | The pith and cortex increased at the internodes of the stem, and the root epidermis and endodermis increased with the formation of lysogenic aerenchyma. | (-) | [35] | |
| Leaf, stem, root | 80 mM | The shoot length and fresh/dry weight, root length/volume, and root fresh/dry weight decreased. | Leaf proline, glycine betaine, hydrogen peroxide, malondialdehyde, APX, SOD, CAT, and POD increased. Leaf protein, soluble sugars, chlorophyll, phenols, flavonoids, DPPH, photosynthetic rate, stomatal conductance, and transpiration rate decreased. | Total phenolic and flavonoid contents decreased in the leaf. | [36] | |
| Seedling root | 100 mM | The primary root length, lateral roots, and biomass increased in tolerant lines as compared to the sensitive one. | Antioxidant enzyme (SOD, APX, and CAT) activities, Na/K ratio, N content, and nitrogen use efficiency increased in salt-tolerant lines. | TCA, glyoxalate, and dicarboxylate metabolites increased. | [37] | |
| Maize | Stem, root | 100 mM | Root lignification increased. | Feruloylation of arabinoxylans in the stem decreased. | Ferulic acid increased | [38] |
| Leaf | 200 mM | The leaf fresh weight and biomass decreased. | Arabitol, glucose, asparagine, and tyrosine increased in the leaf. | Maltitol, raffinose, and cinnamic acid increased. | [39] | |
| Brassica rapa | Root | 150 mM | The root water content and root length decreased. | Hydrogen peroxide and malondialdehyde increased. | (-) | [40] |
| Barley | Seminal roots | 100 mM | There was no significant change in the root length observed in sensitive and tolerant cultivars. | Tyrosine decreased and methionine and beta-alanine increased in the root tips of the sensitive cultivar. Nervonic acid increased and decreased in tolerant and sensitive cultivar root tips, respectively. | Amines increased, and stearic acid decreased. | [41] |
| Sunflower | Leaf, stem | 100 mM | The leaf mass increased and stem mass decreased. | In the leaf, B, Cu, Zn, Mn, and Na ion concentrations increased. In the stem, Fe, N, P, K, S, and the K/Na ratio decreased. | Ionic concentrations of leaf Ca and Mg increased. | [42] |
| Crop | Organ | NaCl Conc. * | Proteomic Technique | No. ** | Major Findings | Ref. *** |
|---|---|---|---|---|---|---|
| Rice | Root | 80 mM | TMT-MS | 200 | Mitochondrial ATPases, SNARE proteins, monosaccharide transporter, and VAMP-like protein increased. | [58] |
| 150 mM | LC-MS/MS | 178 | The protein interaction network displayed connections between proteins involved in cell wall synthesis, transcription, translation, and defense. | [59] | ||
| Shoot | 200 mM | iTRAQ-LC-MS/MS | 149 | Glutamate dehydrogenase, RuBisCO activase, peroxidases, and triose phosphate isomerase increased. | [60] | |
| Leaf | 150 mM | iTRAQ-LC-MS/MS | 332 | Photosynthesis and ROS homeostasis increased. | [61] | |
| Grain | 150 mM | iTRAQ-LC-MS/MS | 279 | HPA1 decreased with impaired chlorophyll metabolism and photosynthesis. | [62] | |
| Wheat | Root | 50 mM | LC-MS/MS | 41 | SOD, malate dehydrogenases, dehydrin proteins, and V-ATPase increased. | [63] |
| 350 mM | LC-MS/MS | 121 | Substrate-recruiting E3 ubiquitin ligases increased BZIP-type transcription factor. | [64] | ||
| Shoot | 200 mM | LC-MS/MS | 234 | Protein folding, quality control, and ER stress-response-related proteins increased. Protein synthesis and translation-related proteins decreased. | [65] | |
| 200 mM | LC-MS/MS | 194 | Chloroplast proteins involved in the light-dependent reaction, Calvin cycle, transcription, amino acid metabolism, and carbon/nitrogen metabolism increased. | [66] | ||
| Soybean | Soybean mash | 18% | TMT-LC-MS/MS | 42 | Hydrolases, dipeptidase, neutral protease 2, leucine aminopeptidase, and beta xylanase decreased. | [67] |
| Radicle | 1.2 M | 2DE-TripleTOF | 12 | Phosphorylated PM18 protein protected lactate dehydrogenase during salt stress acclimation. | [68] | |
| Germinating seeds | 50 mM | LC-MS/MS | 201 | The GABA content and antioxidase activity increased. | [69] | |
| Root | 200 mM | LC-MS/MS | 412 | Dihydroxy B-ring flavonoids increased as anti-oxidants and GmMYB173 was phosphorylated. | [70] | |
| Leaf | 200 mM | iTRAQ-LC-MS/MS | 278 | Carbohydrate/energy metabolism, signaling, membrane/transport, stress/defense, protein synthesis, and redox homeostasis increased. | [71] | |
| Leaf | 0.9% | LC-MS/MS | 2049 | Plastidial JA biosynthesis, phosphatidylinositol production, the TCA cycle, and the glycolysis pathway increased. | [72] | |
| Barley | Germinating seeds | 240 mM | TMT-LC MS/MS | 68 | Melatonin along with proteins related to cellular redox homeostasis, osmotic stress, secondary metabolites, purine degradation, and shikimate pathways increased. | [73] |
| Sweet potato | Root | 150 mM | iTRAQ-LC-MS/MS | 727 | Proteins related to ion accumulation, stress signaling, transcriptional regulation, redox reactions, plant hormone signal transduction, and secondary metabolites increased. | [74] |
| Sugarcane | Leaf | 160 mM | iTRAQ-LC MS/MS | 189 | GDSL-motif lipases, lipoxygenase, and type III chlorophyll a/b binding proteins increased, while phosphoenolpyruvate carboxylase decreased. | [75] |
| Chickpea | Leaf | 100 mM | LC-MS/MS | 364 | Photosynthesis, bioenergy, stress responsiveness, protein synthesis/degradation, and gene transcription/replication increased in the tolerant cultivar. | [76] |
| Crop | Organ | NaCl Conc. * | Proteomic Technique | No. ** | Major Findings | Ref. *** |
|---|---|---|---|---|---|---|
| Rice | Root | 100 mM | MALDI TOF/TOF MS/MS | 27 | The ROS-scavenging system and ABA activation increased in the tolerant cultivar. | [81] |
| Seedling | 200 mM | iTRAQ-LC-MS/MS | 333 | Photosynthesis, ribosome/chloroplast biogenesis, ion transportation, transcription/translation regulation, phytohormones, and secondary metabolite signal transduction increased in the transgenic line of RPL6. | [85] | |
| Leaf | 150 mM | LC-MS/MS | 160 | Antioxidant proteins, RuBisCO, and ribosomal proteins increased. | [82] | |
| Wheat | Leaf | 160 mM | MALDI-TOF/TOF MS | 81 | Stress response/defense, regulatory, folding/assembly, and degradation-related proteins increased. | [86] |
| Shoot, root | 150 mM | iTRAQ/TMT-LC-MS/MS | 1140 | Carbohydrate metabolism, redox metabolism, cell wall, secondary metabolism, and the pentose phosphate pathway increased by ethylene treatment. | [88] | |
| Maize | Root | 180 mM | iTRAQ-LC-MS/MS | 626 | Proteins associated with phenylpropanoid biosynthesis, starch/sucrose metabolism, and mitogen-activated kinase signaling increased in the tolerant cultivar. | [20] |
| Sugar beet | Root | 200 mM | IodoTMT-LC-MS/MS | 462 | Proteins involved in the regulation of ROS homeostasis, carbohydrate and amino acid metabolism, stress/defense, biosynthesis, and signal transduction increased. | [87] |
| Barley | Leaf | 200 mM | MALDI-TOF/TOF MS | 21 | S-adenosylmethionine synthetase 3, photosynthesis, redox homeostasis, defense/stress, and primary metabolism increased in the salt-tolerant cultivar by exogenous ethylene application. | [89] |
| Alfalfa | Leaf | 100 mM | iTRAQ-LC-MS/MS | 438 | In the tolerant cultivar, proteins involved in the antioxidant system, starch/sucrose metabolism, and secondary metabolism increased. In the sensitive cultivar, proteins related to the light-harvesting complex and photosystem II decreased. | [83] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kausar, R.; Komatsu, S. Proteomic Approaches to Uncover Salt Stress Response Mechanisms in Crops. Int. J. Mol. Sci. 2023, 24, 518. https://doi.org/10.3390/ijms24010518
Kausar R, Komatsu S. Proteomic Approaches to Uncover Salt Stress Response Mechanisms in Crops. International Journal of Molecular Sciences. 2023; 24(1):518. https://doi.org/10.3390/ijms24010518
Chicago/Turabian StyleKausar, Rehana, and Setsuko Komatsu. 2023. "Proteomic Approaches to Uncover Salt Stress Response Mechanisms in Crops" International Journal of Molecular Sciences 24, no. 1: 518. https://doi.org/10.3390/ijms24010518
APA StyleKausar, R., & Komatsu, S. (2023). Proteomic Approaches to Uncover Salt Stress Response Mechanisms in Crops. International Journal of Molecular Sciences, 24(1), 518. https://doi.org/10.3390/ijms24010518







