Prophenoloxidase of Odontotermes formosanus (Shiraki) (Blattodea: Termitidae) Is a Key Gene in Melanization and Has a Defensive Role during Bacterial Infection
Abstract
1. Introduction
2. Results
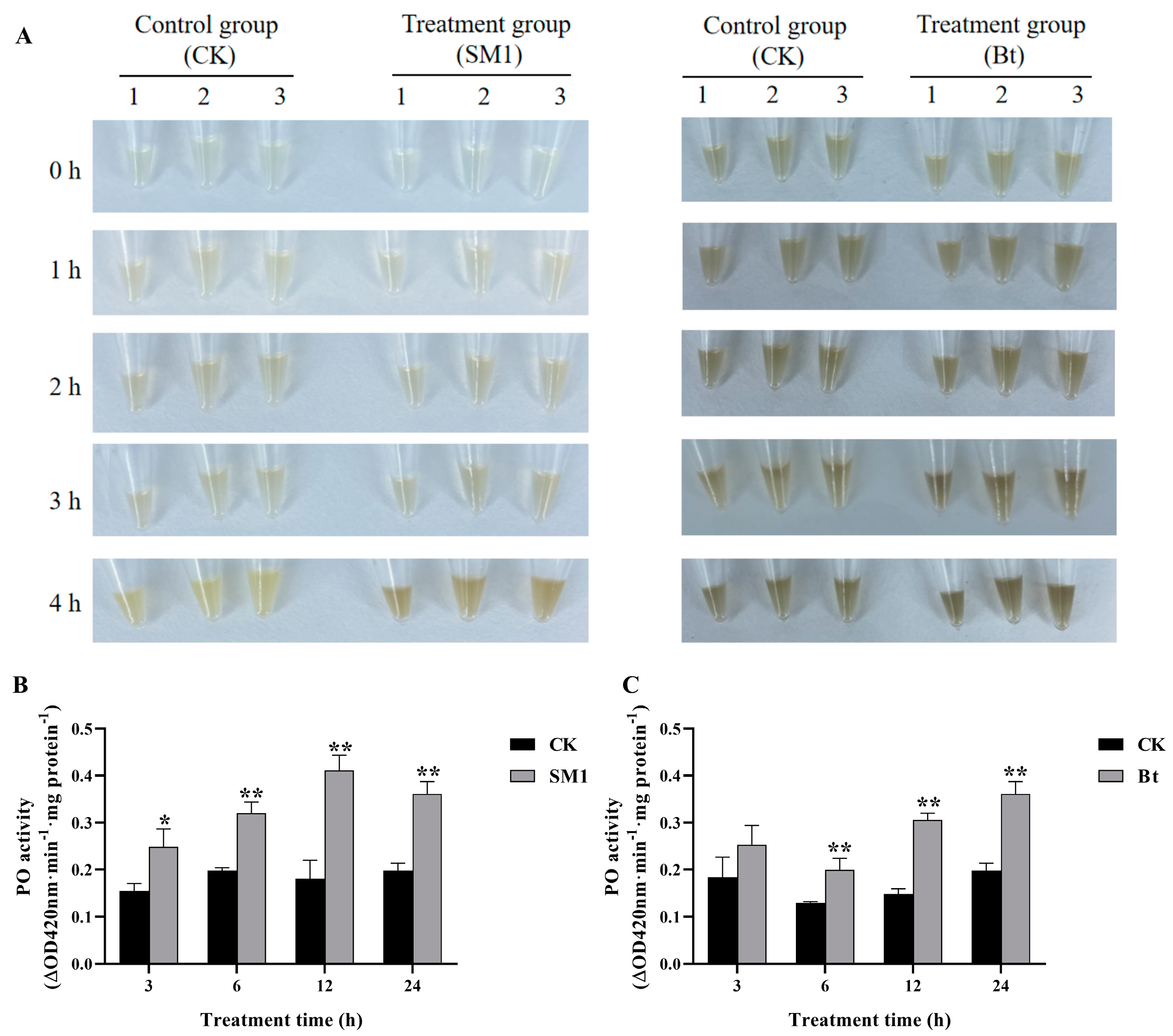
2.1. Melanization Accelerated and PO Activity Induced by SM1 and Bt in O. formosanus
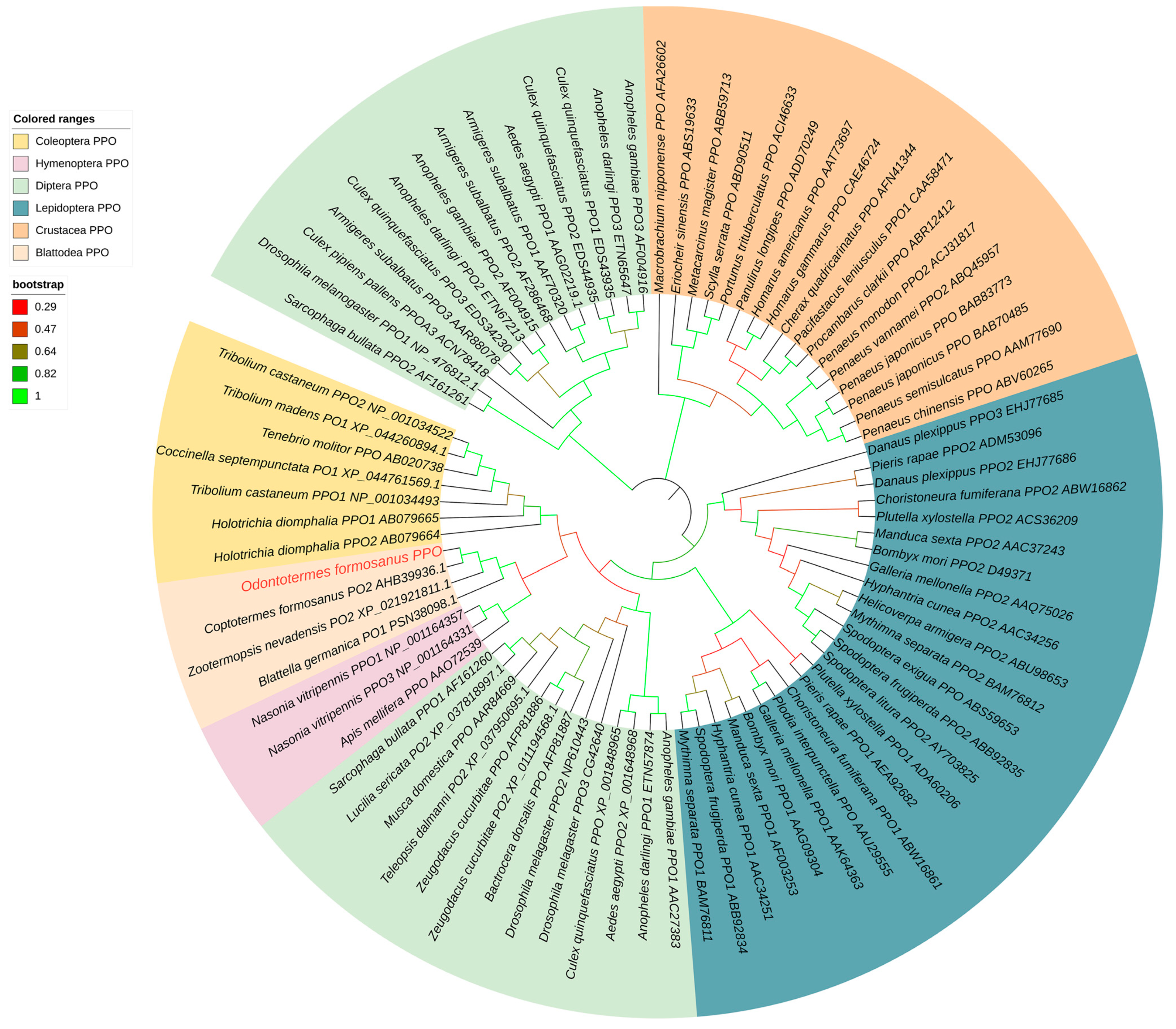
2.2. Cloning and Characterization of OfPPO in O. formosanus
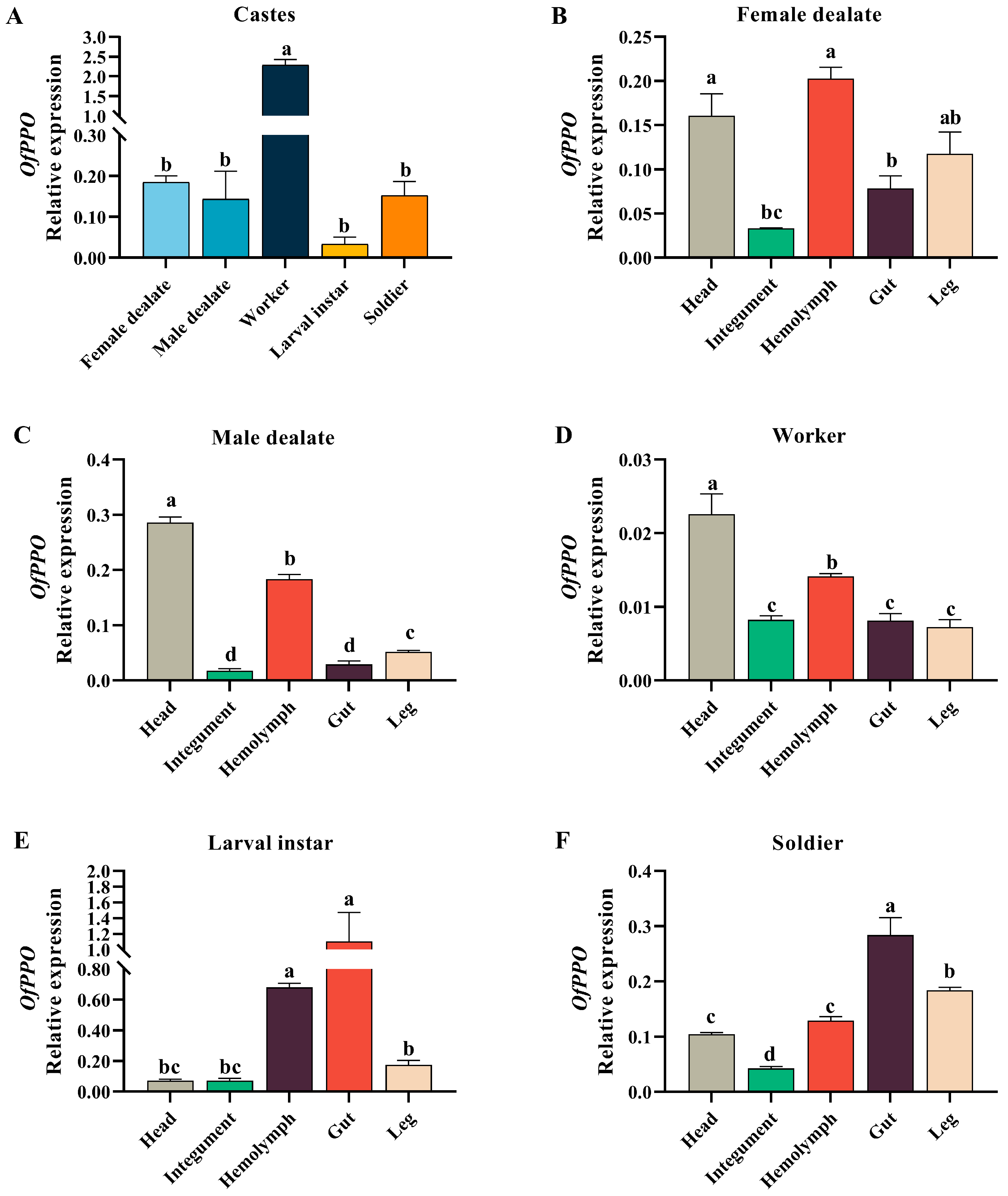
2.3. Caste- and Tissue-Specific Expression Profiles of OfPPO in O. formosanus
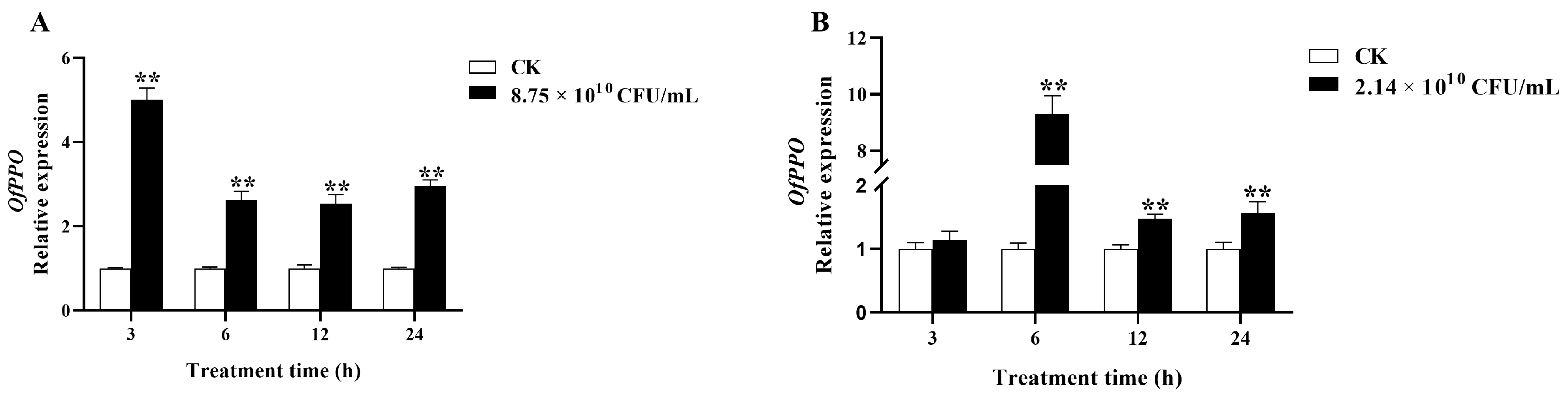
2.4. The OfPPO Expression Induced by SM1 and Bt in O. formosanus
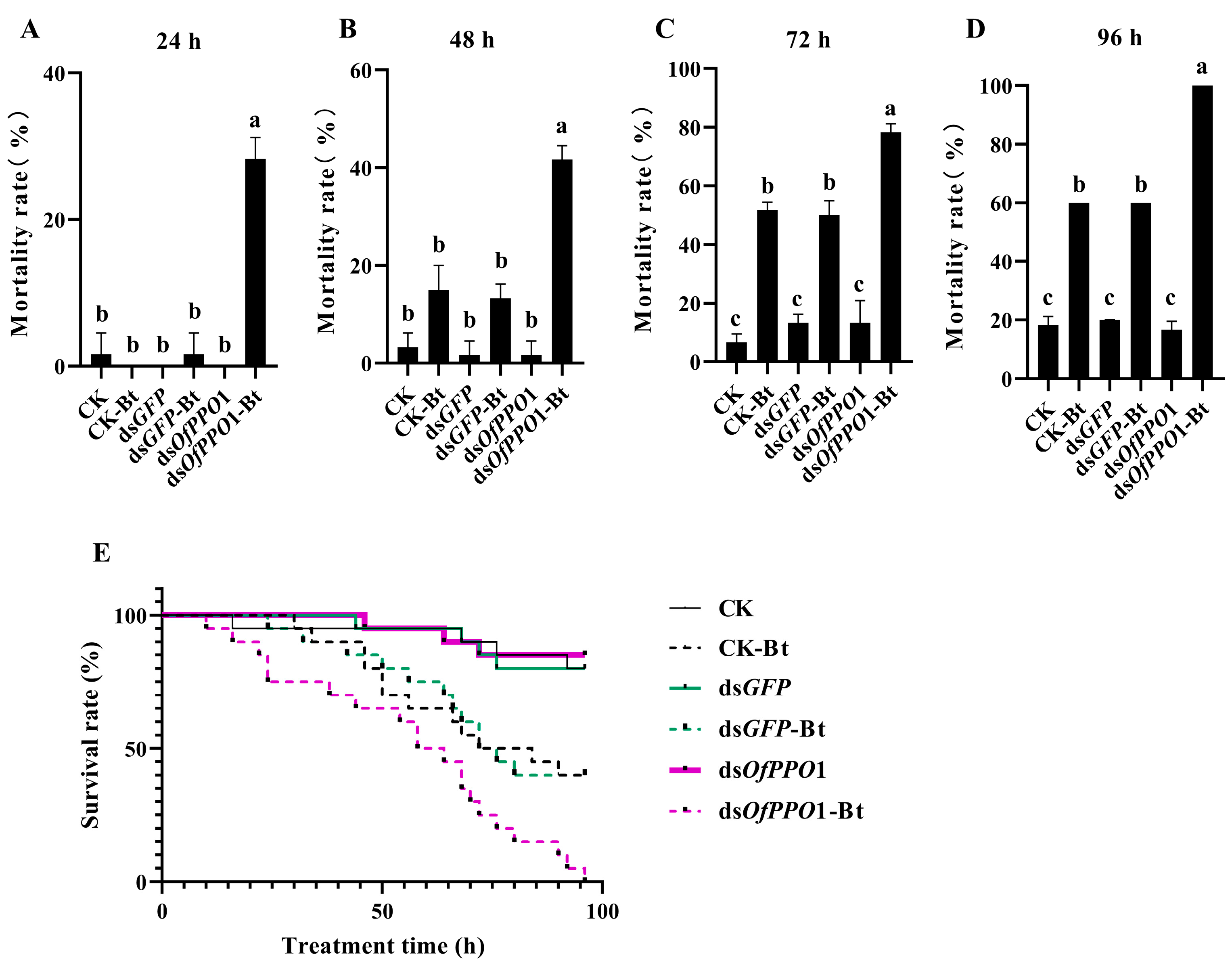
2.5. The Function of OfPPO in Resisting SM1 and Bt Infection in O. formosanus
3. Discussion
4. Materials and Methods
4.1. Insects and Bacteria
4.2. Melanization Assay and PO Activity
4.3. Cloning of the Full-Length Prophenoloxidase Gene of O. formosanus
4.4. Characteristics of OfPPO and Phylogenetic Analysis
4.5. OfPPO Expression in O. formosanus Using qPCR
4.6. In Vitro Synthesis of dsOfPPOs (dsOfPPO1 and dsOfPPO2)
4.7. RNAi Efficiency Evaluation after Treatment with dsRNA
4.8. The Role of OfPPOs in Resisting SM1 and Bt Infection
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rust, M.K.; Su, N.Y. Managing social insects of urban importance. Annu. Rev. Entomol. 2012, 57, 355–375. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, F.; Fouad, H.; Liang, S.Y.; Hu, Y.; Mo, J.C. Termites and Chinese agricultural system: Applications and advances in integrated termite management and chemical control. Insect. Sci. 2021, 28, 2–20. [Google Scholar] [CrossRef] [PubMed]
- Husseneder, C.; Garner, S.P.; Huang, Q.; Booth, W.; Vargo, E.L. Characterization of microsatellites for population genetic analyses of the fungus-growing termite Odontotermes formosanus (Isoptera: Termitidae). Environ. Entomol. 2013, 42, 1092–1099. [Google Scholar] [CrossRef] [PubMed]
- Appel, A.G.; Hu, X.P.; Zhou, J.; Qin, Z.; Zhu, H.; Chang, X.; Wang, Z.; Liu, X.; Liu, M. Observations of the biology and ecology of the black-winged termite, Odontotermes formosanus Shiraki (Termitidae: Isoptera), in camphor, Cinnamomum camphora (L.) (Lauraceae). Psyche 2012, 2012, 123102. [Google Scholar]
- Huang, Q.Y.; Lei, C.L.; Xue, D. Field evaluation of a fipronil bait against subterranean termite Odontotermes formosanus (Isoptera: Termitidae). J. Econ. Entomol. 2006, 99, 455–461. [Google Scholar] [CrossRef]
- Huang, Q.; Sun, P.; Zhou, X.; Lei, C. Characterization of head transcriptome and analysis of gene expression involved in caste differentiation and aggression in Odontotermes formosanus (Shiraki). PLoS ONE 2012, 7, e50383. [Google Scholar] [CrossRef][Green Version]
- Tudi, M.; Daniel, R.H.; Wang, L.; Lyu, J.; Sadler, R.; Connell, D.; Chu, C.; Phung, D.T. Agriculture Development, Pesticide Application and Its Impact on the Environment. Int. J. Environ. Res. Public Health 2021, 18, 1112. [Google Scholar] [CrossRef]
- Aggarwal, C.; Paul, S.; Nain, V.; Tripathi, V.; Paul, B.; Aslam, K.M. Comparative response of Spodoptera litura challenged per os with Serratia marcescens strains differing in virulence. J. Invertebr. Pathol. 2021, 183, 107562. [Google Scholar] [CrossRef]
- Wang, Z.; Feng, K.; Tang, F.; Xu, M. Activation of the host immune response in Hyphantria cunea (Drury) (Lepidoptera: Noctuidae) induced by Serratia marcescens Bizio. Insects 2021, 12, 983. [Google Scholar] [CrossRef]
- Niu, H.; Wang, N.; Liu, B.; Xiao, L.; Wang, L.; Guo, H. Synergistic and additive interactions of Serratia marcescens S-JS1 to the chemical insecticides for controlling Nilaparvata lugens (Hemiptera: Delphacidae). J. Econ. Entomol. 2018, 111, 823–828. [Google Scholar] [CrossRef]
- Wang, C.; Henderson, G.; Gautam, B.K. Lufenuron suppresses the resistance of Formosan subterranean termites (Isoptera: Rhinotermitidae) to entomopathogenic bacteria. J. Econ. Entomol. 2013, 106, 1812–1818. [Google Scholar] [CrossRef] [PubMed]
- Wright, M.S.; Cornelius, M.L. Mortality and repellent effects of microbial pathogens on Coptotermes formosanus (Isoptera: Rhinotermitidae). BMC Microbiol. 2012, 12, 291. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.; Luo, J.; Feng, K.; Lu, X.; Tang, F. Termite-killing components in Serratia marcescens (SM1). J. For. Res. 2021, 32, 1739–1744. [Google Scholar] [CrossRef]
- Hoffmann, J.A. Innate immunity of insects. Curr. Opin. Immunol. 1995, 7, 4–10. [Google Scholar] [CrossRef]
- Hoffmann, J.A. The immune response of Drosophila. Nature 2003, 426, 33–38. [Google Scholar] [CrossRef]
- Ferrandon, D.; Imler, J.L.; Hetru, C.; Hoffmann, J.A. The Drosophila systemic immune response: Sensing and signalling during bacterial and fungal infections. Nat. Rev. Immunol. 2007, 7, 862–874. [Google Scholar] [CrossRef]
- Hillyer, J.F. Insect immunology and hematopoiesis. Dev. Comp. Immunol. 2016, 58, 102–118. [Google Scholar] [CrossRef]
- Caccia, S.; Astarita, F.; Barra, E.; Di Lelio, I.; Varricchio, P.; Pennacchio, F. Enhancement of Bacillus thuringiensis toxicity by feeding Spodoptera littoralis larvae with bacteria expressing immune suppressive dsRNA. J. Pest Sci. 2020, 93, 303–314. [Google Scholar] [CrossRef]
- Fan, Y.; Abbas, M.; Liu, X.; Wang, Y.; Song, H.; Li, T.; Ma, E.; Zhu, K.Y.; Zhang, J. Increased RNAi efficiency by dsEGFP-induced up-regulation of two core RNAi pathway genes (OfDicer2 and OfAgo2) in the Asian Corn Borer (Ostrinia furnacalis). Insects 2022, 13, 274. [Google Scholar] [CrossRef]
- Moreira-Pinto, C.E.; Coelho, R.R.; Leite, A.; Silveira, D.A.; de Souza, D.A.; Lopes, R.B.; Macedo, L.; Silva, M.; Ribeiro, T.P.; Morgante, C.V.; et al. Increasing Anthonomus grandis susceptibility to Metarhizium anisopliae through RNAi-induced AgraRelish knockdown: A perspective to combine biocontrol and biotechnology. Pest. Manag. Sci. 2021, 77, 4054–4063. [Google Scholar] [CrossRef]
- Maximo, W.; Howell, J.L.; Mogilicherla, K.; Basij, M.; Chereddy, S.; Palli, S.R. Inhibitor of apoptosis is an effective target gene for RNAi-mediated control of Colorado potato beetle, Leptinotarsa decemlineata. Arch. Insect. Biochem. Physiol. 2020, 104, e21685. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Keyhani, N.O.; Zhang, H.; Cai, K.; Xia, Y. Inhibitor of apoptosis-1 gene as a potential target for pest control and its involvement in immune regulation during fungal infection. Pest. Manag. Sci. 2020, 76, 1831–1840. [Google Scholar] [CrossRef] [PubMed]
- Bingsohn, L.; Knorr, E.; Billion, A.; Narva, K.E.; Vilcinskas, A. Knockdown of genes in the Toll pathway reveals new lethal RNA interference targets for insect pest control. Insect. Mol. Biol. 2017, 26, 92–102. [Google Scholar] [CrossRef]
- Zhao, P.; Li, J.; Wang, Y.; Jiang, H. Broad-spectrum antimicrobial activity of the reactive compounds generated in vitro by Manduca sexta phenoloxidase. Insect Biochem. Mol. Biol. 2007, 37, 952–959. [Google Scholar] [CrossRef] [PubMed]
- Kanost, M.R.; Jiang, H.; Yu, X.Q. Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol. Rev. 2004, 198, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Lavine, M.D.; Strand, M.R. Surface characteristics of foreign targets that elicit an encapsulation response by the moth Pseudoplusia includens. J. Insect. Physiol. 2001, 47, 965–974. [Google Scholar] [CrossRef]
- Cerenius, L.; Lee, B.L.; Soderhall, K. The proPO-system: Pros and cons for its role in invertebrate immunity. Trends Immunol. 2008, 29, 263–271. [Google Scholar] [CrossRef]
- Prabu, S.; Jing, D.; Shabbir, M.Z.; Yuan, W.; Wang, Z.; He, K. Contribution of phenoloxidase activation mechanism to Bt insecticidal protein resistance in Asian corn borer. Int. J. Biol. Macromol. 2020, 153, 88–99. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, H.; Cheng, Y.; An, C.; Chu, Y.; Raikhel, A.S.; Zou, Z. Activation of Aedes aegypti prophenoloxidase-3 and its role in the immune response against entomopathogenic fungi. Insect Mol. Biol. 2017, 26, 552–563. [Google Scholar] [CrossRef]
- Xu, L.; Ma, L.; Wang, W.; Li, L.; Lu, Z. Phenoloxidases are required for the pea aphid’s defence against bacterial and fungal infection. Insect Mol. Biol. 2019, 28, 176–186. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, W.; Yuan, C.; Lu, Y.; Yang, B.; Wang, C.Y.; Zhang, P.; Dobens, L.; Zou, Z.; Wang, C.; et al. Prophenoloxidase-mediated ex vivo immunity to delay fungal infection after insect ecdysis. Front. Immunol. 2017, 8, 1445. [Google Scholar] [CrossRef] [PubMed]
- Lacey, L.A.; Grzywacz, D.; Shapiro-Ilan, D.I.; Frutos, R.; Brownbridge, M.; Goettel, M.S. Insect pathogens as biological control agents: Back to the future. J. Invertebr. Pathol. 2015, 132, 1–41. [Google Scholar] [CrossRef] [PubMed]
- Chouvenc, T.; Su, N.; Grace, J.K. Fifty years of attempted biological control of termites—Analysis of a failure. Biol. Control 2011, 59, 69–82. [Google Scholar] [CrossRef]
- Rodriguez-Andres, J.; Rani, S.; Varjak, M.; Chase-Topping, M.E.; Beck, M.H.; Ferguson, M.C.; Schnettler, E.; Fragkoudis, R.; Barry, G.; Merits, A.; et al. Phenoloxidase activity acts as a mosquito innate immune response against infection with Semliki Forest virus. PLoS Pathog. 2012, 8, e1002977. [Google Scholar] [CrossRef] [PubMed]
- Yokoi, K.; Hayakawa, Y.; Kato, D.; Minakuchi, C.; Tanaka, T.; Ochiai, M.; Kamiya, K.; Miura, K. Prophenoloxidase genes and antimicrobial host defense of the model beetle, Tribolium castaneum. J. Invertebr. Pathol. 2015, 132, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Yassine, H.; Kamareddine, L.; Osta, M.A. The mosquito melanization response is implicated in defense against the entomopathogenic fungus Beauveria bassiana. PLoS Pathog. 2012, 8, e1003029. [Google Scholar] [CrossRef]
- Liu, L.; Li, G.; Sun, P.; Lei, C.; Huang, Q. Experimental verification and molecular basis of active immunization against fungal pathogens in termites. Sci. Rep. 2015, 5, 15106. [Google Scholar] [CrossRef]
- Kuraishi, T.; Hori, A.; Kurata, S. Host-microbe interactions in the gut of Drosophila melanogaster. Front. Physiol. 2013, 4, 375. [Google Scholar] [CrossRef]
- Bidla, G.; Hauling, T.; Dushay, M.S.; Theopold, U. Activation of insect phenoloxidase after injury: Endogenous versus foreign elicitors. J. Innate. Immun. 2009, 1, 301–308. [Google Scholar] [CrossRef]
- Karlsson, C.; Korayem, A.M.; Scherfer, C.; Loseva, O.; Dushay, M.S.; Theopold, U. Proteomic analysis of the Drosophila larval hemolymph clot. J. Biol. Chem. 2004, 279, 52033–52041. [Google Scholar] [CrossRef]
- Zou, Z.; Shin, S.W.; Alvarez, K.S.; Kokoza, V.; Raikhel, A.S. Distinct melanization pathways in the mosquito Aedes aegypti. Immunity 2010, 32, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Shin, S.W.; Alvarez, K.S.; Bian, G.; Kokoza, V.; Raikhel, A.S. Mosquito RUNX4 in the immune regulation of PPO gene expression and its effect on avian malaria parasite infection. Proc. Natl. Acad. Sci. USA 2008, 105, 18454–18459. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Q.; Song, X.H.; Wang, M.; Wang, S.; Huang, G.H. Melanization induced by Heliothis virescens ascovirus 3h promotes viral replication. Insect Sci. 2021, 28, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Tartar, A.; Wheeler, M.M.; Zhou, X.; Coy, M.R.; Boucias, D.G.; Scharf, M.E. Parallel metatranscriptome analyses of host and symbiont gene expression in the gut of the termite Reticulitermes flavipes. Biotechnol. Biofuels 2009, 2, 25. [Google Scholar] [CrossRef]
- Rosengaus, R.B.; Reichheld, J.L. Phenoloxidase activity in the infraorder Isoptera: Unraveling life-history correlates of immune investment. Naturwissenschaften 2016, 103, 14. [Google Scholar] [CrossRef]
- Yang, N.; Ding, T.; Chu, D. Silencing of the prophenoloxidase gene BtPPO1 increased the ability of acquisition and retention of Tomato chlorosis virus by Bemisia tabaci. Int. J. Mol. Sci. 2022, 23, 6541. [Google Scholar] [CrossRef]
- Lourenco, A.P.; Zufelato, M.S.; Bitondi, M.M.; Simoes, Z.L. Molecular characterization of a cDNA encoding prophenoloxidase and its expression in Apis mellifera. Insect Biochem. Mol. Biol. 2005, 35, 541–552. [Google Scholar] [CrossRef]
- Park, D.S.; Shin, S.W.; Kim, M.G.; Park, S.S.; Lee, W.J.; Brey, P.T.; Park, H.Y. Isolation and characterization of the cDNA encoding the prophenoloxidase of fall webworm, Hyphantria cunea. Insect Biochem. Mol. Biol. 1997, 27, 983–992. [Google Scholar] [CrossRef]
- Muller, H.M.; Dimopoulos, G.; Blass, C.; Kafatos, F.C. A hemocyte-like cell line established from the malaria vector Anopheles gambiae expresses six prophenoloxidase genes. J. Biol. Chem. 1999, 274, 11727–11735. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Li, L.; Xu, Q.; Zou, Q.; Tang, B.; Wang, S. Gene cloning and expression patterns of two prophenoloxidases from Catantops pinguis (Orthoptera: Catantopidae). Bull. Entomol. Res. 2013, 103, 393–405. [Google Scholar] [CrossRef] [PubMed]
- Asano, T.; Ashida, M. Cuticular pro-phenoloxidase of the silkworm, Bombyx mori. Purification and demonstration of its transport from hemolymph. J. Biol. Chem. 2001, 276, 11100–11112. [Google Scholar] [CrossRef] [PubMed]
- Khajuria, C.; Velez, A.M.; Rangasamy, M.; Wang, H.; Fishilevich, E.; Frey, M.L.; Carneiro, N.P.; Gandra, P.; Narva, K.E.; Siegfried, B.D. Parental RNA interference of genes involved in embryonic development of the western corn rootworm, Diabrotica virgifera virgifera LeConte. Insect Biochem. Mol. Biol. 2015, 63, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Terenius, O.; Papanicolaou, A.; Garbutt, J.S.; Eleftherianos, I.; Huvenne, H.; Kanginakudru, S.; Albrechtsen, M.; An, C.; Aymeric, J.L.; Barthel, A.; et al. RNA interference in Lepidoptera: An overview of successful and unsuccessful studies and implications for experimental design. J. Insect Physiol. 2011, 57, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Caccia, S.; Di Lelio, I.; La Storia, A.; Marinelli, A.; Varricchio, P.; Franzetti, E.; Banyuls, N.; Tettamanti, G.; Casartelli, M.; Giordana, B.; et al. Midgut microbiota and host immunocompetence underlie Bacillus thuringiensis killing mechanism. Proc. Natl. Acad. Sci. USA 2016, 113, 9486–9491. [Google Scholar] [CrossRef]
- Joga, M.R.; Zotti, M.J.; Smagghe, G.; Christiaens, O. RNAi Efficiency, systemic properties, and novel delivery methods for pest insect control: What we know so far. Front. Physiol. 2016, 7, 553. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- BENJAMIN, N.D.; Montgomery, M.W. Polyphenoloxidase of Royal Ann cherries: Purification and characterization. J. Food Sci. 1973, 38, 799–806. [Google Scholar] [CrossRef]
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.; Moorman, A.F. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Muller, P.Y.; Janovjak, H.; Miserez, A.R.; Dobbie, Z. Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques 2002, 32, 1372–1374, 1376, 1378–1379. [Google Scholar]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef]
- Newmark, P.A.; Reddien, P.W.; Cebria, F.; Sanchez, A.A. Ingestion of bacterially expressed double-stranded RNA inhibits gene expression in planarians. Proc. Natl. Acad. Sci. USA 2003, 100 (Suppl. 1), 11861–11865. [Google Scholar] [CrossRef] [PubMed]
- Timmons, L.; Court, D.L.; Fire, A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 2001, 263, 103–112. [Google Scholar] [CrossRef] [PubMed]



| Name | OfPPO | CfPO2 | ZnPO2 | BgPO1 | CsPO1 | TcPPO2 |
|---|---|---|---|---|---|---|
| OfPPO | 100.0 | 92.80 | 87.90 | 79.34 | 60.84 | 63.40 |
| CfPO2 | 100.0 | 89.05 | 80.56 | 60.12 | 62.96 | |
| ZnPO2 | 100.0 | 80.56 | 59.68 | 61.93 | ||
| BgPO1 | 100.0 | 59.82 | 63.70 | |||
| CsPO1 | 100 | 76.83 | ||||
| TcPPO2 | 100 |
| Primer Names | Forward Primer Sequences (5′-3′) | Reverse Primer Sequences (5′-3′) | Annealing Temperature | Experiments |
|---|---|---|---|---|
| OfPPO | ATGTCAAGACCAGCACAATC | TCATGAGAGCCGGTTTG | 52 °C | Full-length amplification |
| dsOfPPO1 | CGAGCTCGGTACAACTTCGAACGGCTG | TCCCCCGGGGCTTGCCTCCATCAGGTTAC | 57 °C | RNAi |
| dsOfPPO2 | CGAGCTCTCCAACAACGATGATGATAAGATA | TCCCCCGGGGTTTGGCTCATCTCTACCCTG | 57 °C | |
| dsGFP | ATCGGAGCTCTAGTTGAACGGATCCATCTTCA | CCCAAGCTTAGAACTTTTCACTGGA | 54 °C | |
| OfPPO | GTTGATCTGTCCCGAGGTATTG | GCTGTTCTCAGCCTGAATCTTA | 60 °C | qPCR |
| GAPDH | TCGTATTGGCCGTCTTGTGC | AGCGACCATGGGTGGAATCAT | 60 °C | |
| RPS18 | ATGGCAAACCCCCGTCAGTA | CATACCACGATGCGCACGAA | 60 °C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Luo, J.; Feng, K.; Zhou, Y.; Tang, F. Prophenoloxidase of Odontotermes formosanus (Shiraki) (Blattodea: Termitidae) Is a Key Gene in Melanization and Has a Defensive Role during Bacterial Infection. Int. J. Mol. Sci. 2023, 24, 406. https://doi.org/10.3390/ijms24010406
Wang Z, Luo J, Feng K, Zhou Y, Tang F. Prophenoloxidase of Odontotermes formosanus (Shiraki) (Blattodea: Termitidae) Is a Key Gene in Melanization and Has a Defensive Role during Bacterial Infection. International Journal of Molecular Sciences. 2023; 24(1):406. https://doi.org/10.3390/ijms24010406
Chicago/Turabian StyleWang, Zhiqiang, Jian Luo, Kai Feng, Yujingyun Zhou, and Fang Tang. 2023. "Prophenoloxidase of Odontotermes formosanus (Shiraki) (Blattodea: Termitidae) Is a Key Gene in Melanization and Has a Defensive Role during Bacterial Infection" International Journal of Molecular Sciences 24, no. 1: 406. https://doi.org/10.3390/ijms24010406
APA StyleWang, Z., Luo, J., Feng, K., Zhou, Y., & Tang, F. (2023). Prophenoloxidase of Odontotermes formosanus (Shiraki) (Blattodea: Termitidae) Is a Key Gene in Melanization and Has a Defensive Role during Bacterial Infection. International Journal of Molecular Sciences, 24(1), 406. https://doi.org/10.3390/ijms24010406





