Preclinical and Clinical Applications of Metabolomics and Proteomics in Glioblastoma Research
Abstract
1. Introduction
1.1. Barriers Associated with Current Therapeutic Strategies
1.2. Metabolomics and Proteomics
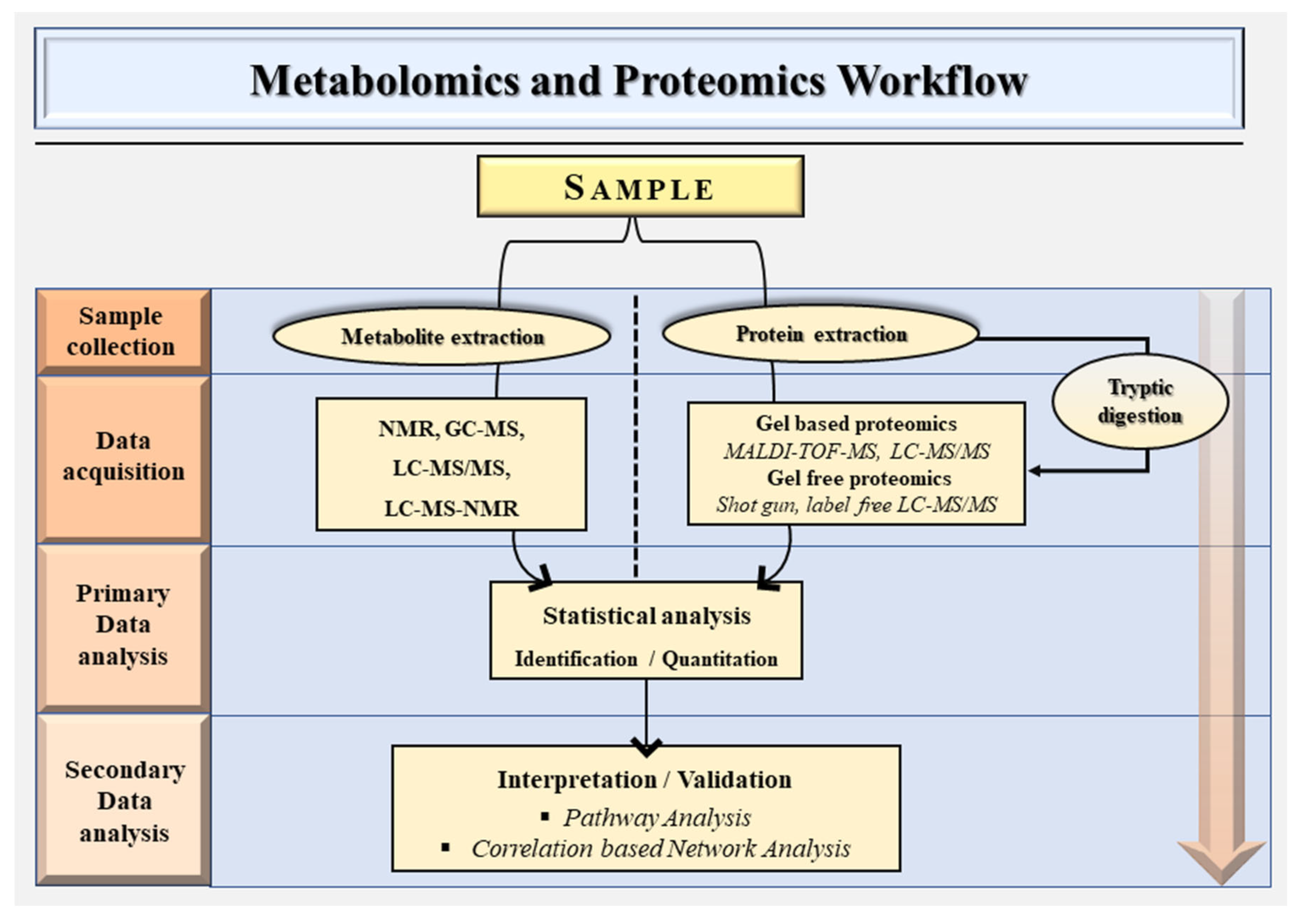
2. Metabolomics and Proteomics Workflow
2.1. Targeted Metabolomics
2.2. Untargeted Metabolomics
| Technique | Characteristics | Advantages | Disadvantages | References |
|---|---|---|---|---|
| NMR | Nuclear magnetic resonance (NMR). 1D/2D 1H NMR usually employed in metabolomics studies. | Brief analysis time. Absolute metabolite quantification. High reproducibility. Intrinsically a quantitative technique since the signal strength is directly correlated with metabolite concentrations. | Its relatively low sensitivity can miss low-abundance metabolites. Signal overlapping due to the lack of a prior separation system. Typically, nonselective analysis is done with NMR. Major complications are presented by peak overlaps from numerous measured metabolites. | [28,29] |
| GC-MS | Gas chromatography–mass spectrometry (GC-MS). Volatile metabolites or those rendered volatile can be analyzed by GC-MS. | Relatively cheap. Better stability and separation efficiency. Being able to monitor highly hydrophobic and volatile compounds not ionized in the ESI source of LCMS/MS. | Unable to directly analyze nonvolatile, polar, or thermally labile drugs. Many biological substances are either too big or too polar to be examined using this method. | [30,31,32,33] |
| LC–MS/MS | liquid chromatography–mass spectrometry (LC–MS). High sensitivity. Biofluids like urine can be introduced directly into the LC system. Multidimensional can study the metabolome and lipidome in the same run. | Multidimensional—can study the metabolome and lipidome in the same run. High sensitivity. Biofluids like urine can be introduced directly into the LC system. | Greater operational costs. Lower concentration sensitivity. More restricted sample throughput. | [34] |
| LC–MS–NMR | Liquid chromatography-(LC)–mass spectrometry (MS)–nuclear magnetic resonance (NMR) Good spectral resolution and excellent metabolite identification ability. | Good spectral resolution. Excellent metabolite identification ability. | Different rates of exchange with deuterium are possible for analytes having exchangeable or “active” hydrogens since NMR uses deuterated solvents. The analyst should be aware of this possibility since it might cause a number of clustered molecular ions. The NMR component has high sample mass requirements. | [35,36,37] |
2.3. Proteomics Workflow
3. Clinical and Preclinical Applications of Metabolomics and Proteomics in Glioblastoma Research
3.1. Metabolomics to Elucidate the Molecular Mechanisms
3.2. Metabolomics to Unravel the Resistance Mechanisms
3.3. Metabolomics and GB Characterization
3.4. Metabolic Phenotyping in Diagnostics
3.5. Pharmacometabolomics
3.6. Proteomics to Elucidate the Molecular Mechanisms
3.7. Proteomics to Unravel the Resistance Mechanisms
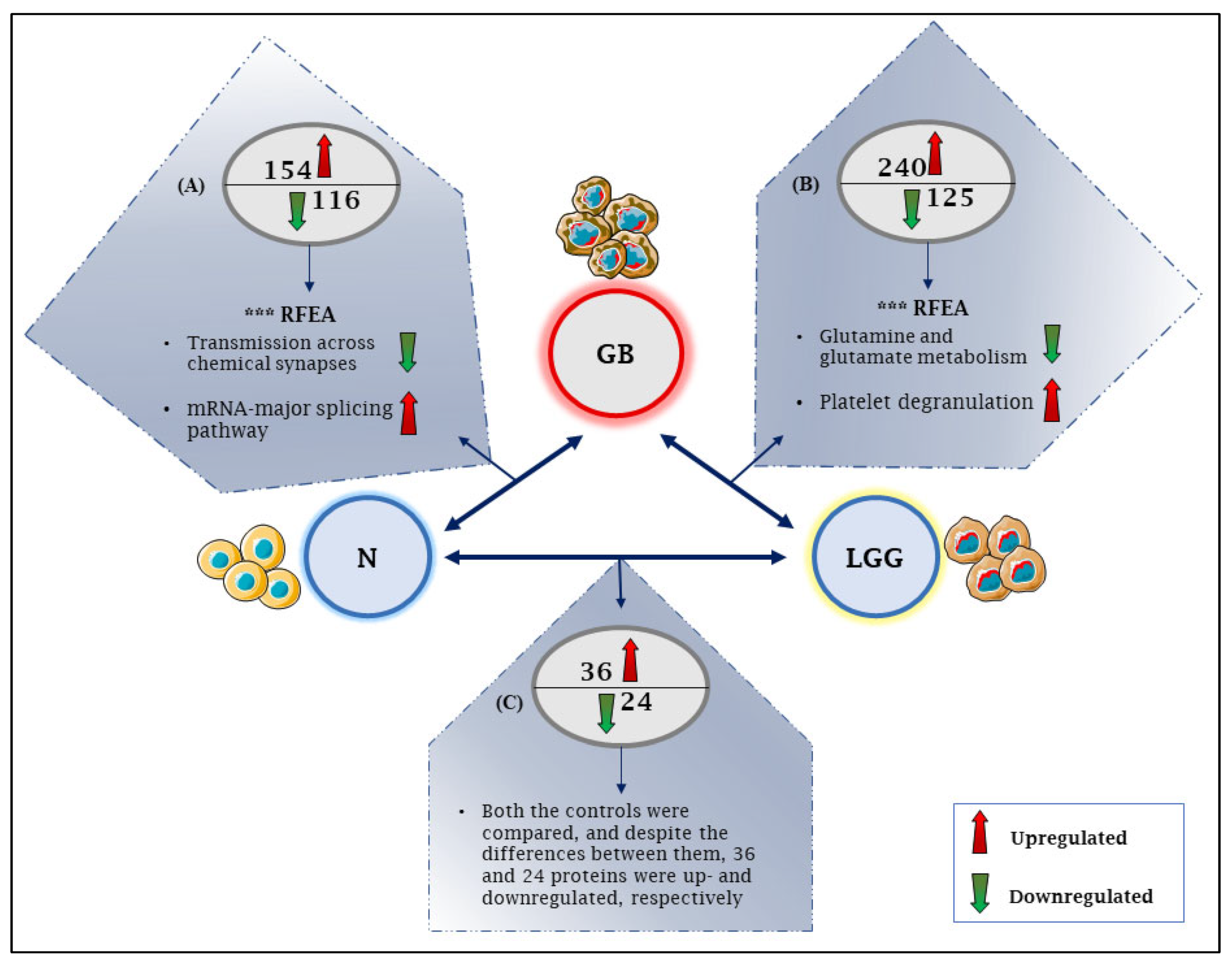
3.8. Proteomics and GB Characterization
3.9. Proteomics for Identifying Biomarkers
3.10. Pharmacoproteomics
4. Integrating Proteomics and Metabolomics
5. Limitations
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2014–2018. Neuro Oncol. 2021, 23, iii1–iii105. [Google Scholar] [CrossRef] [PubMed]
- Chaichana, K.L.; Jusue-Torres, I.; Navarro-Ramirez, R.; Raza, S.M.; Pascual-Gallego, M.; Ibrahim, A.; Hernandez-Hermann, M.; Gomez, L.; Ye, X.; Weingart, J.D.; et al. Establishing Percent Resection and Residual Volume Thresholds Affecting Survival and Recurrence for Patients with Newly Diagnosed Intracranial Glioblastoma. Neuro Oncol. 2014, 16, 113–122. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Cote, D.J.; Ascha, M.; Kruchko, C.; Barnholtz-Sloan, J.S. Adult Glioma Incidence and Survival by Race or Ethnicity in the United States From 2000 to 2014. JAMA Oncol. 2018, 4, 1254–1262. [Google Scholar] [CrossRef] [PubMed]
- Dillekås, H.; Rogers, M.S.; Straume, O. Are 90% of Deaths from Cancer Caused by Metastases? Cancer Med. 2019, 8, 5574–5576. [Google Scholar] [CrossRef] [PubMed]
- Cote, D.J.; Ostrom, Q.T. Epidemiology and Etiology of Glioblastoma. In Precision Molecular Pathology of Glioblastoma; Otero, J.J., Becker, A.P., Eds.; Molecular Pathology Library; Springer International Publishing: Cham, Switzerland, 2021; p. 5. ISBN 978-3-030-69169-1. [Google Scholar]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- van den Bent, M.J. Interobserver Variation of the Histopathological Diagnosis in Clinical Trials on Glioma: A Clinician’s Perspective. Acta Neuropathol. 2010, 120, 297–304. [Google Scholar] [CrossRef]
- Erasimus, H.; Gobin, M.; Niclou, S.; Van Dyck, E. DNA Repair Mechanisms and Their Clinical Impact in Glioblastoma. Mutat. Res. Rev. Mutat. Res. 2016, 769, 19–35. [Google Scholar] [CrossRef]
- Arvanitis, C.D.; Ferraro, G.B.; Jain, R.K. The Blood–Brain Barrier and Blood–Tumour Barrier in Brain Tumours and Metastases. Nat. Rev. Cancer 2020, 20, 26–41. [Google Scholar] [CrossRef]
- Ge, X.; Pan, M.-H.; Wang, L.; Li, W.; Jiang, C.; He, J.; Abouzid, K.; Liu, L.-Z.; Shi, Z.; Jiang, B.-H. Hypoxia-Mediated Mitochondria Apoptosis Inhibition Induces Temozolomide Treatment Resistance through MiR-26a/Bad/Bax Axis. Cell Death Dis. 2018, 9, 1128. [Google Scholar] [CrossRef]
- Yin, J.; Ge, X.; Shi, Z.; Yu, C.; Lu, C.; Wei, Y.; Zeng, A.; Wang, X.; Yan, W.; Zhang, J.; et al. Extracellular Vesicles Derived from Hypoxic Glioma Stem-like Cells Confer Temozolomide Resistance on Glioblastoma by Delivering MiR-30b-3p. Theranostics 2021, 11, 1763–1779. [Google Scholar] [CrossRef] [PubMed]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does It Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-Y.; Kim, Y.-J.; Lee, S.; Park, J.-H. The Critical Role of ERK in Death Resistance and Invasiveness of Hypoxia-Selected Glioblastoma Cells. BMC Cancer 2009, 9, 27. [Google Scholar] [CrossRef]
- Gu, H.; Gowda, G.N.; Raftery, D. Metabolic Profiling: Are We En Route to Better Diagnostic Tests for Cancer? Future Oncol. 2012, 8, 1207–1210. [Google Scholar] [CrossRef] [PubMed]
- Clish, C.B. Metabolomics: An Emerging but Powerful Tool for Precision Medicine. Cold Spring Harb. Mol. Case Stud. 2015, 1, a000588. [Google Scholar] [CrossRef] [PubMed]
- Kuehnbaum, N.L.; Britz-McKibbin, P. New Advances in Separation Science for Metabolomics: Resolving Chemical Diversity in a Post-Genomic Era. Chem. Rev. 2013, 113, 2437–2468. [Google Scholar] [CrossRef] [PubMed]
- Van der Greef, J.; van Wietmarschen, H.; van Ommen, B.; Verheij, E. Looking Back into the Future: 30 Years of Metabolomics at TNO. Mass. Spec. Rev. 2013, 32, 399–415. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0—The Human Metabolome Database in 2013. Nucleic Acids Res. 2013, 41, D801–D807. [Google Scholar] [CrossRef] [PubMed]
- Armitage, E.G.; Barbas, C. Metabolomics in Cancer Biomarker Discovery: Current Trends and Future Perspectives. J. Pharm. Biomed. Anal. 2014, 87, 1–11. [Google Scholar] [CrossRef]
- Contrepois, K.; Liang, L.; Snyder, M. Can Metabolic Profiles Be Used as a Phenotypic Readout of the Genome to Enhance Precision Medicine? Clin. Chem. 2016, 62, 676–678. [Google Scholar] [CrossRef]
- Uzozie, A.C.; Aebersold, R. Advancing Translational Research and Precision Medicine with Targeted Proteomics. J. Proteom. 2018, 189, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Marziali, G.; Signore, M.; Buccarelli, M.; Grande, S.; Palma, A.; Biffoni, M.; Rosi, A.; D’Alessandris, Q.G.; Martini, M.; Larocca, L.M.; et al. Metabolic/Proteomic Signature Defines Two Glioblastoma Subtypes With Different Clinical Outcome. Sci. Rep. 2016, 6, 21557. [Google Scholar] [CrossRef] [PubMed]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Innovation: Metabolomics: The Apogee of the Omics Trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Lelli, V.; Belardo, A.; Maria Timperio, A. From Targeted Quantification to Untargeted Metabolomics. In Metabolomics—Methodology and Applications in Medical Sciences and Life Sciences; Zhan, X., Ed.; IntechOpen: London, UK, 2021; ISBN 978-1-83969-083-9. [Google Scholar]
- Buragohain, L.; Ghosh, M.; Kumar, R.; Dahiya, S.; Malik, Y.S.; Prasad, M.; Prasad, M. Application of Proteomics and Metabolomics in Disease Diagnosis. In Advances in Animal Disease Diagnosis, 1st ed.; Gahlawat, S.K., Maan, S., Eds.; CRC Press: Boca Raton, FL, USA, 2021; pp. 79–102. ISBN 978-1-00-308028-2. [Google Scholar]
- Gowda, G.A.N.; Zhang, S.; Gu, H.; Asiago, V.; Shanaiah, N.; Raftery, D. Metabolomics-Based Methods for Early Disease Diagnostics. Expert Rev. Mol. Diagn. 2008, 8, 617–633. [Google Scholar] [CrossRef]
- Leenders, J.; Frédérich, M.; de Tullio, P. Nuclear Magnetic Resonance: A Key Metabolomics Platform in the Drug Discovery Process. Drug Discov. Today Technol. 2015, 13, 39–46. [Google Scholar] [CrossRef]
- Emwas, A.-H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.A.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR Spectroscopy for Metabolomics Research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef]
- Smart, K.F.; Aggio, R.B.M.; Van Houtte, J.R.; Villas-Bôas, S.G. Analytical Platform for Metabolome Analysis of Microbial Cells Using Methyl Chloroformate Derivatization Followed by Gas Chromatography-Mass Spectrometry. Nat. Protoc. 2010, 5, 1709–1729. [Google Scholar] [CrossRef]
- Rockwood, A.L.; Kushnir, M.M.; Clarke, N.J. Mass Spectrometry. In Principles and Applications of Clinical Mass Spectrometry; Elsevier: Amsterdam, The Netherlands, 2018; pp. 33–65. ISBN 978-0-12-816063-3. [Google Scholar]
- Lynch, K.L. Toxicology: Liquid Chromatography Mass Spectrometry. In Mass Spectrometry for the Clinical Laboratory; Elsevier: Amsterdam, The Netherlands, 2017; pp. 109–130. ISBN 978-0-12-800871-3. [Google Scholar]
- He, P.; Aga, D.S. Comparison of GC-MS/MS and LC-MS/MS for the Analysis of Hormones and Pesticides in Surface Waters: Advantages and Pitfalls. Anal. Methods 2019, 11, 1436–1448. [Google Scholar] [CrossRef]
- Karpievitch, Y.V.; Polpitiya, A.D.; Anderson, G.A.; Smith, R.D.; Dabney, A.R. Liquid Chromatography Mass Spectrometry-Based Proteomics: Biological and Technological Aspects. Ann. Appl. Stat. 2010, 4, 1797–1823. [Google Scholar] [CrossRef]
- Walker, L.R.; Hoyt, D.W.; Walker, S.M.; Ward, J.K.; Nicora, C.D.; Bingol, K. Unambiguous Metabolite Identification in High-Throughput Metabolomics by Hybrid 1D 1 H NMR/ESI MS1 Approach. Magn. Reson. Chem. 2016, 54, 998–1003. [Google Scholar] [CrossRef]
- Silva Elipe, M.V. Advantages and Disadvantages of Nuclear Magnetic Resonance Spectroscopy as a Hyphenated Technique. Anal. Chim. Acta 2003, 497, 1–25. [Google Scholar] [CrossRef]
- Gathungu, R.M.; Kautz, R.; Kristal, B.S.; Bird, S.S.; Vouros, P. The Integration of LC-MS and NMR for the Analysis of Low Molecular Weight Trace Analytes in Complex Matrices. Mass. Spec. Rev. 2020, 39, 35–54. [Google Scholar] [CrossRef] [PubMed]
- Minakshi, P.; Kumar, R.; Ghosh, M.; Saini, H.M.; Ranjan, K.; Brar, B.; Prasad, G. Single-Cell Proteomics: Technology and Applications. In Single-Cell Omics; Elsevier: Amsterdam, The Netherlands, 2019; pp. 283–318. ISBN 978-0-12-814919-5. [Google Scholar]
- Encyclopedia of Spectroscopy and Spectrometry. Volume 4: S-Z. Index, 3rd ed.; Lindon, J.C., Tranter, G.E., Koppenaal, D.W., Eds.; Elsevier AP: Amsterdam, The Netherlands; Academic Press: Boston, FL, USA; Heidelberg, Germany, 2017; ISBN 978-0-12-803224-4. [Google Scholar]
- Catherman, A.D.; Skinner, O.S.; Kelleher, N.L. Top Down Proteomics: Facts and Perspectives. Biochem. Biophys. Res. Commun. 2014, 445, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Soboleva, A.; Schmidt, R.; Vikhnina, M.; Grishina, T.; Frolov, A. Maillard Proteomics: Opening New Pages. Int. J. Mol. Sci. 2017, 18, 2677. [Google Scholar] [CrossRef] [PubMed]
- Dubin, R.F.; Rhee, E.P. Proteomics and Metabolomics in Kidney Disease, Including Insights into Etiology, Treatment, and Prevention. Clin. J. Am. Soc. Nephrol. 2020, 15, 404–411. [Google Scholar] [CrossRef]
- Berger, M.F.; Mardis, E.R. The Emerging Clinical Relevance of Genomics in Cancer Medicine. Nat. Rev. Clin. Oncol. 2018, 15, 353–365. [Google Scholar] [CrossRef]
- Bernini, A.; Masoodi, M.; Solari, D.; Miroz, J.-P.; Carteron, L.; Christinat, N.; Morelli, P.; Beaumont, M.; Abed-Maillard, S.; Hartweg, M.; et al. Modulation of Cerebral Ketone Metabolism Following Traumatic Brain Injury in Humans. J. Cereb. Blood Flow Metab. 2020, 40, 177–186. [Google Scholar] [CrossRef]
- Rusek, M.; Pluta, R.; Ułamek-Kozioł, M.; Czuczwar, S.J. Ketogenic Diet in Alzheimer’s Disease. Int. J. Mol. Sci. 2019, 20, 3892. [Google Scholar] [CrossRef]
- Li, R.-J.; Liu, Y.; Liu, H.-Q.; Li, J. Ketogenic Diets and Protective Mechanisms in Epilepsy, Metabolic Disorders, Cancer, Neuronal Loss, and Muscle and Nerve Degeneration. J. Food Biochem. 2020, 44, e13140. [Google Scholar] [CrossRef]
- Weber, D.D.; Aminzadeh-Gohari, S.; Tulipan, J.; Catalano, L.; Feichtinger, R.G.; Kofler, B. Ketogenic Diet in the Treatment of Cancer—Where Do We Stand? Mol. Metab. 2020, 33, 102–121. [Google Scholar] [CrossRef]
- Damiano, F.; De Benedetto, G.E.; Longo, S.; Giannotti, L.; Fico, D.; Siculella, L.; Giudetti, A.M. Decanoic Acid and Not Octanoic Acid Stimulates Fatty Acid Synthesis in U87MG Glioblastoma Cells: A Metabolomics Study. Front. Neurosci. 2020, 14, 783. [Google Scholar] [CrossRef]
- Schadinger, S.E.; Bucher, N.L.R.; Schreiber, B.M.; Farmer, S.R. PPARgamma2 Regulates Lipogenesis and Lipid Accumulation in Steatotic Hepatocytes. Am. J. Physiol. Endocrinol. Metab. 2005, 288, E1195–E1205. [Google Scholar] [CrossRef] [PubMed]
- Leaver, H.A.; Bell, H.S.; Rizzo, M.T.; Ironside, J.W.; Gregor, A.; Wharton, S.B.; Whittle, I.R. Antitumour and Pro-Apoptotic Actions of Highly Unsaturated Fatty Acids in Glioma. Prostaglandins Leukot. Essent. Fat. Acids 2002, 66, 19–29. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shi, Y.; Wang, X.; Wang, N.; Li, F.-F.; You, Y.-L.; Wang, S.-Q. The Effect of Polysaccharides from Cibotium Barometz on Enhancing Temozolomide–Induced Glutathione Exhausted in Human Glioblastoma U87 Cells, as Revealed by 1H NMR Metabolomics Analysis. Int. J. Biol. Macromol. 2020, 156, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Xie, P.; Fujii, I.; Zhao, J.; Shinohara, M.; Matsukura, M. A Novel Polysaccharide Derived from Algae Extract Induces Apoptosis and Cell Cycle Arrest in Human Gastric Carcinoma MKN45 Cells via ROS/JNK Signaling Pathway. Int. J. Oncol. 2016, 49, 1561–1568. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Wu, S.; Shang, Y.; Li, Z.; Chen, M.; Li, F.; Wang, C. Pleurotus Nebrodensis Polysaccharide(PN50G) Evokes A549 Cell Apoptosis by the ROS/AMPK/PI3K/AKT/MTOR Pathway to Suppress Tumor Growth. Food Funct. 2016, 7, 1616–1627. [Google Scholar] [CrossRef]
- Chowdhury, S.R.; Sengupta, S.; Biswas, S.; Sen, R.; Sinha, T.K.; Basak, R.K.; Adhikari, B.; Bhattacharyya, A. Low Fucose Containing Bacterial Polysaccharide Facilitate Mitochondria-Dependent ROS-Induced Apoptosis of Human Lung Epithelial Carcinoma via Controlled Regulation of MAPKs-Mediated Nrf2/Keap1 Homeostasis Signaling: Controlled Regulation of Homeostasis Signaling. Mol. Carcinog. 2015, 54, 1636–1655. [Google Scholar] [CrossRef]
- Ko, Y.-J.; Jeong, J.-W.; Choi, Y.-H.; Ryu, C.-H. Soy Soluble Polysaccharide Induces Apoptosis in HCT-116 Human Colon Cancer Cells via Reactive Oxygen Species Generation. Mol. Med. Rep. 2013, 8, 1767–1772. [Google Scholar] [CrossRef]
- Hu, M.; Chen, Y.; Wang, C.; Cui, H.; Duan, P.; Zhai, T.; Yang, Y.; Li, S. Induction of Apoptosis in HepG2 Cells by Polysaccharide MEP-II from the Fermentation Broth of Morchella Esculenta. Biotechnol. Lett. 2013, 35, 1–10. [Google Scholar] [CrossRef]
- van Griensven, L.J.; Verhoeven, H.A. Phellinus Linteus Polysaccharide Extracts Increase the Mitochondrial Membrane Potential and Cause Apoptotic Death of THP-1 Monocytes. Chin. Med. 2013, 8, 25. [Google Scholar] [CrossRef]
- Pegg, A.E.; Dolan, M.E.; Moschel, R.C. Structure, Function, and Inhibition of O6-Alkylguanine-DNA Alkyltransferase. Prog. Nucleic Acid Res. Mol. Biol. 1995, 51, 167–223. [Google Scholar] [CrossRef] [PubMed]
- St-Coeur, P.-D.; Poitras, J.J.; Cuperlovic-Culf, M.; Touaibia, M.; Morin, P. Investigating a Signature of Temozolomide Resistance in GB Cell Lines Using Metabolomics. J. Neurooncol. 2015, 125, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Beishline, K.; Azizkhan-Clifford, J. Sp1 and the “Hallmarks of Cancer". FEBS J. 2015, 282, 224–258. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.-B.; Hsu, C.-C.; Hsu, T.-I.; Liou, J.-P.; Chang, K.-Y.; Chen, P.-Y.; Liu, J.-J.; Yang, S.-T.; Wang, J.-Y.; Yeh, S.-H.; et al. Increased Activation of HDAC1/2/6 and Sp1 Underlies Therapeutic Resistance and Tumor Growth in Glioblastoma. Neuro Oncol. 2020, 22, 1439–1451. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.-T.; Lo, W.-L.; Chen, P.-Y.; Ko, C.-Y.; Chuang, J.-Y.; Kao, T.-J.; Yang, W.-B.; Chang, K.-Y.; Hung, C.-Y.; Kikkawa, U.; et al. Reprogramming of Arachidonate Metabolism Confers Temozolomide Resistance to Glioblastoma through Enhancing Mitochondrial Activity in Fatty Acid Oxidation. J. Biomed. Sci. 2022, 29, 21. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Dubois, R.N. Eicosanoids and Cancer. Nat. Rev. Cancer 2010, 10, 181–193. [Google Scholar] [CrossRef]
- Kant, S.; Kesarwani, P.; Prabhu, A.; Graham, S.F.; Buelow, K.L.; Nakano, I.; Chinnaiyan, P. Enhanced Fatty Acid Oxidation Provides Glioblastoma Cells Metabolic Plasticity to Accommodate to Its Dynamic Nutrient Microenvironment. Cell Death Dis. 2020, 11, 253. [Google Scholar] [CrossRef]
- Bailleul, J.; Ruan, Y.; Vlashi, E. Abstract 6058: The Serine Synthesis Pathway Contributes to the Radiation-Induced Metabolic Plasticity in Glioblastoma Multiforme. Cancer Res. 2022, 82, 6058. [Google Scholar] [CrossRef]
- Haufroid, M.; Mirgaux, M.; Leherte, L.; Wouters, J. Crystal Structures and Snapshots along the Reaction Pathway of Human Phosphoserine Phosphatase. Acta Cryst. D Struct. Biol. 2019, 75, 592–604. [Google Scholar] [CrossRef]
- Esteves, L.; Caramelo, F.; Ribeiro, I.P.; Carreira, I.M.; de Melo, J.B. Probability Distribution of Copy Number Alterations along the Genome: An Algorithm to Distinguish Different Tumour Profiles. Sci. Rep. 2020, 10, 14868. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Bao, S.; Wu, Q.; Sathornsumetee, S.; Hao, Y.; Li, Z.; Hjelmeland, A.B.; Shi, Q.; McLendon, R.E.; Bigner, D.D.; Rich, J.N. Stem Cell–like Glioma Cells Promote Tumor Angiogenesis through Vascular Endothelial Growth Factor. Cancer Res. 2006, 66, 7843–7848. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, Y.; Yu, T.-S.; McKay, R.M.; Burns, D.K.; Kernie, S.G.; Parada, L.F. A Restricted Cell Population Propagates Glioblastoma Growth after Chemotherapy. Nature 2012, 488, 522–526. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, J.; Janaki-Raman, S.; Schlicker, L.; Schmitz, W.; Walz, S.; Winkelkotte, A.M.; Herold-Mende, C.; Soares, P.; Schulze, A.; Lima, J. Integrated Metabolomics and Transcriptomics Analysis of Monolayer and Neurospheres from Established Glioblastoma Cell Lines. Cancers 2021, 13, 1327. [Google Scholar] [CrossRef]
- Wang, L.-B.; Karpova, A.; Gritsenko, M.A.; Kyle, J.E.; Cao, S.; Li, Y.; Rykunov, D.; Colaprico, A.; Rothstein, J.H.; Hong, R.; et al. Proteogenomic and Metabolomic Characterization of Human Glioblastoma. Cancer Cell 2021, 39, 509–528.e20. [Google Scholar] [CrossRef]
- Zhang, R.; Hu, P.; Zang, Q.; Yue, X.; Zhou, Z.; Xu, X.; Xu, J.; Li, S.; Chen, Y.; Qiang, B.; et al. LC-MS-Based Metabolomics Reveals Metabolic Signatures Related to Glioma Stem-like Cell Self-Renewal and Differentiation. RSC Adv. 2017, 7, 24221–24232. [Google Scholar] [CrossRef]
- Duarte, T.T.; Spencer, C.T. Personalized Proteomics: The Future of Precision Medicine. Proteomes 2016, 4, 29. [Google Scholar] [CrossRef]
- Rogachev, A.D.; Alemasov, N.A.; Ivanisenko, V.A.; Ivanisenko, N.V.; Gaisler, E.V.; Oleshko, O.S.; Cheresiz, S.V.; Mishinov, S.V.; Stupak, V.V.; Pokrovsky, A.G. Correlation of Metabolic Profiles of Plasma and Cerebrospinal Fluid of High-Grade Glioma Patients. Metabolites 2021, 11, 133. [Google Scholar] [CrossRef]
- Ciocan-Cartita, C.A.; Jurj, A.; Buse, M.; Gulei, D.; Braicu, C.; Raduly, L.; Cojocneanu, R.; Pruteanu, L.L.; Iuga, C.A.; Coza, O.; et al. The Relevance of Mass Spectrometry Analysis for Personalized Medicine through Its Successful Application in Cancer “Omics”. Int. J. Mol. Sci. 2019, 20, 2576. [Google Scholar] [CrossRef]
- Gilard, V.; Ferey, J.; Marguet, F.; Fontanilles, M.; Ducatez, F.; Pilon, C.; Lesueur, C.; Pereira, T.; Basset, C.; Schmitz-Afonso, I.; et al. Integrative Metabolomics Reveals Deep Tissue and Systemic Metabolic Remodeling in Glioblastoma. Cancers 2021, 13, 5157. [Google Scholar] [CrossRef]
- Rajani, K.; Carlstrom, L.; Jacobs, J.; Schroeder, M.; Olson, I.; Hainy, M.; Oh, J.; Elmquist, W.; Sarkaria, J.; Burns, T. BIMG-20. Metabolic Biomarkers in Microdialysate of IDH-1 Mutant Tumors. Neuro-Oncol. Adv. 2021, 3, i5. [Google Scholar] [CrossRef]
- Dhodapkar, M.V.; Steinman, R.M.; Sapp, M.; Desai, H.; Fossella, C.; Krasovsky, J.; Donahoe, S.M.; Dunbar, P.R.; Cerundolo, V.; Nixon, D.F.; et al. Rapid Generation of Broad T-Cell Immunity in Humans after a Single Injection of Mature Dendritic Cells. J. Clin. Investig. 1999, 104, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Hamanishi, J.; Mandai, M.; Matsumura, N.; Abiko, K.; Baba, T.; Konishi, I. PD-1/PD-L1 Blockade in Cancer Treatment: Perspectives and Issues. Int. J. Clin. Oncol. 2016, 21, 462–473. [Google Scholar] [CrossRef] [PubMed]
- Dastmalchi, F.; Karachi, A.; Mehkri, Y.; Khattri, R.; Merritt, M.; Kubilis, P.; Mitchell, D.; Rahman, M. BIOM-36. The unique metabolomics based biomarkers of response to immunotherapy for glioblastoma. Neuro Oncol. 2020, 22, ii9. [Google Scholar] [CrossRef]
- Cuperlovic-Culf, M.; Touaibia, M.; St-Coeur, P.-D.; Poitras, J.; Morin, P.; Culf, A. Metabolic Effects of Known and Novel HDAC and SIRT Inhibitors in Glioblastomas Independently or Combined with Temozolomide. Metabolites 2014, 4, 807–830. [Google Scholar] [CrossRef]
- Falkenberg, K.J.; Johnstone, R.W. Histone Deacetylases and Their Inhibitors in Cancer, Neurological Diseases and Immune Disorders. Nat. Rev. Drug Discov. 2014, 13, 673–691. [Google Scholar] [CrossRef]
- Filippakopoulos, P.; Knapp, S. Targeting Bromodomains: Epigenetic Readers of Lysine Acetylation. Nat. Rev. Drug Discov. 2014, 13, 337–356. [Google Scholar] [CrossRef]
- Was, H.; Krol, S.K.; Rotili, D.; Mai, A.; Wojtas, B.; Kaminska, B.; Maleszewska, M. Histone Deacetylase Inhibitors Exert Anti-Tumor Effects on Human Adherent and Stem-like Glioma Cells. Clin. Epigenet. 2019, 11, 11. [Google Scholar] [CrossRef]
- Kampa, J.M.; Kellner, U.; Marsching, C.; Ramallo Guevara, C.; Knappe, U.J.; Sahin, M.; Giampà, M.; Niehaus, K.; Bednarz, H. Glioblastoma Multiforme: Metabolic Differences to Peritumoral Tissue and IDH-mutated Gliomas Revealed by Mass Spectrometry Imaging. Neuropathology 2020, 40, 546–558. [Google Scholar] [CrossRef]
- Randall, E.C.; Lopez, B.G.C.; Peng, S.; Regan, M.S.; Abdelmoula, W.M.; Basu, S.S.; Santagata, S.; Yoon, H.; Haigis, M.C.; Agar, J.N.; et al. Localized Metabolomic Gradients in Patient-Derived Xenograft Models of Glioblastoma. Cancer Res. 2020, 80, 1258–1267. [Google Scholar] [CrossRef]
- D’Alessandro, G.; Quaglio, D.; Monaco, L.; Lauro, C.; Ghirga, F.; Ingallina, C.; De Martino, M.; Fucile, S.; Porzia, A.; Di Castro, M.A.; et al. 1H-NMR Metabolomics Reveals the Glabrescione B Exacerbation of Glycolytic Metabolism beside the Cell Growth Inhibitory Effect in Glioma. Cell Commun. Signal. 2019, 17, 108. [Google Scholar] [CrossRef] [PubMed]
- Mörén, L.; Perryman, R.; Crook, T.; Langer, J.K.; Oneill, K.; Syed, N.; Antti, H. Metabolomic Profiling Identifies Distinct Phenotypes for ASS1 Positive and Negative GB. BMC Cancer 2018, 18, 167. [Google Scholar] [CrossRef]
- Poore, B.; Yuan, M.; Arnold, A.; Price, A.; Alt, J.; Rubens, J.A.; Slusher, B.S.; Eberhart, C.G.; Raabe, E.H. Inhibition of MTORC1 in Pediatric Low-Grade Glioma Depletes Glutathione and Therapeutically Synergizes with Carboplatin. Neuro Oncol. 2019, 21, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Koch, K.; Hartmann, R.; Tsiampali, J.; Uhlmann, C.; Nickel, A.-C.; He, X.; Kamp, M.A.; Sabel, M.; Barker, R.A.; Steiger, H.-J.; et al. A Comparative Pharmaco-Metabolomic Study of Glutaminase Inhibitors in Glioma Stem-like Cells Confirms Biological Effectiveness but Reveals Differences in Target-Specificity. Cell Death Discov. 2020, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Blandin, A.-F.; Durand, A.; Litzler, M.; Tripp, A.; Guérin, É.; Ruhland, E.; Obrecht, A.; Keime, C.; Fuchs, Q.; Reita, D.; et al. Hypoxic Environment and Paired Hierarchical 3D and 2D Models of Pediatric H3.3-Mutated Gliomas Recreate the Patient Tumor Complexity. Cancers 2019, 11, 1875. [Google Scholar] [CrossRef]
- McBrayer, S.K.; Mayers, J.R.; DiNatale, G.J.; Shi, D.D.; Khanal, J.; Chakraborty, A.A.; Sarosiek, K.A.; Briggs, K.J.; Robbins, A.K.; Sewastianik, T.; et al. Transaminase Inhibition by 2-Hydroxyglutarate Impairs Glutamate Biosynthesis and Redox Homeostasis in Glioma. Cell 2018, 175, 101–116. [Google Scholar] [CrossRef]
- Semreen, A.M.; Alsoud, L.O.; El-Huneidi, W.; Ahmed, M.; Bustanji, Y.; Abu-Gharbieh, E.; El-Awady, R.; Ramadan, W.S.; Alqudah, M.A.Y.; Shara, M.; et al. Metabolomics Analysis Revealed Significant Metabolic Changes in Brain Cancer Cells Treated with Paclitaxel and/or Etoposide. Int. J. Mol. Sci. 2022, 23, 13940. [Google Scholar] [CrossRef]
- Čuperlović-Culf, M.; Khieu, N.H.; Surendra, A.; Hewitt, M.; Charlebois, C.; Sandhu, J.K. Analysis and Simulation of Glioblastoma Cell Lines-Derived Extracellular Vesicles Metabolome. Metabolites 2020, 10, 88. [Google Scholar] [CrossRef]
- Heiland, D.H.; Gaebelein, A.; Börries, M.; Wörner, J.; Pompe, N.; Franco, P.; Heynckes, S.; Bartholomae, M.; hAilín, D.Ó.; Carro, M.S.; et al. Microenvironment-Derived Regulation of HIF Signaling Drives Transcriptional Heterogeneity in Glioblastoma Multiforme. Mol. Cancer Res. 2018, 16, 655–668. [Google Scholar] [CrossRef]
- Shen, J.; Song, R.; Hodges, T.R.; Heimberger, A.B.; Zhao, H. Identification of Metabolites in Plasma for Predicting Survival in Glioblastoma. Mol. Carcinog. 2018, 57, 1078–1084. [Google Scholar] [CrossRef]
- Hvinden, I.C.; Berg, H.E.; Sachse, D.; Skaga, E.; Skottvoll, F.S.; Lundanes, E.; Sandberg, C.J.; Vik-Mo, E.O.; Rise, F.; Wilson, S.R. Nuclear Magnetic Resonance Spectroscopy to Identify Metabolite Biomarkers of Nonresponsiveness to Targeted Therapy in Glioblastoma Tumor Stem Cells. J. Proteome Res. 2019, 18, 2012–2020. [Google Scholar] [CrossRef] [PubMed]
- Buser, D.P.; Ritz, M.-F.; Moes, S.; Tostado, C.; Frank, S.; Spiess, M.; Mariani, L.; Jenö, P.; Boulay, J.-L.; Hutter, G. Quantitative Proteomics Reveals Reduction of Endocytic Machinery Components in Gliomas. EBioMedicine 2019, 46, 32–41. [Google Scholar] [CrossRef]
- Mayor, S.; Parton, R.G.; Donaldson, J.G. Clathrin-Independent Pathways of Endocytosis. Cold Spring Harb. Perspect. Biol. 2014, 6, a016758. [Google Scholar] [CrossRef]
- Gollapalli, K.; Ghantasala, S.; Atak, A.; Rapole, S.; Moiyadi, A.; Epari, S.; Srivastava, S. Tissue Proteome Analysis of Different Grades of Human Gliomas Provides Major Cues for Glioma Pathogenesis. OMICS J. Integr. Biol. 2017, 21, 275–284. [Google Scholar] [CrossRef]
- Tribe, A.K.W.; McConnell, M.J.; Teesdale-Spittle, P.H. The Big Picture of Glioblastoma Malignancy: A Meta-Analysis of Glioblastoma Proteomics to Identify Altered Biological Pathways. ACS Omega 2021, 6, 24535–24544. [Google Scholar] [CrossRef]
- Lin, R.; Xu, Y.; Xie, S.; Zhang, Y.; Wang, H.; Yi, G.-Z.; Huang, G.; Ni, B.; Song, H.; Wang, Z.; et al. Recycling of SLC38A1 to the Plasma Membrane by DSCR3 Promotes Acquired Temozolomide Resistance in Glioblastoma. J. Neurooncol. 2022, 157, 15–26. [Google Scholar] [CrossRef]
- Feldman, L.S.; Fuchshuber, P.R.; Jones, D.B. The SAGES Manual on the Fundamental Use of Surgical Energy (FUSE); Springer: New York, NY, USA, 2012; ISBN 978-1-4614-2074-3. [Google Scholar]
- La Rocca, G.L.; Simboli, G.A.; Vincenzoni, F.; Rossetti, D.V.; Urbani, A.; Ius, T.; Della Pepa, G.M.; Olivi, A.; Sabatino, G.; Desiderio, C. Glioblastoma CUSA Fluid Protein Profiling: A Comparative Investigation of the Core and Peripheral Tumor Zones. Cancers 2020, 13, 30. [Google Scholar] [CrossRef]
- Buehler, M.; Yi, X.; Ge, W.; Blattmann, P.; Rushing, E.; Reifenberger, G.; Felsberg, J.; Yeh, C.; Corn, J.E.; Regli, L.; et al. Quantitative Proteomic Landscapes of Primary and Recurrent Glioblastoma Reveal a Protumorigeneic Role for FBXO2-Dependent Glioma-Microenvironment Interactions. Neuro Oncol. 2022, noac169. [Google Scholar] [CrossRef]
- Digregorio, M.; Coppieters, N.; Lombard, A.; Lumapat, P.N.; Scholtes, F.; Rogister, B. The Expression of B7-H3 Isoforms in Newly Diagnosed Glioblastoma and Recurrence and Their Functional Role. Acta Neuropathol. Commun. 2021, 9, 59. [Google Scholar] [CrossRef]
- Steinberger, P.; Majdic, O.; Derdak, S.V.; Pfistershammer, K.; Kirchberger, S.; Klauser, C.; Zlabinger, G.; Pickl, W.F.; Stöckl, J.; Knapp, W. Molecular Characterization of Human 4Ig-B7-H3, a Member of the B7 Family with Four Ig-like Domains. J. Immunol. 2004, 172, 2352–2359. [Google Scholar] [CrossRef]
- Seaman, S.; Zhu, Z.; Saha, S.; Zhang, X.M.; Yang, M.Y.; Hilton, M.B.; Morris, K.; Szot, C.; Morris, H.; Swing, D.A.; et al. Eradication of Tumors through Simultaneous Ablation of CD276/B7-H3-Positive Tumor Cells and Tumor Vasculature. Cancer Cell 2017, 31, 501–515. [Google Scholar] [CrossRef]
- Maas, S.L.N.; Breakefield, X.O.; Weaver, A.M. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef]
- Pekcan Erkan, E.; Saydam, O. Extracellular Vesicles as Novel Delivery Tools for Cancer Treatment. CCDT 2015, 16, 34–42. [Google Scholar] [CrossRef]
- Erkan, E.P.; Senfter, D.; Madlener, S.; Jungwirth, G.; Ströbel, T.; Saydam, N.; Saydam, O. Extracellular Vesicle-Mediated Suicide MRNA/Protein Delivery Inhibits Glioblastoma Tumor Growth in Vivo. Cancer Gene 2017, 24, 38–44. [Google Scholar] [CrossRef]
- Zhang, W.; Zhou, X.; Zhang, H.; Yao, Q.; Liu, Y.; Dong, Z. Extracellular Vesicles in Diagnosis and Therapy of Kidney Diseases. Am. J. Physiol. Ren. Physiol. 2016, 311, F844–F851. [Google Scholar] [CrossRef]
- Kang, H.; Kim, J.; Park, J. Methods to Isolate Extracellular Vesicles for Diagnosis. Micro Nano Syst. Lett. 2017, 5, 15. [Google Scholar] [CrossRef]
- Jansen, F.; Nickenig, G.; Werner, N. Extracellular Vesicles in Cardiovascular Disease: Potential Applications in Diagnosis, Prognosis, and Epidemiology. Circ. Res. 2017, 120, 1649–1657. [Google Scholar] [CrossRef]
- Casella, G.; Munk, R.; Kim, K.M.; Piao, Y.; De, S.; Abdelmohsen, K.; Gorospe, M. Transcriptome Signature of Cellular Senescence. Nucleic Acids Res. 2019, 47, 7294–7305. [Google Scholar] [CrossRef]
- Tambe, Y.; Yoshioka-Yamashita, A.; Mukaisho, K.-I.; Haraguchi, S.; Chano, T.; Isono, T.; Kawai, T.; Suzuki, Y.; Kushima, R.; Hattori, T.; et al. Tumor Prone Phenotype of Mice Deficient in a Novel Apoptosis-Inducing Gene, Drs. Carcinogenesis 2006, 28, 777–784. [Google Scholar] [CrossRef]
- Bastola, S.; Pavlyukov, M.S.; Yamashita, D.; Ghosh, S.; Cho, H.; Kagaya, N.; Zhang, Z.; Minata, M.; Lee, Y.; Sadahiro, H.; et al. Glioma-Initiating Cells at Tumor Edge Gain Signals from Tumor Core Cells to Promote Their Malignancy. Nat. Commun. 2020, 11, 4660. [Google Scholar] [CrossRef]
- Ampudia-Mesias, E.; El-Hadad, S.; Cameron, C.S.; Wöhrer, A.; Ströbel, T.; Saydam, N.; Saydam, O. SRPX Emerges as a Potential Tumor Marker in the Extracellular Vesicles of Glioblastoma. Cancers 2022, 14, 1984. [Google Scholar] [CrossRef]
- Dahlberg, D.; Rummel, J.; Distante, S.; De Souza, G.A.; Stensland, M.E.; Mariussen, E.; Rootwelt, H.; Voie, Ø.; Hassel, B. Glioblastoma Microenvironment Contains Multiple Hormonal and Non-Hormonal Growth-Stimulating Factors. Fluids Barriers CNS 2022, 19, 45. [Google Scholar] [CrossRef]
- Lane, R.; Cilibrasi, C.; Chen, J.; Shah, K.; Messuti, E.; Mazarakis, N.K.; Stebbing, J.; Critchley, G.; Song, E.; Simon, T.; et al. PDGF-R Inhibition Induces Glioblastoma Cell Differentiation via DUSP1/P38MAPK Signalling. Oncogene 2022, 41, 2749–2763. [Google Scholar] [CrossRef]
- Roberts, W.G.; Whalen, P.M.; Soderstrom, E.; Moraski, G.; Lyssikatos, J.P.; Wang, H.-F.; Cooper, B.; Baker, D.A.; Savage, D.; Dalvie, D.; et al. Antiangiogenic and Antitumor Activity of a Selective PDGFR Tyrosine Kinase Inhibitor, CP-673,451. Cancer Res. 2005, 65, 957–966. [Google Scholar] [CrossRef]
- Arima, Y.; Nobusue, H.; Saya, H. Targeting of Cancer Stem Cells by Differentiation Therapy. Cancer Sci. 2020, 111, 2689–2695. [Google Scholar] [CrossRef]
- Tapon, N.; Harvey, K.F.; Bell, D.W.; Wahrer, D.C.R.; Schiripo, T.A.; Haber, D.A.; Hariharan, I.K. Salvador Promotes Both Cell Cycle Exit and Apoptosis in Drosophila and Is Mutated in Human Cancer Cell Lines. Cell 2002, 110, 467–478. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, X. The Hippo Pathway in Chemotherapeutic Drug Resistance: Hippo Pathway and Drug Resistance. Int. J. Cancer 2015, 137, 2767–2773. [Google Scholar] [CrossRef]
- Harvey, K.F.; Zhang, X.; Thomas, D.M. The Hippo Pathway and Human Cancer. Nat. Rev. Cancer 2013, 13, 246–257. [Google Scholar] [CrossRef]
- Zanconato, F.; Cordenonsi, M.; Piccolo, S. YAP/TAZ at the Roots of Cancer. Cancer Cell 2016, 29, 783–803. [Google Scholar] [CrossRef]
- Yu, F.-X.; Guan, K.-L. The Hippo Pathway: Regulators and Regulations. Genes Dev. 2013, 27, 355–371. [Google Scholar] [CrossRef]
- Zhang, J. Hub. In Encyclopedia of Systems Biology; Dubitzky, W., Wolkenhauer, O., Cho, K.-H., Yokota, H., Eds.; Springer: New York, NY, USA, 2013; pp. 926–927. ISBN 978-1-4419-9862-0. [Google Scholar]
- Lim, M.; Xia, Y.; Bettegowda, C.; Weller, M. Current State of Immunotherapy for Glioblastoma. Nat. Rev. Clin. Oncol. 2018, 15, 422–442. [Google Scholar] [CrossRef]
- Filley, A.C.; Dey, M. Immune System, Friend or Foe of Oncolytic Virotherapy? Front. Oncol. 2017, 7, 106. [Google Scholar] [CrossRef]
- Iorgulescu, J.B.; Reardon, D.A.; Chiocca, E.A.; Wu, C.J. Immunotherapy for Glioblastoma: Going Viral. Nat. Med. 2018, 24, 1094–1096. [Google Scholar] [CrossRef]
- Godlewski, J.; Farhath, M.; Ricklefs, F.L.; Passaro, C.; Kiel, K.; Nakashima, H.; Chiocca, E.A.; Bronisz, A. Oncolytic Virus Therapy Alters the Secretome of Targeted Glioblastoma Cells. Cancers 2021, 13, 1287. [Google Scholar] [CrossRef]
- Jain, K.K. Role of Pharmacoproteomics. In Textbook of Personalized Medicine; Springer International Publishing: Cham, Switzerland, 2021; pp. 167–175. ISBN 978-3-030-62079-0. [Google Scholar]
- González-Morales, A.; Zabaleta, A.; Guruceaga, E.; Alonso, M.M.; García-Moure, M.; Fernández-Irigoyen, J.; Santamaría, E. Spatial and Temporal Proteome Dynamics of Glioma Cells during Oncolytic Adenovirus Delta-24-RGD Infection. Oncotarget 2018, 9, 31045–31065. [Google Scholar] [CrossRef]
- Zhu, C.; Mustafa, D.A.M.; Krebber, M.M.; Chrifi, I.; Leenen, P.J.M.; Duncker, D.J.; Dekker, L.; Luider, T.M.; Kros, J.M.; Cheng, C. Comparative Proteomic Analysis of Cat Eye Syndrome Critical Region Protein 1- Function in Tumor-Associated Macrophages and Immune Response Regulation of Glial Tumors. Oncotarget 2018, 9, 33500–33514. [Google Scholar] [CrossRef]
- Choi, D.; Montermini, L.; Kim, D.-K.; Meehan, B.; Roth, F.P.; Rak, J. The Impact of Oncogenic EGFRvIII on the Proteome of Extracellular Vesicles Released from Glioblastoma Cells. Mol. Cell. Proteom. 2018, 17, 1948–1964. [Google Scholar] [CrossRef]
- Neidert, M.C.; Kowalewski, D.J.; Silginer, M.; Kapolou, K.; Backert, L.; Freudenmann, L.K.; Peper, J.K.; Marcu, A.; Wang, S.S.-Y.; Walz, J.S.; et al. The Natural HLA Ligandome of Glioblastoma Stem-like Cells: Antigen Discovery for T Cell-Based Immunotherapy. Acta Neuropathol. 2018, 135, 923–938. [Google Scholar] [CrossRef]
- Almeida, J.; Costa, J.; Coelho, P.; Cea, V.; Galesio, M.; Noronha, J.P.; Diniz, M.S.; Prudêncio, C.; Soares, R.; Sala, C.; et al. Adipocyte Proteome and Secretome Influence Inflammatory and Hormone Pathways in Glioma. Metab. Brain Dis. 2019, 34, 141–152. [Google Scholar] [CrossRef]
- Miyauchi, E.; Furuta, T.; Ohtsuki, S.; Tachikawa, M.; Uchida, Y.; Sabit, H.; Obuchi, W.; Baba, T.; Watanabe, M.; Terasaki, T.; et al. Identification of Blood Biomarkers in Glioblastoma by SWATH Mass Spectrometry and Quantitative Targeted Absolute Proteomics. PLoS ONE 2018, 13, e0193799. [Google Scholar] [CrossRef]
- Cilibrasi, C.; Simon, T.; Vintu, M.; Tolias, C.; Samuels, M.; Mazarakis, N.K.; Eravci, M.; Stewart, N.; Critchley, G.; Giamas, G. Definition of an Inflammatory Biomarker Signature in Plasma-Derived Extracellular Vesicles of Glioblastoma Patients. Biomedicines 2022, 10, 125. [Google Scholar] [CrossRef]
- Rose, M.; Cardon, T.; Aboulouard, S.; Hajjaji, N.; Kobeissy, F.; Duhamel, M.; Fournier, I.; Salzet, M. Surfaceome Proteomic of Glioblastoma Revealed Potential Targets for Immunotherapy. Front. Immunol. 2021, 12, 746168. [Google Scholar] [CrossRef]
- D’Souza, R.C.J.; Offenhäuser, C.; Straube, J.; Baumgartner, U.; Kordowski, A.; Li, Y.; Stringer, B.W.; Alexander, H.; Lwin, Z.; Inglis, P.-L.; et al. Q-Cell Glioblastoma Resource: Proteomics Analysis Reveals Unique Cell-States Are Maintained in 3D Culture. Cells 2020, 9, 267. [Google Scholar] [CrossRef]
- Anastasi, F.; Greco, F.; Dilillo, M.; Vannini, E.; Cappello, V.; Baroncelli, L.; Costa, M.; Gemmi, M.; Caleo, M.; McDonnell, L.A. Proteomics Analysis of Serum Small Extracellular Vesicles for the Longitudinal Study of a Glioblastoma Multiforme Mouse Model. Sci. Rep. 2020, 10, 20498. [Google Scholar] [CrossRef]
- Yi, G.; Xiang, W.; Feng, W.; Chen, Z.; Li, Y.; Deng, S.; Guo, M.; Zhao, L.; Sun, X.; He, M.; et al. Identification of Key Candidate Proteins and Pathways Associated with Temozolomide Resistance in Glioblastoma Based on Subcellular Proteomics and Bioinformatical Analysis. BioMed Res. Int. 2018, 2018, 5238760. [Google Scholar] [CrossRef]
- Lam, K.H.B.; Leon, A.J.; Hui, W.; Lee, S.C.-E.; Batruch, I.; Faust, K.; Klekner, A.; Hutóczki, G.; Koritzinsky, M.; Richer, M.; et al. Topographic Mapping of the Glioblastoma Proteome Reveals a Triple-Axis Model of Intra-Tumoral Heterogeneity. Nat. Commun. 2022, 13, 116. [Google Scholar] [CrossRef]
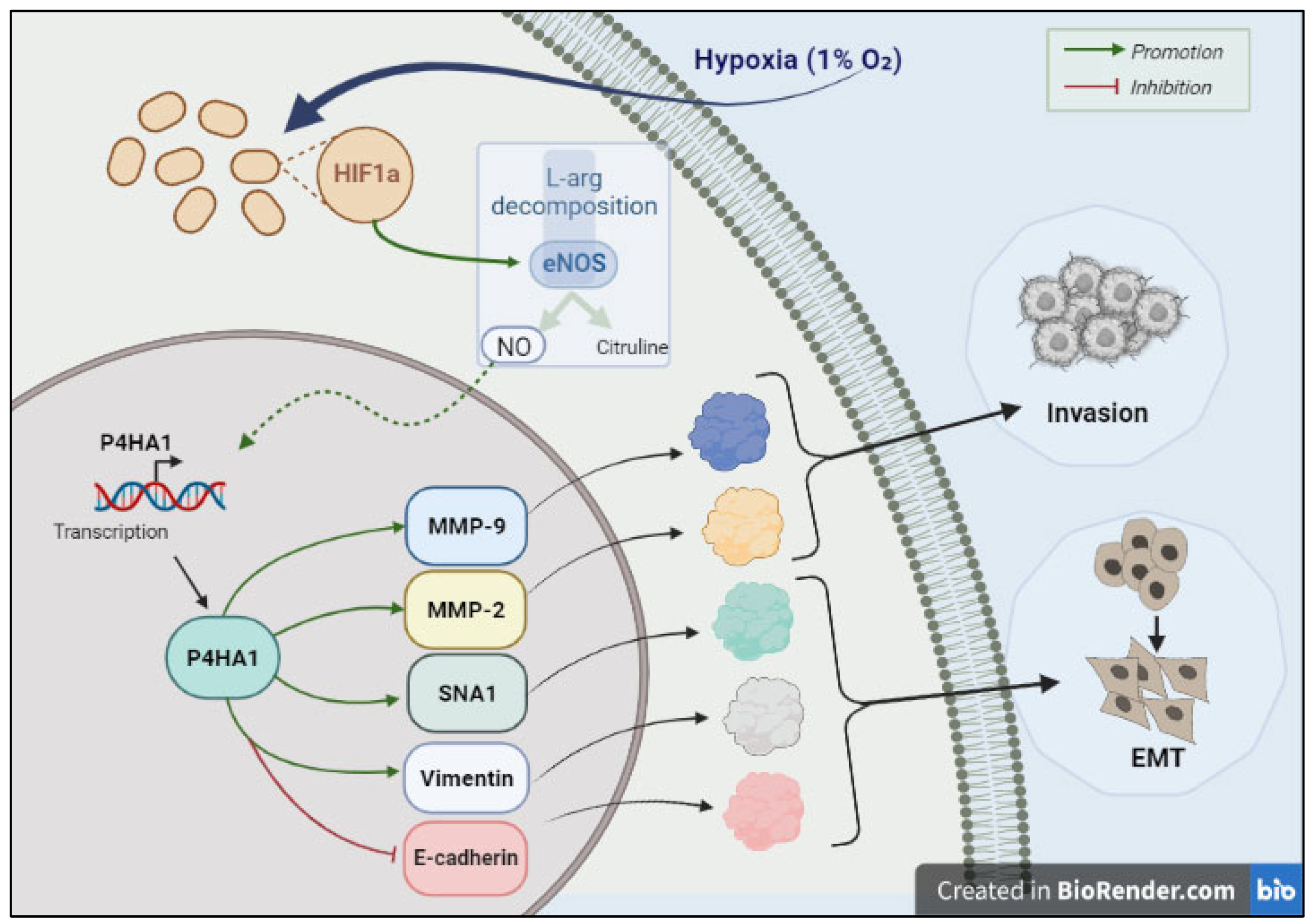
- Zhu, X.; Liu, S.; Yang, X.; Wang, W.; Shao, W.; Ji, T. P4HA1 as an Unfavorable Prognostic Marker Promotes Cell Migration and Invasion of Glioblastoma via Inducing EMT Process under Hypoxia Microenvironment. Am. J. Cancer Res. 2021, 11, 590–617. [Google Scholar] [PubMed]
- Colwell, N.; Larion, M.; Giles, A.J.; Seldomridge, A.N.; Sizdahkhani, S.; Gilbert, M.R.; Park, D.M. Hypoxia in the Glioblastoma Microenvironment: Shaping the Phenotype of Cancer Stem-like Cells. Neuro Oncol. 2017, 19, 887–896. [Google Scholar] [CrossRef]
- Papale, M.; Buccarelli, M.; Mollinari, C.; Russo, M.A.; Pallini, R.; Ricci-Vitiani, L.; Tafani, M. Hypoxia, Inflammation and Necrosis as Determinants of Glioblastoma Cancer Stem Cells Progression. Int. J. Mol. Sci. 2020, 21, 2660. [Google Scholar] [CrossRef]
- Chédeville, A.L.; Madureira, P.A. The Role of Hypoxia in Glioblastoma Radiotherapy Resistance. Cancers 2021, 13, 542. [Google Scholar] [CrossRef]
- Spence, A.M.; Muzi, M.; Swanson, K.R.; O’Sullivan, F.; Rockhill, J.K.; Rajendran, J.G.; Adamsen, T.C.H.; Link, J.M.; Swanson, P.E.; Yagle, K.J.; et al. Regional Hypoxia in Glioblastoma Multiforme Quantified with [18F]Fluoromisonidazole Positron Emission Tomography before Radiotherapy: Correlation with Time to Progression and Survival. Clin. Cancer Res. 2008, 14, 2623–2630. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, Y.; Shingu, T.; Feng, L.; Chen, Z.; Ogasawara, M.; Keating, M.J.; Kondo, S.; Huang, P. Metabolic Alterations in Highly Tumorigenic Glioblastoma Cells. J. Biol. Chem. 2011, 286, 32843–32853. [Google Scholar] [CrossRef]
- Marampon, F.; Gravina, G.L.; Zani, B.M.; Popov, V.M.; Fratticci, A.; Cerasani, M.; Di Genova, D.; Mancini, M.; Ciccarelli, C.; Ficorella, C.; et al. Hypoxia Sustains Glioblastoma Radioresistance through ERKs/DNA-PKcs/HIF-1α Functional Interplay. Int. J. Oncol. 2014, 44, 2121–2131. [Google Scholar] [CrossRef]
- Chédeville, A.L.; Lourdusamy, A.; Monteiro, A.R.; Hill, R.; Madureira, P.A. Investigating Glioblastoma Response to Hypoxia. Biomedicines 2020, 8, 310. [Google Scholar] [CrossRef]
- Grimes, D.R.; Jansen, M.; Macauley, R.J.; Scott, J.G.; Basanta, D. Evidence for Hypoxia Increasing the Tempo of Evolution in Glioblastoma. Br J. Cancer 2020, 123, 1562–1569. [Google Scholar] [CrossRef]
- Semenza, G.L. Oxygen Sensing, Homeostasis, and Disease. N. Engl. J. Med. 2011, 365, 537–547. [Google Scholar] [CrossRef]
- van Valkengoed, I.G.M.; Argmann, C.; Ghauharali-van der Vlugt, K.; Aerts, J.M.F.G.; Brewster, L.M.; Peters, R.J.G.; Vaz, F.M.; Houtkooper, R.H. Ethnic Differences in Metabolite Signatures and Type 2 Diabetes: A Nested Case–Control Analysis among People of South Asian, African and European Origin. Nutr. Diabetes 2017, 7, 300. [Google Scholar] [CrossRef]
- Lusczek, E.R.; Myers, C.; Popovsky, K.; Mulier, K.; Beilman, G.; Sawyer, R. Plasma Metabolomics Pilot Study Suggests Age and Sex-Based Differences in the Metabolic Response to Traumatic Injury. Injury 2018, 49, 2178–2185. [Google Scholar] [CrossRef]
- Bell, J.A.; Santos Ferreira, D.L.; Fraser, A.; Soares, A.L.G.; Howe, L.D.; Lawlor, D.A.; Carslake, D.; Davey Smith, G.; O’Keeffe, L.M. Sex Differences in Systemic Metabolites at Four Life Stages: Cohort Study with Repeated Metabolomics. BMC Med. 2021, 19, 58. [Google Scholar] [CrossRef]
- Lenting, K.; Verhaak, R.; ter Laan, M.; Wesseling, P.; Leenders, W. Glioma: Experimental Models and Reality. Acta Neuropathol. 2017, 133, 263–282. [Google Scholar] [CrossRef]
| Application | Aim | Tissue Type /Cell Line | Technique | Key Metabolites | Key Metabolic Pathways | Principle Insights | Reference |
|---|---|---|---|---|---|---|---|
| Biochemical characterization | Differentiate glioblastoma subtypes; define infiltrative tumor boundaries; potential utility in evaluating treatment effects. | Tumor and peritumoral GB tissue | MALDI-TOF-MSI (matrix-assisted laser desorption/ionization-time-of-flight-mass spectrometry imaging) | Antioxidants Fatty acids Purine and pyrimidine metabolites Reduced N-acetylaspartate abundance, etc. | Purine and pyrimidine metabolism, arachidonic acid synthesis, TCA cycle. | Metabolic information obtained could enhance and customize therapy methods The study underlines MSI’s appropriateness for GB research | [86] |
| Biochemical characterization | Create a xenograft for GB therapeutic testing; investigate the link between treatment efficacy and tumor metabolism. | Glioblastoma xenograft tissue | MALDI –Fourier transform ion cyclotron resonance (FT-ICR)-MSI | Heme ATP Acylcarnitine | Glycolysis, fatty acid metabolism, antioxidant, and anti-apoptotic functions. | Cells in the tumor’s core and edge experience distinct fatty acid metabolism, leading to different chemical microenvironments within the tumor.This can impact medication distribution via changes in tissue drug affinity or transport and is an essential consideration for therapeutic options in the treatment of GB. | [87] |
| Pharmacometabolomic approach | To investigate, for the first time, the influence of glabrescione B (GlaB), a known Hedgehog (Hh) pathway inhibitor, on glioma cell proliferation and metabolism in in vivo and in vitro models. | Murine glioma cells (GL261) | 1H-NMR, HPLC–MS | Lactate Glycine Tyrosine Phenylalanine Histidine Alanine Leucine Isoleucine Valine | Glycolytic metabolism. | The endo- and exo-metabolomes of GlaB-treated and untreated cells exhibited changes in metabolite levels over time. GlaB, a direct inhibitor of the transcription factor Gli1, suppresses glioma cell proliferation while exacerbating the Warburg effect. | [88] |
| Comparative biomarker discovery | Because altered tumor metabolism is one of the hallmarks of cancer, the aim was to explore if the rate-limiting enzyme argininosuccinate synthetase (ASS1) positive and negative GB cell lines had distinct metabolic profiles that may allow for non-invasive diagnosis and reveal new treatment prospects. | GAMG LN229 SNB19 T98G U118 U87 Normal Human Astrocytes (NHA) | One and two-dimensional gas chromatography-time-of-flight mass spectrometry (1D/2D GC-TOFMS), LC-TOFMS. | Mannose Galactose Glucose Pyruvic acid Citrate α-ketoglutaric acid | Not detected. | The metabolome contains systematic information distinguishing between ASS1 positive and negative GB cell lines. There is a possibility of identifying metabolite biomarkers for the non-invasive detection from these subtypes, as well as the identification of novel treatment targets. | [89] |
| Pharmacometabolomic approach | The goal of this trial was to see if carboplatin worked in tandem with the mTOR complex 1 inhibitor (everolimus) in pediatric low-grade glioma (pLGG). | pLGG cell line BT66 JHH-NF1-PA1 Res259 Res18 | LC–MS | Glutathione Glutamine Glutamate | Comparable pathways were discovered in patient-derived xenograft in mice. | The combination of everolimus and carboplatin works synergistically in pLGG. The study confirms a novel therapy regimen that may be promptly pushed into pediatric phase I/II clinical trials. This work presents a justification for novel mTORC1-based inhibitor therapy combinations in brain malignancies. | [90] |
| Pharmacometabolomic approach | To investigate the effect of glutaminase (GLS) inhibition on GSCs, which have been implicated in the development of medication resistance and tumor recurrence. | 1H-NMR JHH520 GBM1 268, 407, 23, 233, 349 SF188 NCH644 | 1H-NMR | Alanine Aspartate Glutamine Glutamate Glycine Glutathione Lactate Myo-inositol Succinate Tricarboxylic acid Total choline | Not detected | The findings demonstrate the use of in vitro pharmaco-metabolomics for therapeutic effectiveness evaluation and compound risk assessment. It emphasizes the importance of GLS as a druggable and prospective therapeutic target in our desire to enhance the management of GB medication resistance and tumor relapse by focusing on GSCs subpopulation. | [91] |
| Developmental therapeutics | To develop standardized pediatric high-grade gliomas (pHGGs) models for drug testing and to generate an exact physiological brain environment in vitro. | Primary glioblastoma | NMR | Acetate Alanine Beta-glucose Choline Creatine Glutamate Glycerophosphocholine Glycine Lactate Myo-inositol N Acetylaspartate, Serine Taurine Valine | Some pathways were altered in the 2D/3D cell cultures pathways in patient tumor relapse. | A hypoxic environment helps to preserve the original patient tumor metabolism and characteristics. The multi-step effort may be regarded as a standard for developing therapeutically relevant models. | [92] |
| Developmental therapeutics | Researchers hypothesized that the branched-chain α-ketoacids (BCKA) depletion is caused by the (R) enantiomer of 2-hydroxyglutarate((R)-2HG)’s direct, competitive suppression of branched chain amino acids transaminases (BCAT) activity. | GSC lines: TS603, TS516, MGG152, TS676, BT054 BT260NHA HT1080 HOG IDH1 R132H mutant IDH2 R172K mutant HCT116 HEK293T NCI-H82 | GC-MS, hybrid triple quadrupole mass spectrometer, Hydrophilic interaction liquid chromatography(HILIC) | Alpha-Keto-beta-methylvalerate Alpha-Ketoisocaproate Glutamate 2-hydroxyglutarate Leucine Valine isoleucine | Increased BCAT activity in vitro and in vivo. | BCAT suppression produces metabolic vulnerabilities that can be leveraged therapeutically to sensitize IDH mutant gliomas. ((R)-2HG is overproduced in IDH mutant GBs). Gliomas with IDH mutations are more sensitive to radiation when combined with glutaminase inhibition, suggesting a novel way to treating these tumors. | [93] |
| Developmental therapeutics | To assess the effect of paclitaxel and/or etoposide on the molecular changes in GB cells | U87 U373 | Ultra-high-performance liquid chromatography-electrospray ionization quadrupole time-of-flight mass spectrometry (UHPLC-ESI-QTOF-MS) | Nutriacholic acid L-phenylalanine L-arginine Guanosine ADP Hypoxanthine guanine | Urea and citric acid cycles Metabolism of polyamines and amino acids | The results can be used to map the anticancer activity of paclitaxel and/or etoposide within the cancer cells under investigation. | [94] |
| Biomarker discovery | To use NMR spectroscopy to characterize the metabolome of tiny EVs or exosomes produced from distinct GB cells and compare them to the metabolic profile of their parental cells. | NHA U118 LN-18 A172 | 1H-NMR | Asparagine Acetone Carnitine Ethanol Formate Glycerol malate GSSG GSH GABA G6P Glucose Isoleucine Taurocholic acid Niacinamide lactate 5-oxoproline Citrate Proline succinate Homoserine Glycine | Not detected | The findings revealed a distinct divergence in the metabolic profiles of GB cells, EVs, and medium. The findings are reviewed in relation to new GB diagnostics and therapy monitoring. | [95] |
| Biomarker discovery | To describe a transcriptional adaption regulatory system that is influenced by environmental factors. | Primary GB | 1H-NMR | Alpha-ketoglutarate Arginine Caproic acid Choline Dodecanoic acid Fructose Fumarate Glyceraldehyde Glutathione Glycine Guaiacol Glucose-6-phosphateLysine Succinic acid Serine Selenomethionine | Comparable metabolic environment spatial disparities | A multi-regional examination of a glioblastoma patient biopsy indicated complex metabolic landscape with varied degrees of hypoxia and creatine enrichment. In creatine-enriched settings, the glycine cleavage system, and hypoxia-inducible factor-1α (HIF1A) destabilization were changed, resulting in transcriptional adaptability. | [96] |
| Biomarker discovery | To test the hypothesis that GB plasma metabolite profiles may predict clinical outcomes. | Primary and recurrent glioblastoma | LC- triple quadrupole- MS | Arginine Kynurenate Methionine | N/A | The study discovered numerous plasma metabolites that are predictive in glioblastoma patients. | [97] |
| Biomarker discovery | To investigate the effects of a survivin inhibitor (pro-apoptotic effect) on the metabolome of primary GSCs to look for treatment response signals. | GSCs cultures established from IDH-wildtype GB tumor | NMR spectroscopy | Citrate Lactate | N/A | In comparison to spectrometry-based proteomics, the metabolomics technique used generated alternative biomarker possibilities, highlighting the benefits of complementary approaches. Citrate and lactate are magnetic resonance spectroscopy (MRS) -visible, therefore, these first findings provide the groundwork for further research into in vivo MRS of brain malignancies. NMR metabolomics, when combined, is a technique for tackling glioblastoma. | [98] |
| Application | Aim | Tissue Type /Cell Line | Technique | Key Proteins | Key Pathways | Principle Insights | Reference |
|---|---|---|---|---|---|---|---|
| Pharmacoproteomics | To understand the extensive regulation of glioma metabolism in response to Delta-24-RGD (an E1A mutant oncolytic adenovirus) infection, by performing a cell-wide study of cytosolic, nuclear, and secreted proteomes during the early time course of the infection. | U87 | Using iTRAQ, a shotgun comparative proteomic analysis of cytosolic fractions | Cytosolic proteins: Serine hydroxymethyltransferase, mitochondrial lactotransferrin Alpha-2-macroglobulin Proliferating cell nuclear antigen etc. Nuclear proteins: Programmed cell death protein 6 Replication factor C subunit 2 Annexin A2 Ribosome-binding protein 1 etc. | Proteostasis pathway Protein kinase C, ERK1/2, and p38 MAPK pathways | The findings assist in understanding the methods through which Delta-24-RGD exploits glioma proteome organization. Further exploration of this proteomic resource may lead to the development of complementary adenoviral-based vectors with increased specificity and efficacy against glioma. | [135] |
| Unraveling GB pathophysiology | To define the role of cat eye syndrome critical region protein 1 (CECR1) in tumor associated macrophages (TAMs) through a proteomic investigation of siRNA-mediated CECR1 silencing in THP-1-derived macrophages co-cultured with or without glial tumor cells. | U87 THP-1 cells (human monocytic cell line) | Mass spectrometry | ISG15 HLA-A HLA-B HLA-C TAP1 TAP2 TAPBP TIMP-1 WDFY1 SEPT7 S100A9 PLAU LAT2 | MHC I antigen presenting pathway, Phagosome maturation, caveolin-mediated endocytosis, and type I interferon signaling pathways | CECR1-mediated molecular pathways and essential molecules operate in macrophages and glial TAMs (Tyro3, Axl, Mer, family of receptor tyrosine kinases). The proteome dataset might be used to create novel therapeutic targets for future immunotherapy research in the treatment of malignant (glial) cancers and autoimmune illnesses. | [136] |
| Unraveling GB pathophysiology | The constitutively active epidermal growth factor receptor (EGFRvIII) is an oncogenic factor that fuels GB aggressiveness and is ascribable to the release of extracellular vesicles (EVs). Researchers aimed to examine the effect of this oncogene on the profile of glioma EVs. | U373 U373vIII | MS | CD44/BSG TSPAN8 CD151 CD81 CD9 SDCB1 Actin, GAPDH CD44 ITGA6, ITGB4 TGFB1 Laminins, collagens | Vesiculation pathways | CD44/BSG were co-localized in cellular filopodia and EVs generated by EGFRvIII-expressing cells were double positive for these proteins. Oncogenic EGFRvIII alters the proteome and uptake of EVs related to GB. | [137] |
| Developmental therapeutics | To perform a comparison of the ligandomes of GSC and patient samples with the goal of discovering GSC-associated targets that are also present on primary patient malignancies. | Primary GB Peripheral blood mononuclear cells (PBMC) GSC cell lines: GS-2 GS-5 GS-9 | LC–MS/MS | Cancer testis antigen (CTA) SERPINE1 FABP7 PTGFRN ALFPERITV (ATAT1) RLAPFVYLL (HEPACAM) SILDIVTKV (RFTN2) | Not available | The study identified a novel panel of T cell antigens characterized by the exclusive identification of malignant specimens, a substantial prevalence of presentation, and presence on the GSC compartment by mapping the HLA peptidome of glioblastoma and GSC. Epitopes of functional CD8 T-cell responses were identified, making them excellent candidates for immunotherapy. | [138] |
| Biomarker discovery | To investigate, in a cellular rat model, the line GL261. | 3T3-L1 adipocyte cell line GL-261 murine model | MALDI-TOF-MS | Arbonic anhydrase Aldose reductase Endocan HGF IGF-I IL-6 IL-11 LIF PAI-1 SerpinE1 TNF-α TIMP-1 VEGF | Not available | STI1, hnRNPs, and PGK1, overexpressed otherwise in cancer, were under expressed. Carbonic anhydrase and aldose reductase, both of which play significant roles in inflammation, and cancer metabolism, are also reduced in glioma cells cultured in an adipokine-enriched environment, displaying a paradoxical association of a protective function between fat and cancer. | [139] |
| Biomarker discovery | To utilize SWATH-MS and quantitative targeted absolute proteomics to find plasma biomarker candidates for GB patients (QTAP). | Cyst fluid samples of IDH wildtype GB Non-cancerous brain tissue samples | SWATH-MS LC–MS/MS | LRG1 C9 CRP SERPINA3 APOB GSN IGHA1 APOA4 | Not detected | To evaluate the links between biomarker candidates and GB Biology, the study looked at associations between biomarker candidate plasma concentrations and clinical presentation (tumor size, overall survival time, etc.) in patients. LRG1, CRP, and C9 plasma concentrations all revealed strong positive relationships with tumor growth. | [140] |
| Biomarker discovery | To identify and describe the effective biomarkers present in the small extracellular vesicles (sEVs) to improve GB diagnosis, and ultimately, patient prognosis. | Blood samples from GB patients and controls | MS, MS/MS | VWF FCGBP C3 PROS1 SERPINA1 | B-cell receptor signaling pathway Pathways involved in complement activation, innate immune response, and platelet degranulation | Overall, the development of a non-invasive liquid biopsy method for the discovery of valuable biomarkers that could considerably enhance GB diagnosis and, as a result, patients’ prognosis, and quality of life, is promoted through this study. | [141] |
| Developmental therapeutics | To study GB- associated surfaceome by comparing it to the surfaceome of astrocyte cell lines in order to find new GB-specific targets. | NCH82 U-87 MG | MALDI-mass spectrometry | Plaur B41 alpha chain (HLA-b) A-24 alpha chain (HLA-a) DP beta 1 chain (hla-dpb1). CADM3 CADM4 NRCAM | Cell contact, cell adhesion, vascularization, and proliferation pathways | 11 distinct potential GB targets were discovered, including 5 altered proteins such as MHC I, CYBA, EGFR, and RELL1. | [142] |
| Optimize GB datasets | To boost the translational importance of the Q-Cell datasets and to create a platform for academics to conduct rigorous preclinical neuro-oncology research. | Primary GB characterized cell line (Q-Cell) | LC–MS | BAH1 GSN JK2 MMK1 MN1 NNMT PLP2 PRDX6 RN1 SB2b SB2 SOD2 SERPINE1 WK1 | PI3K and mTOR signaling pathways TCA cycle, NFKB, and MAPK signaling. | In-depth proteomic characterization of the GB Q-Cell resource was obtained, serving as a dataset for future biological and preclinical research. | [143] |
| Establishing research methodology | To develop methods to analyze the proteome of small extracellular vesicles (sEVs) from low serum volume that is obtained from mice, to perform a longitudinal analysis of disease models. | Adult C57BL/6J mice murine glioma GL261 cell line | LC–MS/MS | Tetraspanins integrins Sdcbp Hspa8 Cd9 Itga2 Anxa4, Anxa5, Anxa7 Vamp8 Lrp1 Cpn1 Mhy9 Tln1 Tfrc, CD71 Apoc4 | PI3K/AKT pathway | The methodology allowed for the identification and quantification of 274 protein groups. The longitudinal study discovered 25 altered proteins in GB serum sEVs, including proteins previously linked to GB development and metastasis. | [144] |
| Identification of resistance mechanism | To investigate the cytoplasmic proteome of U87 GB cells treated with TMZ, using bioinformatic approaches to thoroughly evaluate the raw data. | U87 | Liquid chromatography–electrospray ionization–tandem mass spectrometry (LC–ESI–MS/MS) | DHX9 HNRNPR RPL3 HNRNPA3 SF1 DDX5 EIF5B BTF RPL8 | Thyroid hormone, p53 and the PI3K-Akt signaling pathways Regulation of actin cytoskeleton | Dysregulation of spliceosome-related proteins SF-1, DDX5, and HNRNPR may all contribute to a disruption in DHX9 synthesis, eventually leading to GB TMZ resistance. | [145] |
| Morphoproteomics | To create a spatially conserved proteomic atlas of GB by meticulous microdissection and LC–MS/MS profiling of the traditional histomorphologic characteristics of the malignancy. | MYC-enriched cell lines (3-CI-AHPC, CD-437) | LC–MS/MS | Immunoglobulin CD276 (B7-H3) AKAP12 PTPRZ1 | Hypoxia pathway axis | Various glioblastoma locations may be divided into subpopulations in which glioma cells favor migration and infiltration above proliferation and growth. | [146] |
| Disease | Intervention/Treatment | Country | ID | Status | Outcome to Assess (in Relation to Metabolomics and Proteomics) |
|---|---|---|---|---|---|
| Prostate cancer | N/A | Taiwan | NCT03237026 | Recruiting (as of September 2022). | Biochemical recurrence or progression. |
| Hepatocellular Carcinoma | Procedure: surgical resection drug: adjuvant atezolizumab–bevacizumab Therapy | Singapore | NCT05516628 | Recruiting (as of September 2022). | Biomarkers based on multi-omics (epigenomics, genomics, transcriptomics, immunomics, proteomics, and metabolomics) and spatial tumor microenvironment profiles of both tissue and peripheral blood that predict therapy response. |
| Urothelial Carcinoma | Biomarkers and proteomics | Italy | NCT04770974 | Not yet recruiting (as of September 2022). | Bladder cancer’s metabolomic profile. |
| Pancreatic Neoplasms | Diagnostic test: soluble biomarkers dosage | France | NCT04370574 | Recruiting (as of September 2022). | To identify biomarkers that seem to be prognostically significant on overall survival or disease independent in pancreatic cancer patients. |
| Metastatic Urothelial Carcinoma | N/A | Taiwan | NCT04641936 | Recruiting (as of September 2022). | To discover possible metabolite and protein indicators capable of predicting the success and side effects of immuno–oncology-based therapies. |
| Adrenal Neoplasm Endocrine Tumors Neuroblastoma Parathyroid Neoplasms Thyroid Neoplasms | N/A | Unites States | NCT01005654 | Recruiting (as of September 2022). | To create a metabolomic, proteomic, genetic, and epigenetic profile of endocrine neoplasm that would allow discriminating between benign and malignant tumors in each of the endocrine histologies under investigation. |
| Metastatic Renal Cell Carcinoma | N/A | Taiwan | NCT04712305 | Recruiting (as of September 2022). | To discover possible metabolite and protein indicators capable of predicting the success and side effects of immuno–oncology-based therapies. |
| Significant Prostate Cancer | Dietary supplement: multi-carotenoids (MCS) | Taiwan | NCT03237702 | Recruiting (as of September 2022). | Evaluating the effect of urine omics tests (metabolomics and proteomics) in participants undergoing or having undergone prostate biopsy and/or subsequent MCS supplementing |
| Colorectal Cancer (CRC) | N/A | United States | NCT00898378 | Completed | Utilize biological samples from patients with CRC or colorectal adenomatous polyps, as well as those without polyps, to perform genomic, metabolomic, lipidomic, glycoproteomic, and proteomic profiling to create an omic profile. |
| Sarcoma Endocrine Tumors Neuroblastoma Retinoblastoma Renal Cancer | N/A | United States | NCT01109394 | Recruiting (as of September 2022). | To conduct on tumor and normal tissues, systematic molecular, genomic, proteomic, metabolomic, and other high throughput (Omics) profiling. |
| Extrapulmonary Small Cell Cancer Non-Small Cell Lung Cancer Small Cell Lung Cancer Pulmonary Neuroendocrine Tumors Thymic Epithelial Tumors | N/A | United States | NCT02146170 | Recruiting (as of September 2022). | Conduct genomic, proteomic, and immunological investigations on blood, tumor, bodily fluid, and normal tissue to identify new therapeutic drugs, innovative treatment techniques, and new prognostic and diagnostic markers. |
| Thyroid Nodule Thyroid Cancer | Diagnostic test: multi-omic analyses of blood and surgical specimens | Italy | NCT05428371 | Not yet recruiting (as of September 2022). | Identification of biomarkers of thyroid carcinoma. |
| Cutaneous Squamous Cell Carcinoma Basal Cell Carcinomas | Biopsy | France | NCT04389112 | Recruiting (as of September 2022). | Metabolic profiling of the carcinogenesis stages of glycolysis, oxidative phosphorylation. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, M.; Semreen, A.M.; El-Huneidi, W.; Bustanji, Y.; Abu-Gharbieh, E.; Alqudah, M.A.Y.; Alhusban, A.; Shara, M.; Abuhelwa, A.Y.; Soares, N.C.; et al. Preclinical and Clinical Applications of Metabolomics and Proteomics in Glioblastoma Research. Int. J. Mol. Sci. 2023, 24, 348. https://doi.org/10.3390/ijms24010348
Ahmed M, Semreen AM, El-Huneidi W, Bustanji Y, Abu-Gharbieh E, Alqudah MAY, Alhusban A, Shara M, Abuhelwa AY, Soares NC, et al. Preclinical and Clinical Applications of Metabolomics and Proteomics in Glioblastoma Research. International Journal of Molecular Sciences. 2023; 24(1):348. https://doi.org/10.3390/ijms24010348
Chicago/Turabian StyleAhmed, Munazza, Ahlam M. Semreen, Waseem El-Huneidi, Yasser Bustanji, Eman Abu-Gharbieh, Mohammad A. Y. Alqudah, Ahmed Alhusban, Mohd Shara, Ahmad Y. Abuhelwa, Nelson C. Soares, and et al. 2023. "Preclinical and Clinical Applications of Metabolomics and Proteomics in Glioblastoma Research" International Journal of Molecular Sciences 24, no. 1: 348. https://doi.org/10.3390/ijms24010348
APA StyleAhmed, M., Semreen, A. M., El-Huneidi, W., Bustanji, Y., Abu-Gharbieh, E., Alqudah, M. A. Y., Alhusban, A., Shara, M., Abuhelwa, A. Y., Soares, N. C., Semreen, M. H., & Alzoubi, K. H. (2023). Preclinical and Clinical Applications of Metabolomics and Proteomics in Glioblastoma Research. International Journal of Molecular Sciences, 24(1), 348. https://doi.org/10.3390/ijms24010348






