Evolution and Phylogeny of Soybean Mosaic Virus Based on 143 Complete Genomes
Abstract
1. Introduction
2. Results
2.1. Complete Genome Sequences of Seven SMV Isolates from Soybean Germplasms Obtained via RNA Sequencing
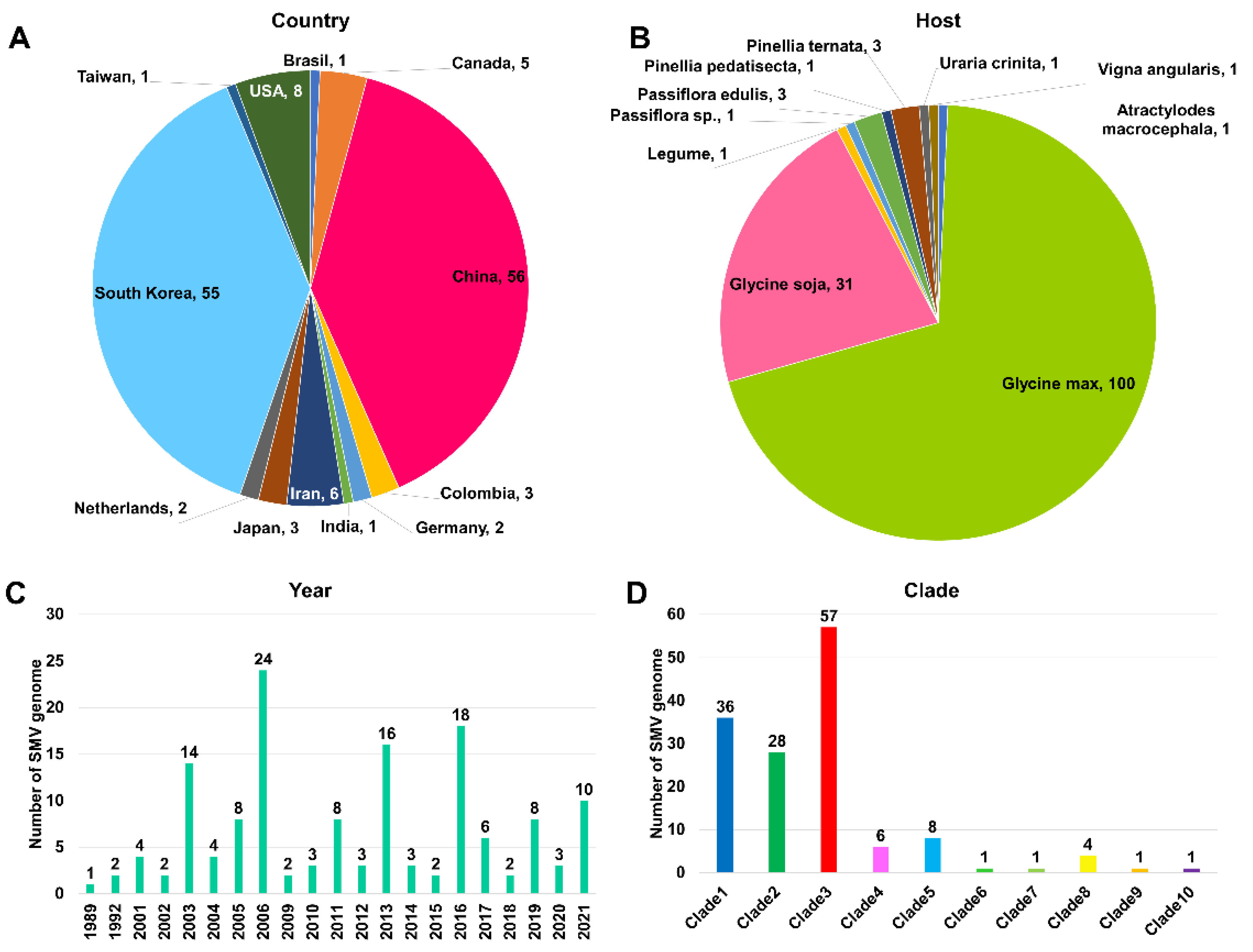
2.2. Examination of 143 Complete SMV Genomes Based on Country, Plant Host, and Year of Identification
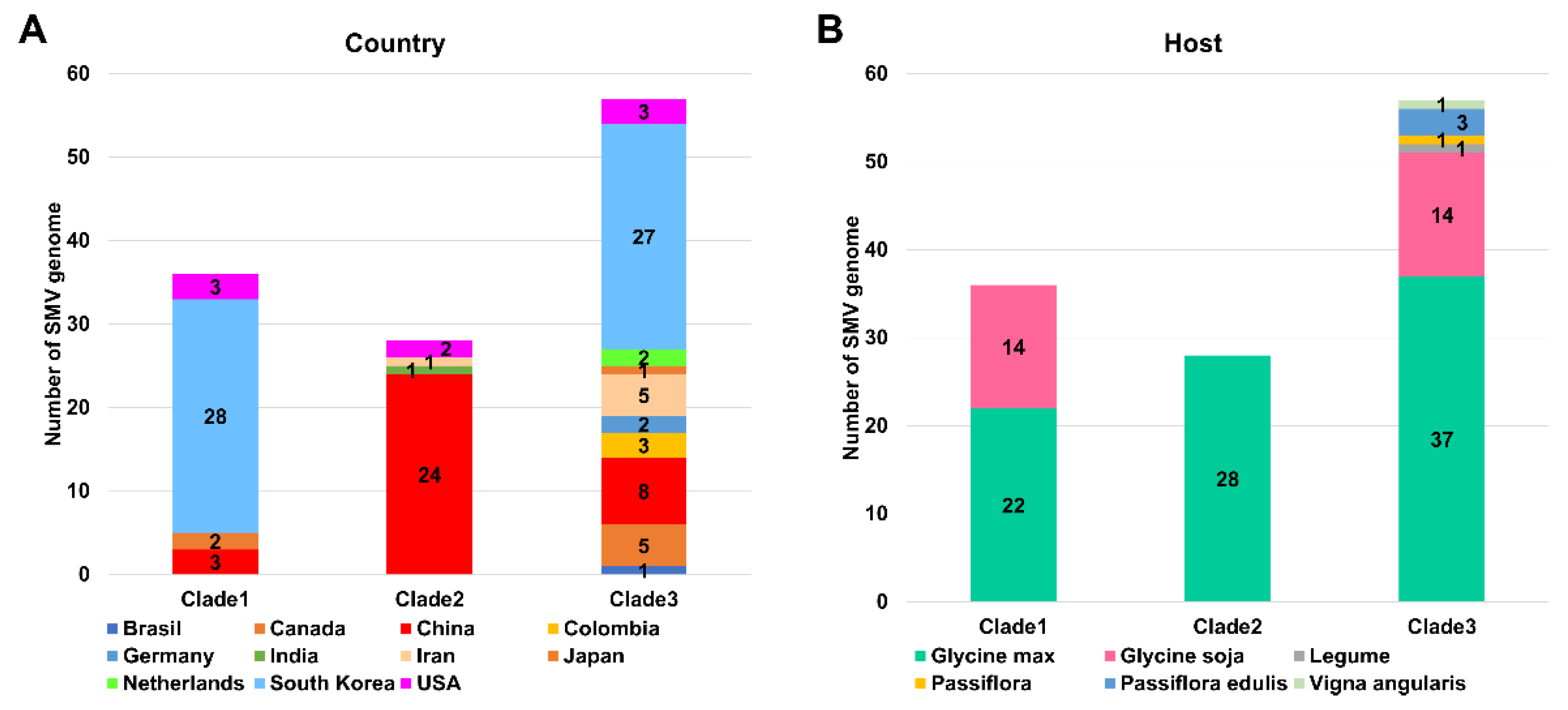
2.3. Phylogenetic Analysis of 143 SMV Genomes
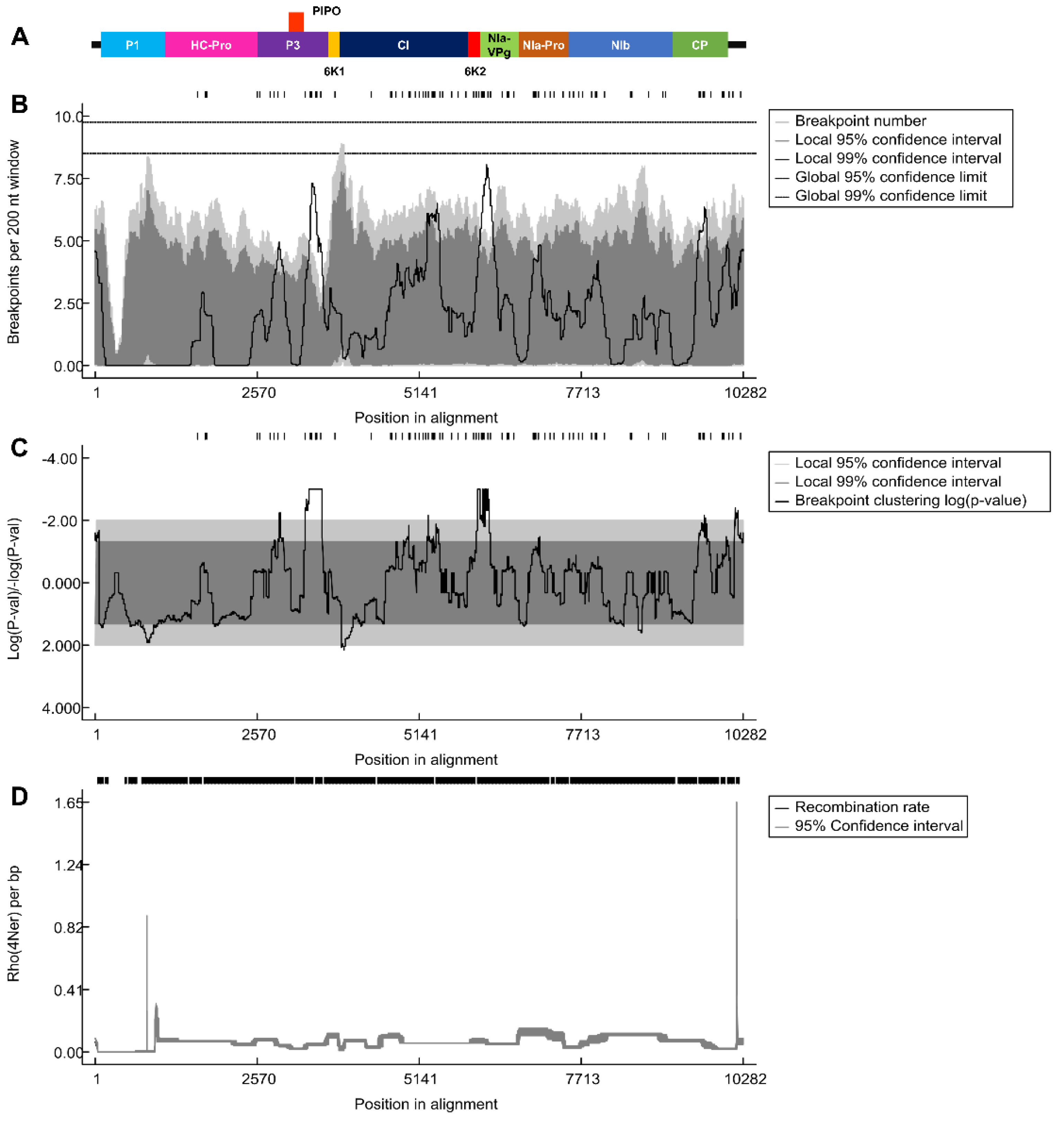
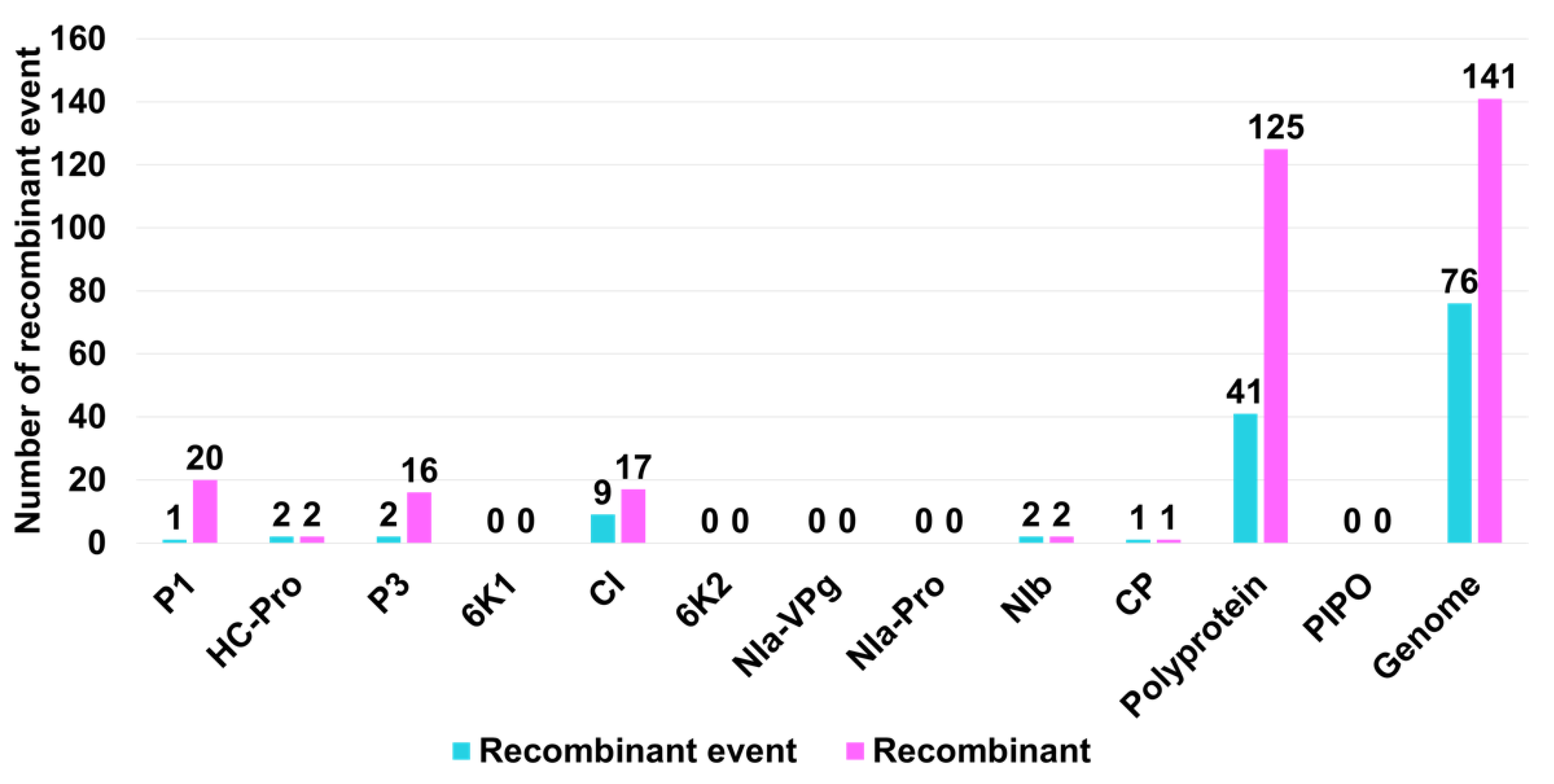
2.4. Recombination Analysis of 143 SMV Isolates/Strains
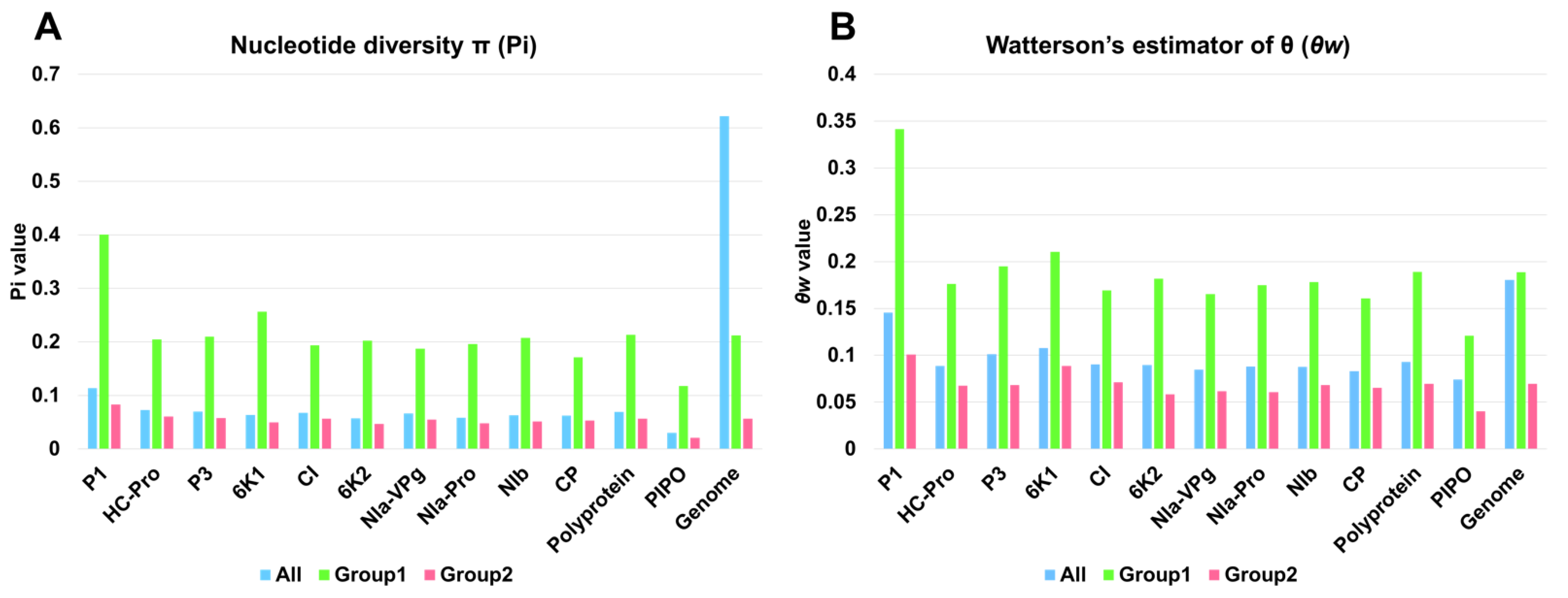
2.5. Genetic Diversity Analysis of 143 SMV Genomes
2.6. Estimation of the Nonsynonymous to Synonymous Rate Ratio of all SMV Protein Sequences
3. Discussion
4. Materials and Methods
4.1. Soybean Germplasms
4.2. Total RNA Extraction and Library Zpreparation
4.3. RNA-Sequencing and Viral Genome Assembly
4.4. Collection of SMV Genomes from GenBank
4.5. Generation of Phylogenetic Trees
4.6. Recombination Analysis and Network Tree for 143 SMV Genomes
4.7. Analyses of Genetic Diversity and Selection Pressure for Individual SMV Proteins
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, J.-Z.; Fang, Y.; Pang, H. The current status of the soybean-Soybean mosaic virus (SMV) pathosystem. Front. Microbiol. 2016, 7, 1906. [Google Scholar] [CrossRef] [PubMed]
- Hajimorad, M.; Domier, L.; Tolin, S.; Whitham, S.; Saghai Maroof, M. Soybean mosaic virus: A successful potyvirus with a wide distribution but restricted natural host range. Mol. Plant Pathol. 2018, 19, 1563–1579. [Google Scholar] [CrossRef]
- Domier, L.L.; Hobbs, H.A.; McCoppin, N.K.; Bowen, C.R.; Steinlage, T.A.; Chang, S.; Wang, Y.; Hartman, G.L. Multiple loci condition seed transmission of Soybean mosaic virus (SMV) and SMV-induced seed coat mottling in soybean. Phytopathology 2011, 101, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Wrather, J.; Anderson, T.; Arsyad, D.; Tan, Y.; Ploper, L.D.; Porta-Puglia, A.; Ram, H.; Yorinori, J. Soybean disease loss estimates for the top ten soybean-producing counries in 1998. Can. J. Plant Pathol. 2001, 23, 115–121. [Google Scholar] [CrossRef]
- Domier, L.L.; Steinlage, T.A.; Hobbs, H.A.; Wang, Y.; Herrera-Rodriguez, G.; Haudenshield, J.S.; McCoppin, N.K.; Hartman, G.L. Similarities in seed and aphid transmission among Soybean mosaic virus isolates. Plant Dis. 2007, 91, 546–550. [Google Scholar] [CrossRef] [PubMed]
- Jossey, S.; Hobbs, H.A.; Domier, L.L. Role of Soybean mosaic virus–encoded proteins in seed and aphid transmission in soybean. Phytopathology 2013, 103, 941–948. [Google Scholar] [CrossRef] [PubMed]
- Kiihl, R.A.; Hartwig, E. Inheritance of reaction to Soybean mosaic virus in soybeans 1. Crop. Sci. 1979, 19, 372–375. [Google Scholar] [CrossRef]
- Hayes, A.; Jeong, S.; Gore, M.; Yu, Y.; Buss, G.; Tolin, S.; Maroof, M.S. Recombination within a nucleotide-binding-site/leucine-rich-repeat gene cluster produces new variants conditioning resistance to Soybean mosaic virus in soybeans. Genetics 2004, 166, 493–503. [Google Scholar] [CrossRef]
- Jain, R.; McKern, N.M.; Tolin, S.; Hill, J.; Barnett, O.; Tosic, M.; Ford, R.; Beachy, R.; Yu, M.; Ward, C. Confirmation that fourteen potyvirus isolates from soybean are strains of one virus by comparing coat protein peptide profiles. Phytopathology 1992, 82, 294–299. [Google Scholar] [CrossRef]
- Jayaram, C.; Hill, J.H.; Miller, W.A. Nucleotide sequences of the coat protein genes of two aphid-transmissible strains of Soybean mosaic virus. J. Gen. Virol. 1991, 72, 1001–1003. [Google Scholar] [CrossRef]
- Jayaram, C.; Hill, J.H.; Miller, W.A. Complete nucleotide sequences of two Soybean mosaic virus strains differentiated by response of soybean containing the Rsv resistance gene. J. Gen. Virol. 1992, 73, 2067–2077. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.-K.; Ohshima, K.; Lee, H.-G.; Son, M.; Choi, H.-S.; Lee, S.-H.; Sohn, S.-H.; Kim, K.-H. Molecular variability and genetic structure of the population of Soybean mosaic virus based on the analysis of complete genome sequences. Virology 2009, 393, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.-C.; Shao, Z.-Q.; Ma, F.-F.; Wu, P.; Wu, X.-Y.; Xie, Z.-Y.; Yu, D.-Y.; Cheng, H.; Liu, Z.-H.; Jiang, Z.-F. The evolution of Soybean mosaic virus: An updated analysis by obtaining 18 new genomic sequences of Chinese strains/isolates. Virus Res. 2015, 208, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Ahangaran, A.; Habibi, M.K.; Mohammadi, G.-H.M.; Winter, S.; García-Arenal, F. Analysis of Soybean mosaic virus genetic diversity in Iran allows the characterization of a new mutation resulting in overcoming Rsv4-resistance. J. Gen. Virol. 2013, 94, 2557–2568. [Google Scholar] [CrossRef]
- Hajimorad, M.; Eggenberger, A.; Hill, J. Evolution of Soybean mosaic virus-G7 molecularly cloned genome in Rsv1-genotype soybean results in emergence of a mutant capable of evading Rsv1-mediated recognition. Virology 2003, 314, 497–509. [Google Scholar] [CrossRef]
- Massart, S.; Olmos, A.; Jijakli, H.; Candresse, T. Current impact and future directions of high throughput sequencing in plant virus diagnostics. Virus Res. 2014, 188, 90–96. [Google Scholar] [CrossRef]
- Nakasato, K.; Fujioka, S.; Sugawara, Y.; Ono, T.; Nishio, T.; Tsuda, S. First detection of two potyviruses, uraria mosaic virus and passiflora mosaic virus Y, from passionfruit in Japan. J. Gen. Plant Pathol. 2020, 86, 401–404. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Jeffares, D.C.; Tomiczek, B.; Sojo, V.; Reis, M.d. A beginners guide to estimating the non-synonymous to synonymous rate ratio of all protein-coding genes in a genome. In Parasite Genomics Protocols; Springer: Cham, Switzerland, 2015; pp. 65–90. [Google Scholar]
- Domingo, E.; Perales, C. Viral quasispecies. PLoS Genet. 2019, 15, e1008271. [Google Scholar] [CrossRef]
- Bowers, G., Jr.; Goodman, R. Soybean mosaic virus: Infection of soybean seed parts and seed transmission. Phytopathology 1979, 69, 569–572. [Google Scholar] [CrossRef]
- Jo, Y.; Choi, H.; Bae, M.; Kim, S.-M.; Kim, S.-L.; Lee, B.C.; Cho, W.K.; Kim, K.-H. De novo genome assembly and single nucleotide variations for Soybean mosaic virus using soybean seed transcriptome data. Plant Pathol. J. 2017, 33, 478. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Losada, M.; Arenas, M.; Galán, J.C.; Palero, F.; González-Candelas, F. Recombination in viruses: Mechanisms, methods of study, and evolutionary consequences. Infect. Genet. Evol. 2015, 30, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Gagarinova, A.G.; Babu, M.; Strömvik, M.V.; Wang, A. Recombination analysis of Soybean mosaic virus sequences reveals evidence of RNA recombination between distinct pathotypes. Virol. J. 2008, 5, 143. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Lin, J.; Zheng, G.; Zhang, M.; Zhi, H. Recombinant Soybean mosaic virus is prevalent in Chinese soybean fields. Arch. Virol. 2014, 159, 1793–1796. [Google Scholar] [CrossRef]
- Cho, E.-K.; Goodman, R.M. Strains of Soybean mosaic virus: Classification based on virulence in resistant soybean cultivars. Phytopathology 1979, 69, 467–470. [Google Scholar] [CrossRef]
- Li, K.; Yang, Q.; Zhi, H.; Gai, J. Identification and distribution of Soybean mosaic virus strains in southern China. Plant Dis. 2010, 94, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Wenqing, L.; Minghou, Z.; Peiwen, W.; Shuyi, X. Classification and distribution of strains of Soybean mosaic virus in northeast China. Acta Phytophylacica Sin. 1985, 15, 225–229. [Google Scholar]
- Gao, L.; Wu, Y.; An, J.; Huang, W.; Liu, X.; Xue, Y.; Luan, X.; Lin, F.; Sun, L. Pathogenicity and genome-wide sequence analysis reveals relationships between Soybean mosaic virus strains. Arch. Virol. 2022, 167, 517–529. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Shan, S.; Huang, Y.; Jiang, C.; Zhang, H.; Li, Y.; Chen, J.; Wei, Z.; Sun, Z. The hypervariable N-terminal of Soybean mosaic virus P1 protein influences its pathogenicity and host defense responses. Phytopathol. Res. 2022, 4, 10. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Wheeler, D.L.; Chappey, C.; Lash, A.E.; Leipe, D.D.; Madden, T.L.; Schuler, G.D.; Tatusova, T.A.; Rapp, B.A. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2000, 28, 10–14. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]



| Protein | Sites | S | Eta | H | Hd | Pi | θw | Tajima’s D |
|---|---|---|---|---|---|---|---|---|
| P1 | 1436 | 393 | 641 | 99 | 0.98 ± 0.003 | 0.11 ± 0.01 | 0.14 ± 0.03 | −1.71 |
| HC-Pro | 1374 | 668 | 948 | 124 | 0.99 ± 0.001 | 0.07 ± 0.005 | 0.08 ± 0.02 | −1.38 |
| P3 | 1041 | 581 | 808 | 126 | 0.99 ± 0.001 | 0.06 ± 0.005 | 0.1 ± 0.02 | −1.65 |
| 6K1 | 156 | 93 | 137 | 76 | 0.97 ± 0.005 | 0.06 ± 0.006 | 0.1 ± 0.02 | −1.92 |
| CI | 1902 | 949 | 1335 | 131 | 0.99 ± 0.001 | 0.06 ± 0.004 | 0.09 ± 0.02 | −1.55 |
| 6K2 | 159 | 79 | 114 | 66 | 0.95 ± 0.01 | 0.05 ± 0.004 | 0.08 ± 0.02 | −1.78 |
| NIa-VPg | 576 | 264 | 360 | 103 | 0.99 ± 0.002 | 0.06 ± 0.004 | 0.08 ± 0.01 | −1.38 |
| NIa-Pro | 729 | 335 | 461 | 112 | 0.99 ± 0.001 | 0.05 ± 0.004 | 0.08 ± 0.02 | −1.70 |
| NIb | 1551 | 648 | 894 | 125 | 0.99 ± 0.001 | 0.06 ± 0.004 | 0.08 ± 0.02 | −1.58 |
| CP | 873 | 365 | 483 | 118 | 0.99 ± 0.001 | 0.06 ± 0.004 | 0.08 ± 0.01 | −1.41 |
| Polyprotein | 9803 | 4375 | 6181 | 142 | 0.99 ± 0.0008 | 0.06 ± 0.005 | 0.09 ± 0.02 | −1.57 |
| PIPO | 225 | 92 | 106 | 72 | 0.96 ± 0.01 | 0.02 ± 0.003 | 0.07 ± 0.01 | −2.07 |
| Genome | 10,044 | 8653 | 25,946 | 143 | 1.000 ± 0.0008 | 0.62 ± 0.01 | 0.18 ± 0.04 | 0.49 |
| Protein | Number of Sites | Average | Min | Max | Synonymous Site | Nonsynonymous Sites |
|---|---|---|---|---|---|---|
| P1 | 1436 | 1.066121 | 0 | 6.822368 | 99.16 | 356.84 |
| HC-Pro | 1374 | 0.050681 | 0 | 1.117647 | 294.73 | 1064.28 |
| P3 | 1041 | 0.08778 | 0 | 1.326087 | 217.4 | 817.6 |
| 6K1 | 156 | 0.062452 | 0 | 1.373193 | 30.8 | 125.2 |
| CI | 1902 | 0.0342 | 0 | 1.173913 | 431.7 | 1467.3 |
| 6K2 | 159 | 0.023262 | 0 | 0.754717 | 32.24 | 26.76 |
| Nia-VPg | 576 | 0.025513 | 0 | 0.560976 | 121.22 | 439.78 |
| Nia-Pro | 729 | 0.012687 | 0 | 0.823529 | 147.79 | 536.21 |
| Nib | 1551 | 0.04367 | 0 | 2.2 | 288.16 | 1046.84 |
| CP | 873 | 0.0326 | 0 | 0.830508 | 169.69 | 619.31 |
| Polyprotein | 9803 | 1.568258 | 0 | 10 | 1702.53 | 6721.47 |
| PIPO | 225 | 0.429524 | 0 | 4.219048 | 47.26 | 174.74 |
| Genome | 10,044 | 1.004237 | 0 | 3.685714 | 1776.3 | 6854.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, H.; Jo, Y.; Choi, S.Y.; Kim, S.-M.; Choi, Y.M.; Hong, J.-S.; Lee, B.C.; Cho, W.K. Evolution and Phylogeny of Soybean Mosaic Virus Based on 143 Complete Genomes. Int. J. Mol. Sci. 2023, 24, 22. https://doi.org/10.3390/ijms24010022
Choi H, Jo Y, Choi SY, Kim S-M, Choi YM, Hong J-S, Lee BC, Cho WK. Evolution and Phylogeny of Soybean Mosaic Virus Based on 143 Complete Genomes. International Journal of Molecular Sciences. 2023; 24(1):22. https://doi.org/10.3390/ijms24010022
Chicago/Turabian StyleChoi, Hoseong, Yeonhwa Jo, Soo Yeon Choi, Sang-Min Kim, Yu Mi Choi, Jin-Sung Hong, Bong Choon Lee, and Won Kyong Cho. 2023. "Evolution and Phylogeny of Soybean Mosaic Virus Based on 143 Complete Genomes" International Journal of Molecular Sciences 24, no. 1: 22. https://doi.org/10.3390/ijms24010022
APA StyleChoi, H., Jo, Y., Choi, S. Y., Kim, S.-M., Choi, Y. M., Hong, J.-S., Lee, B. C., & Cho, W. K. (2023). Evolution and Phylogeny of Soybean Mosaic Virus Based on 143 Complete Genomes. International Journal of Molecular Sciences, 24(1), 22. https://doi.org/10.3390/ijms24010022








