A Novel Molecular Signature of Cancer-Associated Fibroblasts Predicts Prognosis and Immunotherapy Response in Pancreatic Cancer
Abstract
1. Introduction
2. Results
2.1. α-SMA+ CAFs Accumulate in PC Tissues with Worse Prognoses
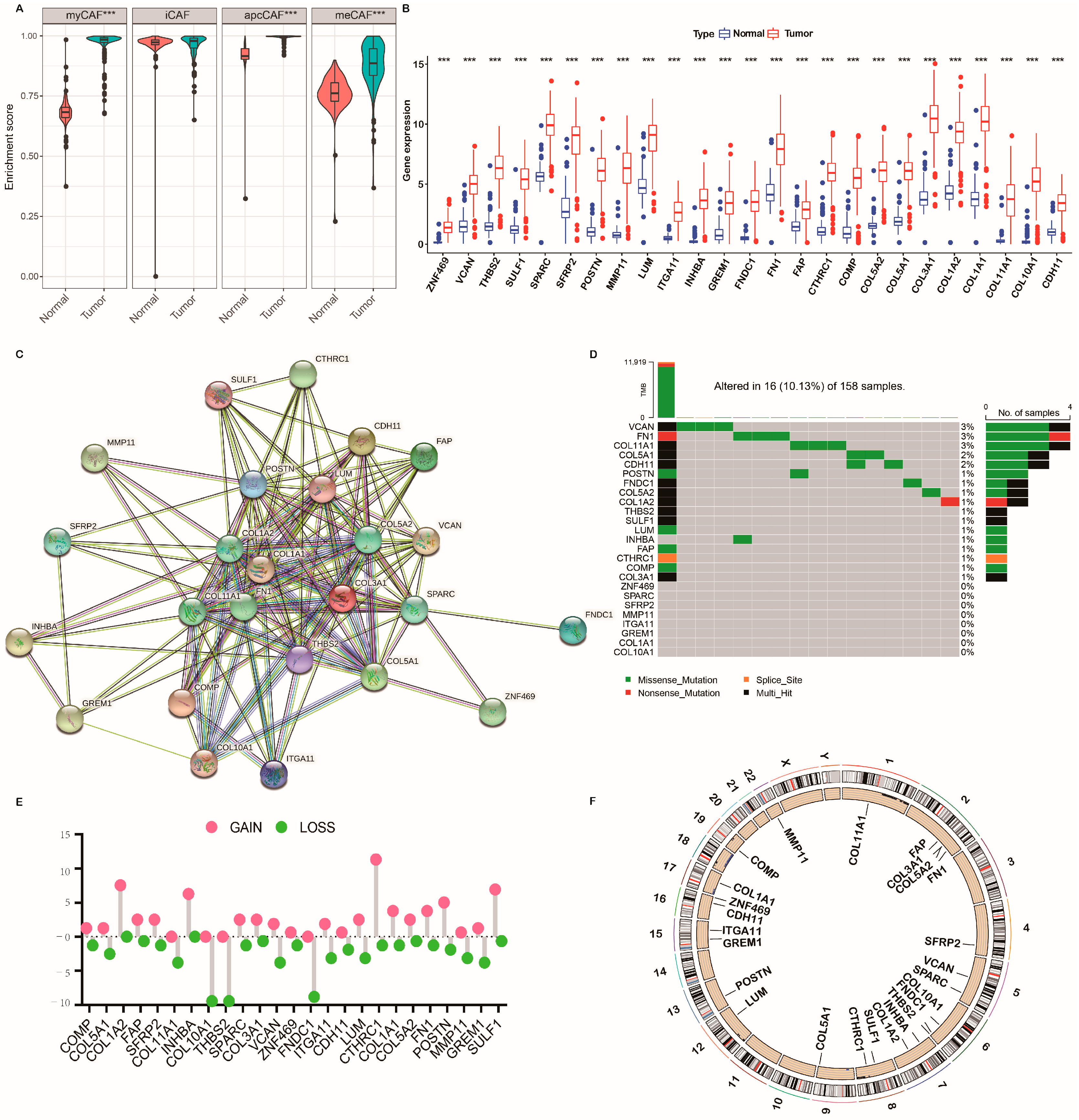
2.2. Genetic Mutation Landscape of CRGs in PC
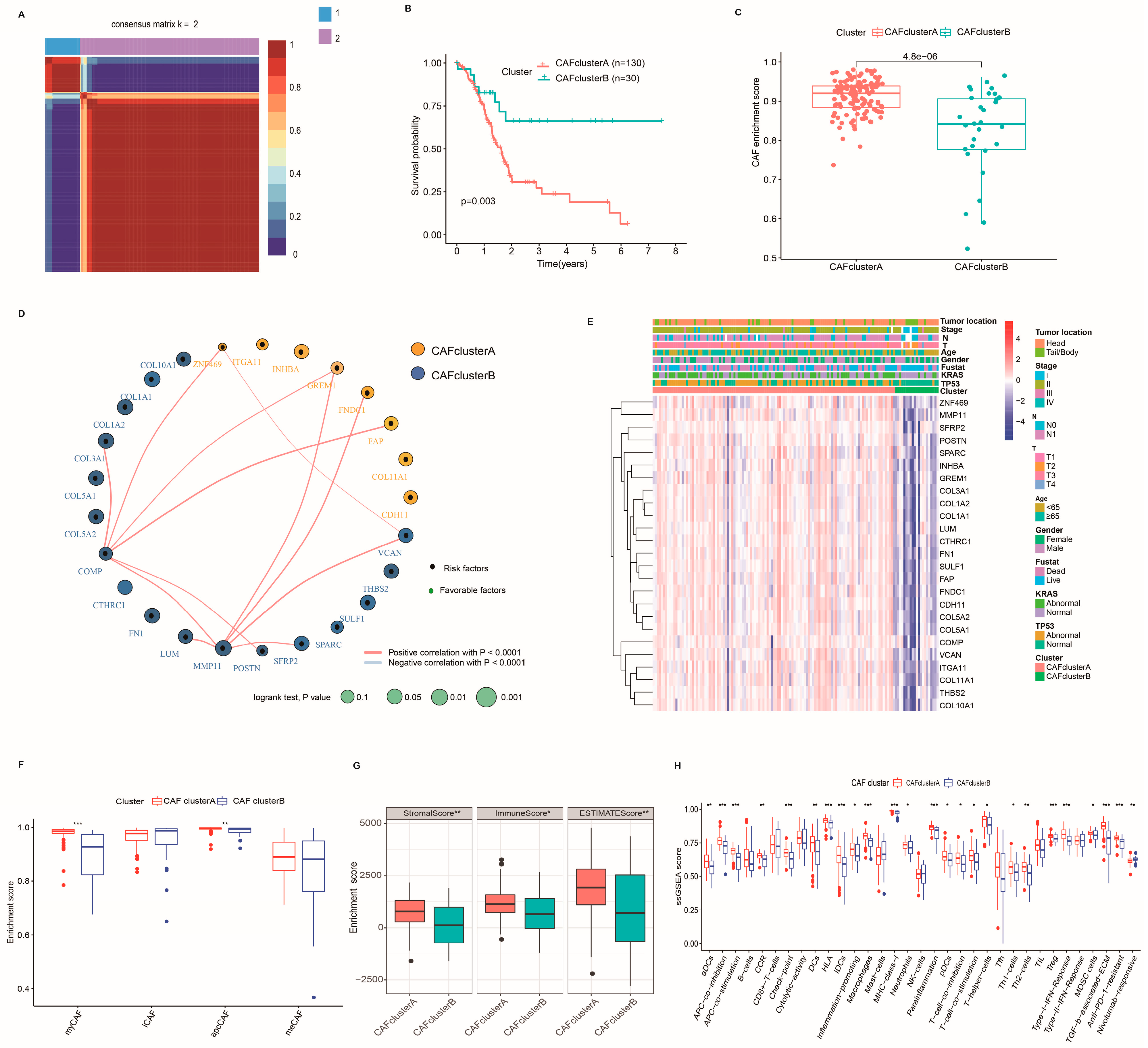
2.3. Identification of CAF Subtypes and Characteristics of the TME in PC
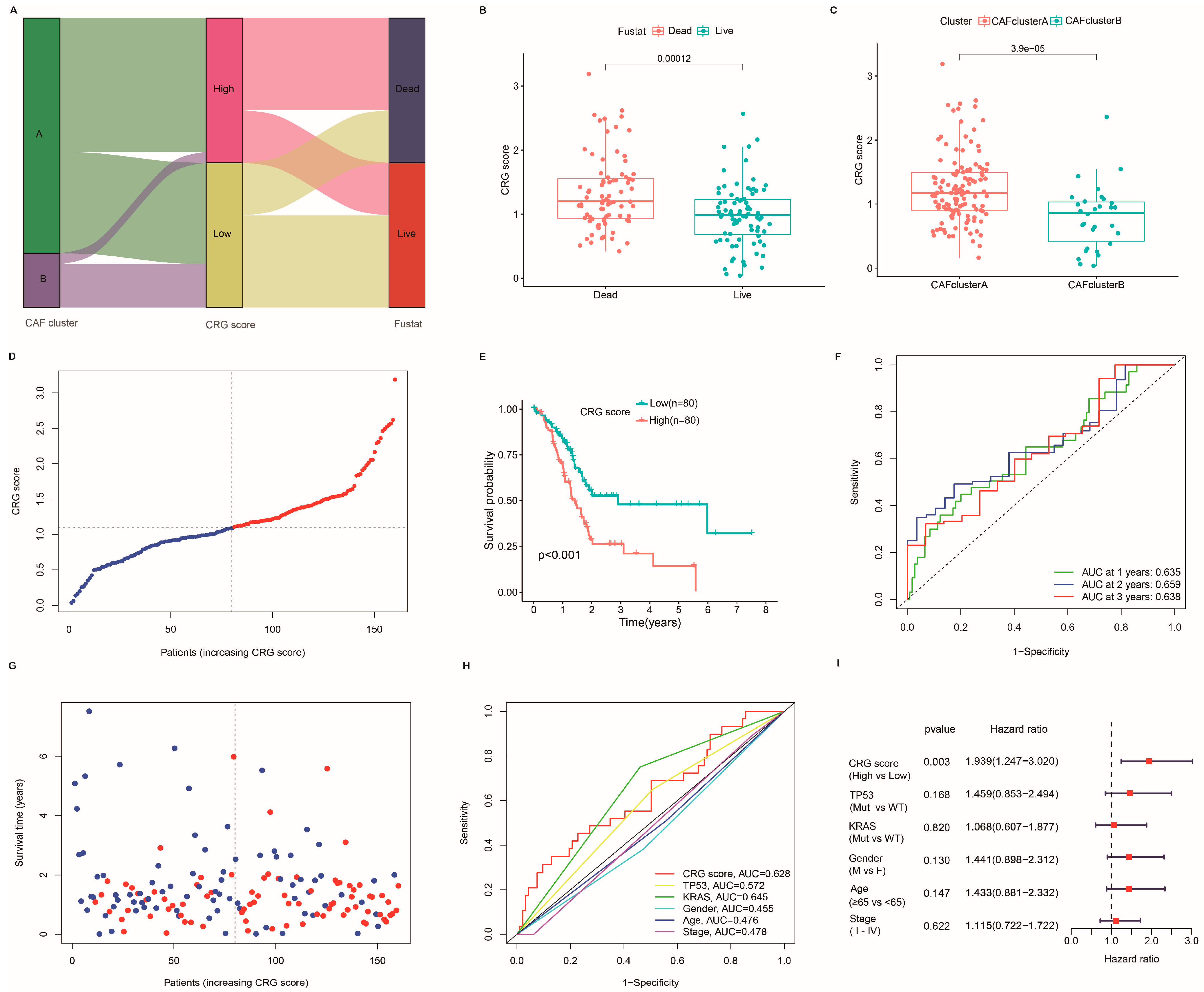
2.4. Establishment and Verification of the Prognostic CRG Score
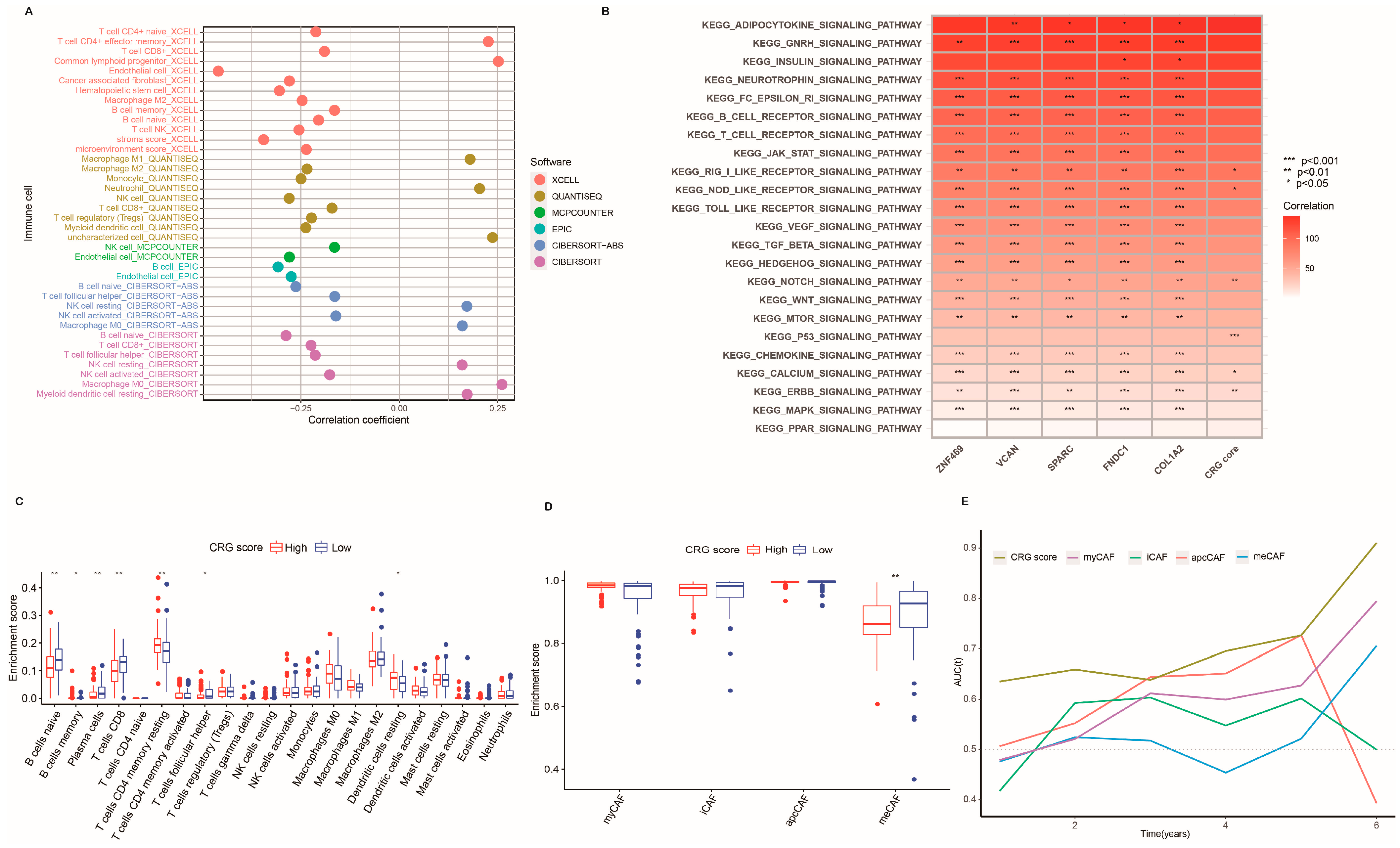
2.5. Characteristics of the TME and Function Enrichment in Distinct Subgroups
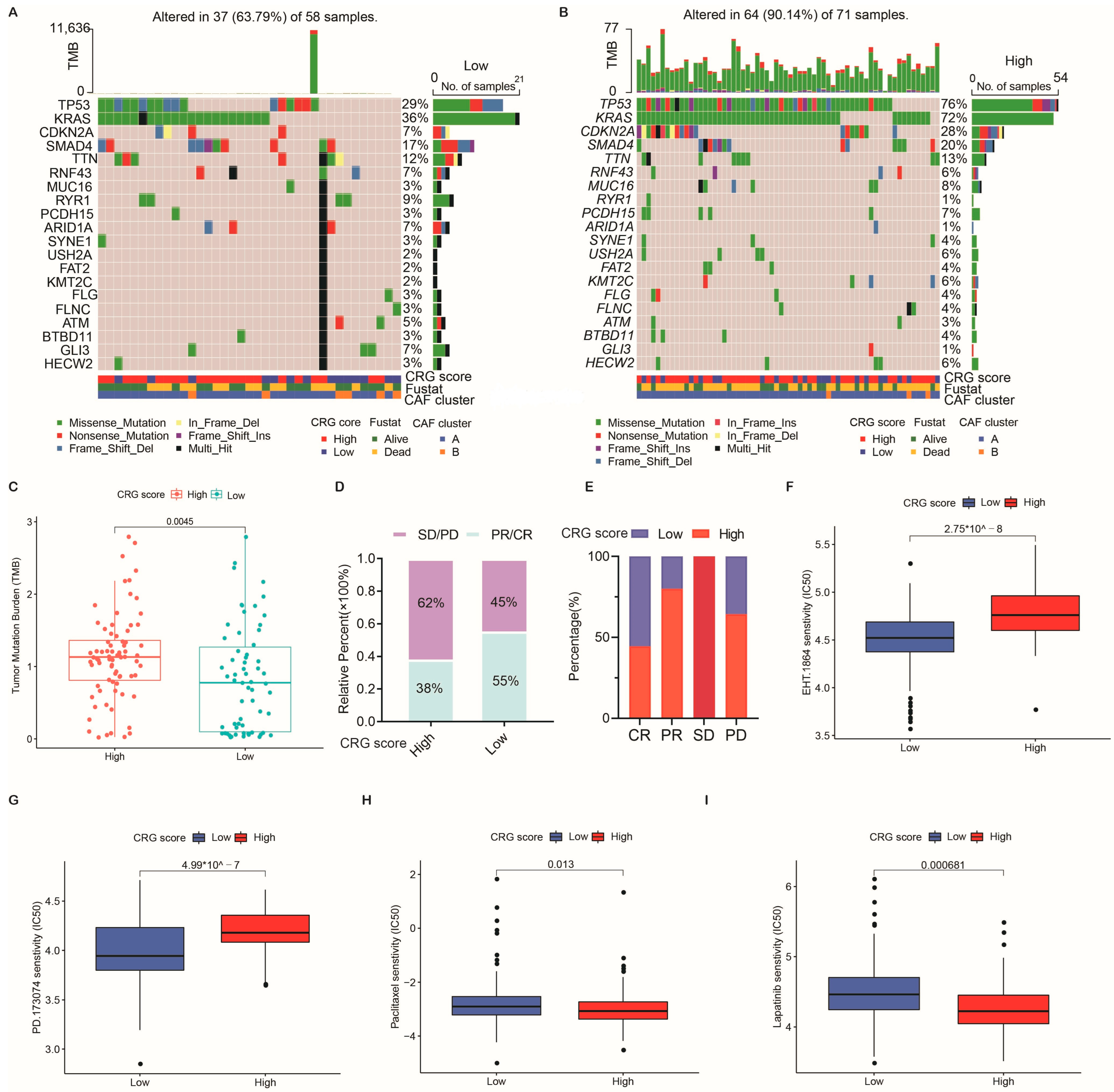
2.6. Association of the CRG Score with Tumor Mutation Burden (TMB) and Mutation
2.7. Clinical Outcomes and Drug Susceptibility Analysis
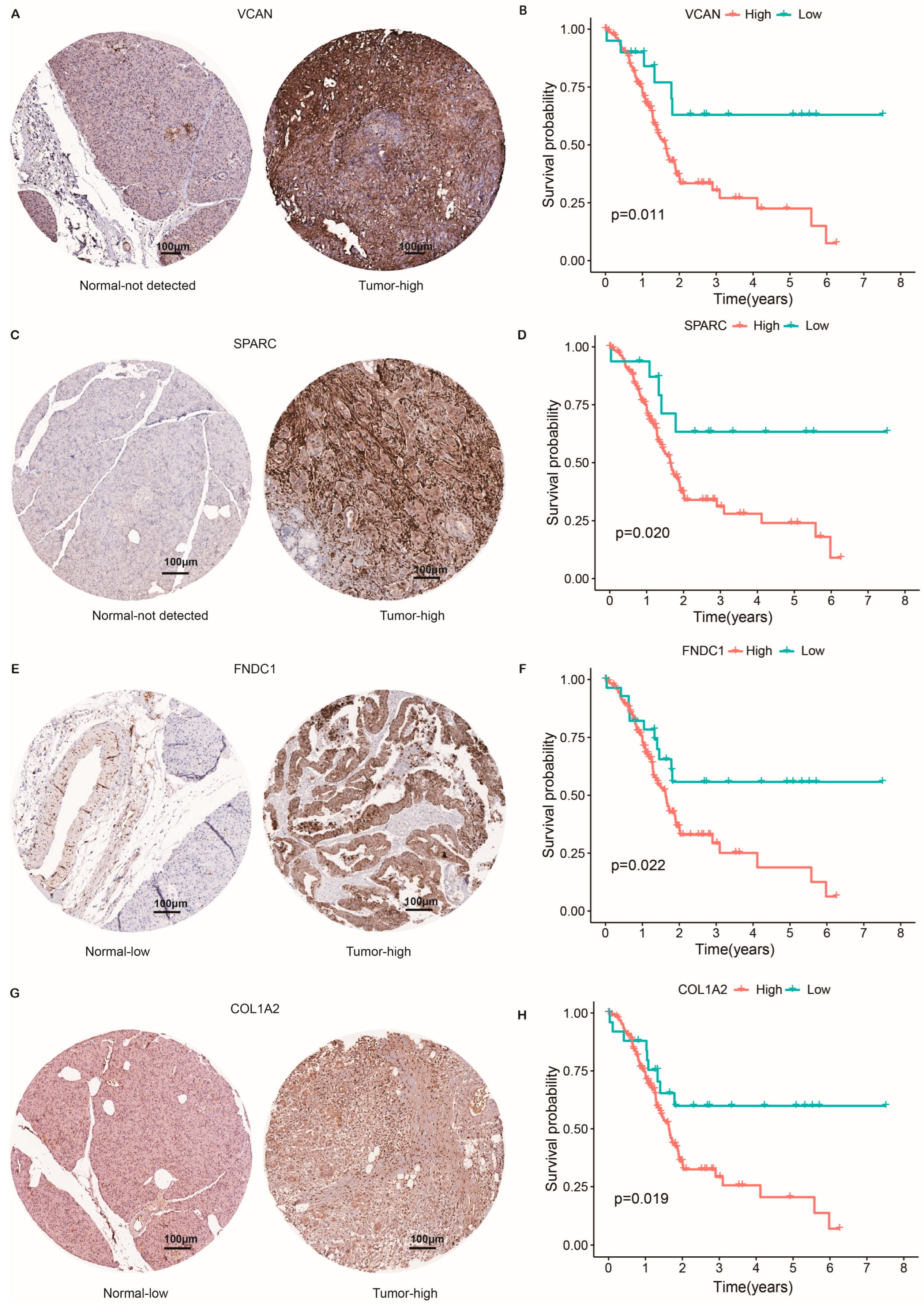
2.8. Protein Expression Level of CAF-Related Risk Genes and Survival Analysis
3. Discussion
4. Materials and Methods
4.1. Human Tissue Specimens
4.2. Cell Lines and Mouse Pancreatic Cancer Model
4.3. Masson’s Trichrome Staining
4.4. Immunohistochemistry and Tissue Microarrays
4.5. Flow Cytometry Analysis
4.6. PC Transcriptome Data Sources
4.7. Consensus Clustering Analysis of CRGs
4.8. Correlations of Molecular Patterns with the Clinical Features and Prognosis of PC
4.9. Association of Molecular Subtypes with Tumor Immune Microenvironment of PC
4.10. Relationship of CAF Subtypes with Immunotherapy Responses in PC
4.11. Development of the CAF-Related Gene Risk Signature
4.12. Expression Level Validation of CAF-Related Risk Gene Expression
4.13. Identification of Immune Microenvironment Affected by CRGs
4.14. Correlation of CRG Score Signature with Signal Pathways, Tumor Mutation, and Chemosensitivity
4.15. Additional Bioinformatics and Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Helms, E.; Onate, M.K.; Sherman, M.H. Fibroblast Heterogeneity in the Pancreatic Tumor Microenvironment. Cancer Discov. 2020, 10, 648–656. [Google Scholar] [CrossRef] [PubMed]
- Neuzillet, C.; Tijeras-Raballand, A.; Ragulan, C.; Cros, J.; Patil, Y.; Martinet, M.; Erkan, M.; Kleeff, J.; Wilson, J.; Apte, M.; et al. Inter- and intra-tumoural heterogeneity in cancer-associated fibroblasts of human pancreatic ductal adenocarcinoma. J. Pathol. 2019, 248, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Erez, N.; Truitt, M.; Olson, P.; Hanahan, D. Cancer-Associated Fibroblasts Are Activated in Incipient Neoplasia to Orchestrate Tumor-Promoting Inflammation in an NF-kappa B-Dependent Manner. Cancer Cell 2010, 17, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Fearon, D.T. The Carcinoma-Associated Fibroblast Expressing Fibroblast Activation Protein and Escape from Immune Surveillance. Cancer Immunol. Res. 2014, 2, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Whittle, M.C.; Hingorani, S.R. Fibroblasts in Pancreatic Ductal Adenocarcinoma: Biological Mechanisms and Therapeutic Targets. Gastroenterology 2019, 156, 2085–2096. [Google Scholar] [CrossRef]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef]
- Kraman, M.; Bambrough, P.J.; Arnold, J.N.; Roberts, E.W.; Magiera, L.; Jones, J.O.; Gopinathan, A.; Tuveson, D.A.; Fearon, D.T. Suppression of Antitumor Immunity by Stromal Cells Expressing Fibroblast Activation Protein-alpha. Science 2010, 330, 827–830. [Google Scholar] [CrossRef]
- Costa, A.; Kieffer, Y.; Scholer-Dahirel, A.; Pelon, F.; Bourachot, B.; Cardon, M.; Sirven, P.; Magagna, I.; Fuhrmann, L.; Bernard, C.; et al. Fibroblast Heterogeneity and Immunosuppressive Environment in Human Breast Cancer. Cancer Cell 2018, 33, 463–479. [Google Scholar] [CrossRef]
- Moncada, R.; Barkley, D.; Wagner, F.; Chiodin, M.; Devlin, J.C.; Baron, M.; Hajdu, C.H.; Simeone, D.M.; Yanai, I. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat. Biotechnol. 2020, 38, 333–342. [Google Scholar] [CrossRef]
- Peng, J.Y.; Sun, B.F.; Chen, C.Y.; Zhou, J.Y.; Chen, Y.S.; Chen, H.; Liu, L.L.; Huang, D.; Jiang, J.L.; Cui, G.S.; et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019, 29, 725–738. [Google Scholar] [CrossRef]
- Elyada, E.; Bolisetty, M.; Laise, P.; Flynn, W.F.; Courtois, E.T.; Burkhart, R.A.; Teinor, J.A.; Belleau, P.; Biffi, G.; Lucito, M.S.; et al. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discov. 2019, 9, 1102–1123. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liang, Y.Y.; Xu, H.Y.; Zhang, X.; Mao, T.B.; Cui, J.J.; Yao, J.Y.; Wang, Y.C.; Jiao, F.; Xiao, X.Y.; et al. Single-cell analysis of pancreatic ductal adenocarcinoma identifies a novel fibroblast subtype associated with poor prognosis but better immunotherapy response. Cell Discov. 2021, 7, 36. [Google Scholar] [CrossRef] [PubMed]
- Toullec, A.; Gerald, D.; Despouy, G.; Bourachot, B.; Cardon, M.; Lefort, S.; Richardson, M.; Rigaill, G.; Parrini, M.-C.; Lucchesi, C.; et al. Oxidative stress promotes myofibroblast differentiation and tumour spreading. EMBO Mol. Med. 2010, 2, 211–230. [Google Scholar] [CrossRef] [PubMed]
- Benyahia, Z.; Dussault, N.; Cayol, M.; Sigaud, R.; Berenguer-Daize, C.; Delfino, C.; Tounsi, A.; Garcia, S.; Martin, P.-M.; Mabrouk, K.; et al. Stromal fibroblasts present in breast carcinomas promote tumor growth and angiogenesis through adrenomedullin secretion. Oncotarget 2017, 8, 15744–15762. [Google Scholar] [CrossRef]
- Feig, C.; Jones, J.O.; Kraman, M.; Wells, R.J.B.; Deonarine, A.; Chan, D.S.; Connell, C.M.; Roberts, E.W.; Zhao, Q.; Caballero, O.L.; et al. Targeting CXCL12 from FAP-expressing carcinomaassociated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 20212–20217. [Google Scholar] [CrossRef]
- Straussman, R.; Morikawa, T.; Shee, K.; Barzily-Rokni, M.; Qian, Z.R.; Du, J.; Davis, A.; Mongare, M.M.; Gould, J.; Frederick, D.T.; et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 2012, 487, 500–504. [Google Scholar] [CrossRef]
- Paulsson, J.; Ryden, L.; Strell, C.; Frings, O.; Tobin, N.P.; Fornander, T.; Bergh, J.; Landberg, G.; Stal, O.; Ostman, A. High expression of stromal PDGFR beta is associated with reduced benefit of tamoxifen in breast cancer. J. Pathol. Clin. Res. 2017, 3, 38–43. [Google Scholar] [CrossRef]
- Gillen, S.; Schuster, T.; Bueschenfelde, C.M.Z.; Friess, H.; Kleeff, J. Preoperative/Neoadjuvant Therapy in Pancreatic Cancer: A Systematic Review and Meta-analysis of Response and Resection Percentages. PLoS Med. 2010, 7, e1000267. [Google Scholar] [CrossRef]
- Hosein, A.N.; Brekken, R.A.; Maitra, A. Pancreatic cancer stroma: An update on therapeutic targeting strategies. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 487–505. [Google Scholar] [CrossRef]
- Oezdemir, B.C.; Pentcheva-Hoang, T.; Carstens, J.L.; Zheng, X.; Wu, C.-C.; Simpson, T.R.; Laklai, H.; Sugimoto, H.; Kahlert, C.; Novitskiy, S.V.; et al. Depletion of Carcinoma-Associated Fibroblasts and Fibrosis Induces Immunosuppression and Accelerates Pancreas Cancer with Reduced Survival. Cancer Cell 2014, 25, 719–734. [Google Scholar] [CrossRef]
- Yoshida, G.J. Regulation of heterogeneous cancer-associated fibroblasts: The molecular pathology of activated signaling pathways. J. Exp. Clin. Cancer Res. 2020, 39, 15. [Google Scholar] [CrossRef]
- Geng, X.L.; Chen, H.Z.; Zhao, L.; Hu, J.S.; Yang, W.B.; Li, G.Q.; Cheng, C.D.; Zhao, Z.J.; Zhang, T.; Li, L.; et al. Cancer-Associated Fibroblast (CAF) Heterogeneity and Targeting Therapy of CAFs in Pancreatic Cancer. Front. Cell Dev. Biol. 2021, 9, 655152. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Bai, X.; Wang, J.; Tang, X.-R.; Wu, D.-H.; Du, S.-S.; Du, X.-J.; Zhang, Y.-W.; Zhu, H.-B.; Fang, Y.; et al. Combination of TMB and CNA Stratifies Prognostic and Predictive Responses to Immunotherapy Across Metastatic Cancer. Clin. Cancer Res. 2019, 25, 7413–7423. [Google Scholar] [CrossRef] [PubMed]
- Moffitt, R.A.; Marayati, R.; Flate, E.L.; Volmar, K.E.; Loeza, S.G.H.; Hoadley, K.A.; Rashid, N.U.; Williams, L.A.; Eaton, S.C.; Chung, A.H.; et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 2015, 47, 1168–1178. [Google Scholar] [CrossRef]
- Junttila, M.R.; de Sauvage, F.J. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 2013, 501, 346–354. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, K.; Shahmoradgoli, M.; Martinez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Trevino, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef]
- Kalluri, R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 2016, 16, 582–598. [Google Scholar] [CrossRef]
- Mhaidly, R.; Mechta-Grigoriou, F. Fibroblast heterogeneity in tumor micro-environment: Role in immunosuppression and new therapies. In Seminars in Immunology; Academic Press: Cambridge, MA, USA, 2020; Volume 48. [Google Scholar] [CrossRef]
- Xie, J.D.; Zheng, S.Q.; Zou, Y.T.; Tang, Y.H.; Tian, W.W.; Wong, C.W.; Wu, S.; Ou, X.Q.; Zhao, W.Z.; Cai, M.B.; et al. Turning up a new pattern: Identification of cancer-associated fibroblast-related clusters in TNBC. Front. Immunol. 2022, 13, 1022147. [Google Scholar] [CrossRef]
- Wang, F.; Wei, X.L.; Wang, F.H.; Xu, N.; Shen, L.; Dai, G.H.; Yuan, X.L.; Chen, Y.; Yang, S.J.; Shi, J.H.; et al. Safety, efficacy and tumor mutational burden as a biomarker of overall survival benefit in chemo-refractory gastric cancer treated with toripalimab, a PD-1 antibody in phase Ib/II clinical trial NCT02915432. Ann. Oncol. 2019, 30, 1479–1486. [Google Scholar] [CrossRef]
- Chalabi, M.; Fanchi, L.F.; Dijkstra, K.K.; Van den Berg, J.G.; Aalbers, A.G.; Sikorska, K.; Lopez-Yurda, M.; Grootscholten, C.; Beets, G.L.; Snaebjornsson, P.; et al. Neoadjuvant immunotherapy leads to pathological responses in MMR-proficient and MMR-deficient early-stage colon cancers. Nat. Med. 2020, 26, 566–576. [Google Scholar] [CrossRef]
- Ribas, A.; Wolchok, J.D. Cancer immunotherapy using checkpoint blockade. Science 2018, 359, 1350–1355. [Google Scholar] [CrossRef]
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.R.; Gettinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Carvajal, R.D.; Sosman, J.A.; Atkins, M.B.; et al. Safety, Activity, and Immune Correlates of Anti-PD-1 Antibody in Cancer. N. Engl. J. Med. 2012, 366, 2443–2454. [Google Scholar] [CrossRef]
- O’Reilly, E.M.; Oh, D.-Y.; Dhani, N.; Renouf, D.J.; Lee, M.A.; Sun, W.; Fisher, G.; Hezel, A.; Chang, S.-C.; Vlahovic, G.; et al. Durvalumab With or Without Tremelimumab for Patients With Metastatic Pancreatic Ductal Adenocarcinoma: A Phase 2 Randomized Clinical Trial. Jama Oncol. 2019, 5, 1431–1438. [Google Scholar] [CrossRef]
- Kieffer, Y.; Hocine, H.R.; Gentric, G.; Pelon, F.; Bernard, C.; Bourachot, B.; Lameiras, S.; Albergante, L.; Bonneau, C.; Guyard, A.; et al. Single-Cell Analysis Reveals Fibroblast Clusters Linked to Immunotherapy Resistance in Cancer. Cancer Discov. 2020, 10, 1330–1351. [Google Scholar] [CrossRef]
- Collisson, E.A.; Bailey, P.; Chang, D.K.; Biankin, A.V. Molecular subtypes of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 207–220. [Google Scholar] [CrossRef]
- Samstein, R.M.; Lee, C.-H.; Shoushtari, A.N.; Hellmann, M.D.; Shen, R.; Janjigian, Y.Y.; Barron, D.A.; Zehir, A.; Jordan, E.J.; Omuro, A.; et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019, 51, 202–206. [Google Scholar] [CrossRef]
- Sun, R.; Shen, J.; Gao, Y.; Zhou, Y.; Yu, Z.; Hornicek, F.; Kan, Q.; Duan, Z. Overexpression of EZH2 is associated with the poor prognosis in osteosarcoma and function analysis indicates a therapeutic potential. Oncotarget 2016, 7, 38333–38346. [Google Scholar] [CrossRef]
- Seiler, M.; Huang, C.C.; Szalma, S.; Bhanot, G. ConsensusCluster: A Software Tool for Unsupervised Cluster Discovery in Numerical Data. Omics-J. Integr. Biol. 2010, 14, 109–113. [Google Scholar] [CrossRef]
- Sabah, A.; Tiun, S.; Sani, N.S.; Ayob, M.; Taha, A.Y. Enhancing web search result clustering model based on multiview multirepresentation consensus cluster ensemble (mmcc) approach. PLoS ONE 2021, 16, e0245264. [Google Scholar] [CrossRef]
- Rich, J.T.; Neely, J.G.; Paniello, R.C.; Voelker, C.C.J.; Nussenbaum, B.; Wang, E.W. A practical guide to understanding Kaplan-Meier curves. Otolaryngol.-Head Neck Surg. 2010, 143, 331–336. [Google Scholar] [CrossRef]
- Meng, Z.; Ren, D.; Zhang, K.; Zhao, J.; Jin, X.; Wu, H. Using ESTIMATE algorithm to establish an 8-mRNA signature prognosis prediction system and identify immunocyte infiltration-related genes in Pancreatic adenocarcinoma. Aging-Us 2020, 12, 5048–5070. [Google Scholar] [CrossRef]
- Huang, L.; Wu, C.; Xu, D.; Cui, Y.; Tang, J. Screening of Important Factors in the Early Sepsis Stage Based on the Evaluation of ssGSEA Algorithm and ceRNA Regulatory Network. Evol. Bioinform. 2021, 17, 11769343211058463. [Google Scholar] [CrossRef]
- Chen, D.S.; Mellman, I. Elements of cancer immunity and the cancer-immune set point. Nature 2017, 541, 321–330. [Google Scholar] [CrossRef]
- Chen, Y.P.; Lv, J.W.; Mao, Y.P.; Li, X.M.; Li, J.Y.; Wang, Y.Q.; Xu, C.; Li, Y.Q.; He, Q.M.; Yang, X.J.; et al. Unraveling tumour microenvironment heterogeneity in nasopharyngeal carcinoma identifies biologically distinct immune subtypes predicting prognosis and immunotherapy responses. Mol. Cancer 2021, 20, 14. [Google Scholar] [CrossRef]
- Mayakonda, A.; Lin, D.-C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef]
- Geeleher, P.; Cox, N.; Huang, R.S. pRRophetic: An R Package for Prediction of Clinical Chemotherapeutic Response from Tumor Gene Expression Levels. PLoS ONE 2014, 9, e107468. [Google Scholar] [CrossRef]
- Geeleher, P.; Cox, N.J.; Huang, R.S. Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol. 2014, 15, R47. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ge, W.; Yue, M.; Wang, Y.; Wang, Y.; Xue, S.; Shentu, D.; Mao, T.; Zhang, X.; Xu, H.; Li, S.; et al. A Novel Molecular Signature of Cancer-Associated Fibroblasts Predicts Prognosis and Immunotherapy Response in Pancreatic Cancer. Int. J. Mol. Sci. 2023, 24, 156. https://doi.org/10.3390/ijms24010156
Ge W, Yue M, Wang Y, Wang Y, Xue S, Shentu D, Mao T, Zhang X, Xu H, Li S, et al. A Novel Molecular Signature of Cancer-Associated Fibroblasts Predicts Prognosis and Immunotherapy Response in Pancreatic Cancer. International Journal of Molecular Sciences. 2023; 24(1):156. https://doi.org/10.3390/ijms24010156
Chicago/Turabian StyleGe, Weiyu, Ming Yue, Yanling Wang, Yongchao Wang, Shengbai Xue, Daiyuan Shentu, Tiebo Mao, Xiaofei Zhang, Haiyan Xu, Shumin Li, and et al. 2023. "A Novel Molecular Signature of Cancer-Associated Fibroblasts Predicts Prognosis and Immunotherapy Response in Pancreatic Cancer" International Journal of Molecular Sciences 24, no. 1: 156. https://doi.org/10.3390/ijms24010156
APA StyleGe, W., Yue, M., Wang, Y., Wang, Y., Xue, S., Shentu, D., Mao, T., Zhang, X., Xu, H., Li, S., Ma, J., Wang, L., & Cui, J. (2023). A Novel Molecular Signature of Cancer-Associated Fibroblasts Predicts Prognosis and Immunotherapy Response in Pancreatic Cancer. International Journal of Molecular Sciences, 24(1), 156. https://doi.org/10.3390/ijms24010156




