Saprophytic to Pathogenic Mycobacteria: Loss of Cytochrome P450s Vis a Vis Their Prominent Involvement in Natural Metabolite Biosynthesis
Abstract
1. Introduction
2. Results and Discussion
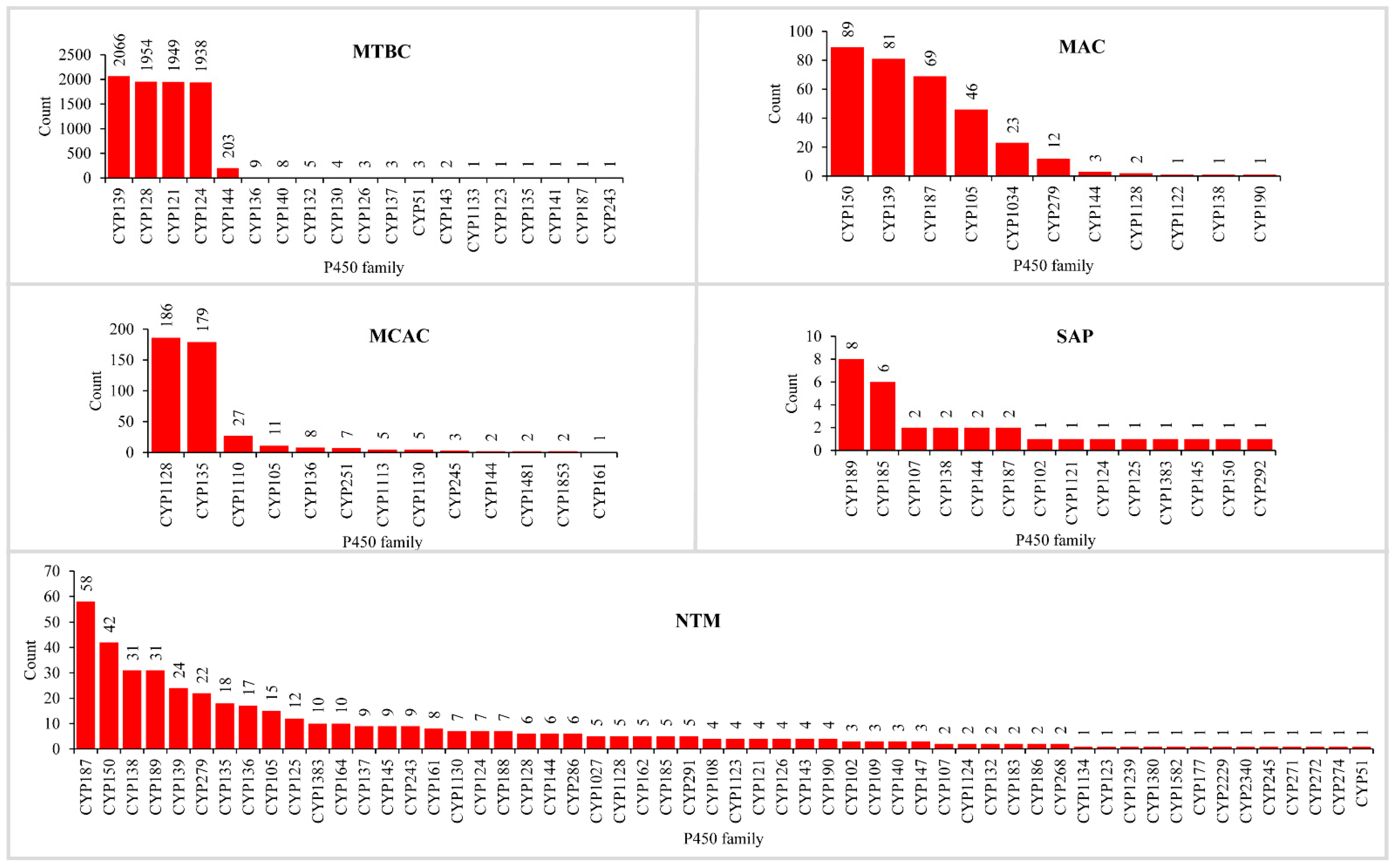
2.1. Saprophytic to Pathogen Life Style Led to the Loss of P450s in Mycobacterial Species
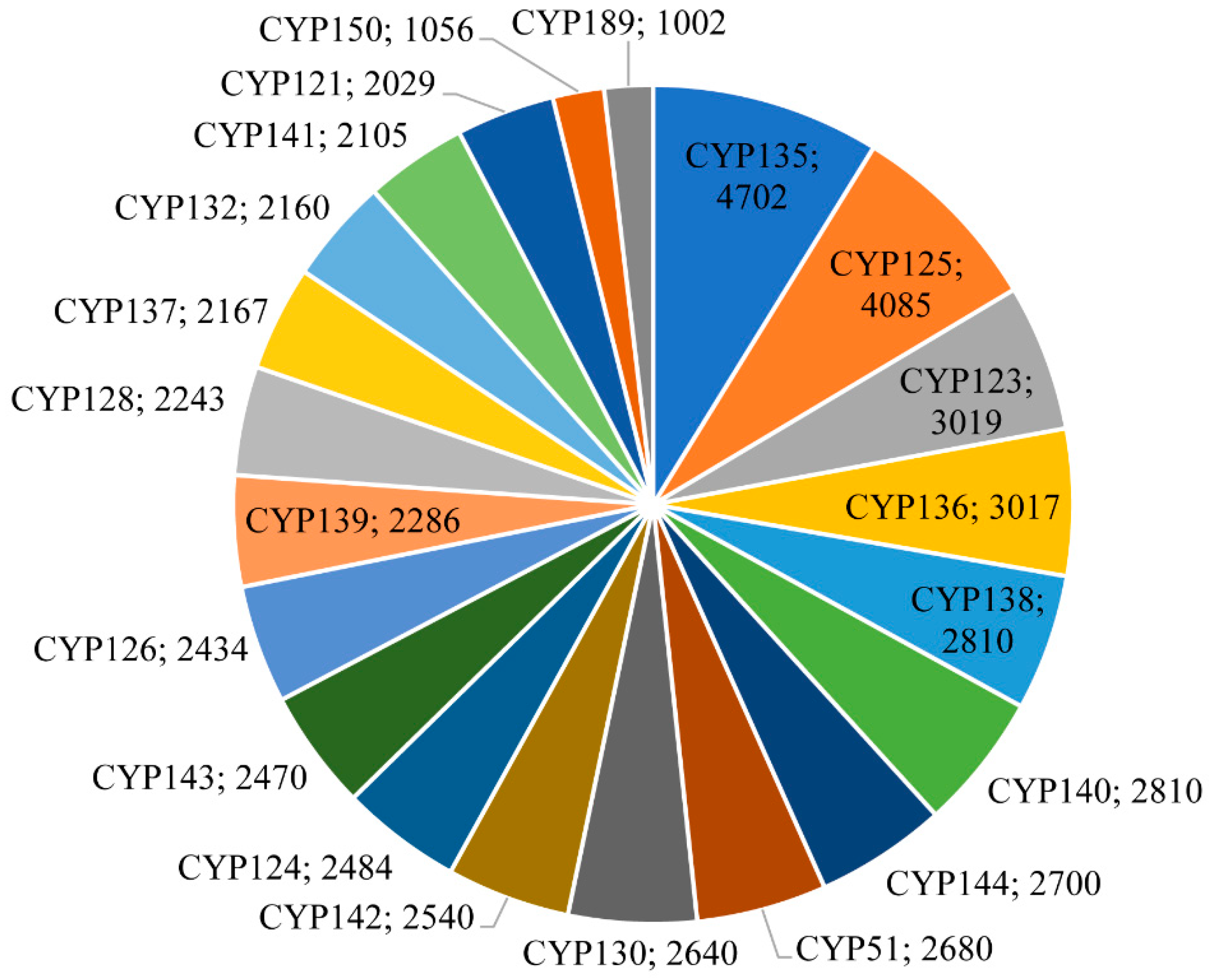
2.2. P450 Family and Subfamily Blooming/Expansion in Mycobacterial Species
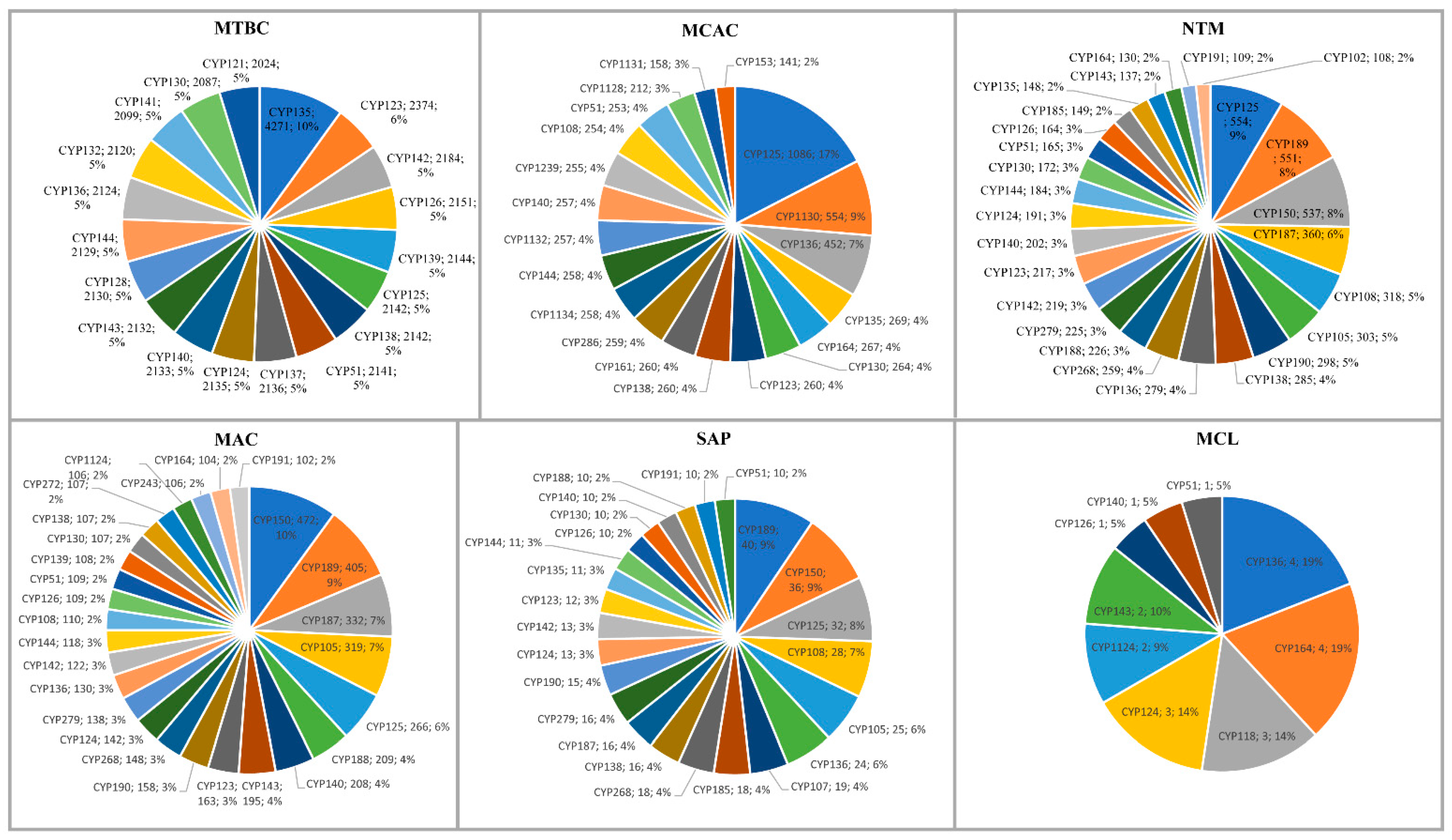
2.3. Different Mycobacterial Categories Have Distinct P450 Profiles
2.4. CYP121, CYP124, and CYP128 P450s Are Part of the Same Biosynthetic Gene Cluster
2.5. More P450s Are Involved in Natural Metabolite Biosynthesis in Pathogenic Mycobacterial Species
3. Materials and Methods
3.1. Species and Their Genome Database Information
3.2. Grouping of Mycobacterial Species
3.3. Genome Data Mining and Annotation of P450s
3.4. Identification of P450s Part of BGCs
3.5. P450 Key Features Analysis
3.6. Comparative Analysis of P450s and BGCs Data
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yamazaki, H. Fifty Years of Cytochrome P450 Research; Springer Japan: Tokyo, Japan, 2014; p. IX. [Google Scholar]
- McLean, K.J.; Leys, D.; Munro, A.W. Microbial cytochrome P450s. In Cytochrome P450: Structure, Mechanism, and Biochemistry, 4th ed.; Oritz de Montellano, P.R., Ed.; Springer International Publishing: Berlin/Heidelberg, Germany, 2015; Chapter 6; pp. 261–407. [Google Scholar]
- Nelson, D.R. Cytochrome P450 diversity in the tree of life. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Garfinkel, D. Studies on pig liver microsomes. I. Enzymic and pigment composition of different microsomal fractions. Arch. Biochem. Biophys. 1958, 77, 493–509. [Google Scholar] [CrossRef] [PubMed]
- Klingenberg, M. Pigments of rat liver microsomes. Arch. Biochem. Biophys. 1958, 75, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Omura, T. Recollection of the early years of the research on cytochrome P450. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2011, 87, 617–640. [Google Scholar] [CrossRef]
- Omura, T.; Sato, R. A new cytochrome in liver microsomes. J. Biol. Chem. 1962, 237, 1375–1376. [Google Scholar] [CrossRef]
- White, R.E.; Coon, M.J. Oxygen activation by cytochrome P-450. Annu. Rev. Biochem. 1980, 49, 315–356. [Google Scholar] [CrossRef]
- Bernhardt, R. Cytochromes P450 as versatile biocatalysts. J. Biotechnol. 2006, 124, 128–145. [Google Scholar] [CrossRef]
- Sono, M.; Roach, M.P.; Coulter, E.D.; Dawson, J.H. Heme-containing oxygenases. Chem. Rev. 1996, 96, 2841–2888. [Google Scholar] [CrossRef]
- Yan, Y.; Wu, J.; Hu, G.; Gao, C.; Guo, L.; Chen, X.; Liu, L.; Song, W. Current state and future perspectives of cytochrome P450 enzymes for C–H and C= C oxygenation. Synth. Syst. Biotechnol. 2022, 7, 887–899. [Google Scholar] [CrossRef]
- Li, Z.; Jiang, Y.; Guengerich, F.P.; Ma, L.; Li, S.; Zhang, W. Engineering cytochrome P450 enzyme systems for biomedical and biotechnological applications. J. Biol. Chem. 2020, 295, 833–849. [Google Scholar] [CrossRef]
- Urlacher, V.B.; Girhard, M. Cytochrome P450 Monooxygenases in Biotechnology and Synthetic Biology. Trends Biotechnol. 2019, 37, 882–897. [Google Scholar] [CrossRef] [PubMed]
- Girvan, H.M.; Munro, A.W. Applications of microbial cytochrome P450 enzymes in biotechnology and synthetic biology. Curr. Opin. Chem. Biol. 2016, 31, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Kelly, S.L.; Kelly, D.E. Microbial cytochromes P450: Biodiversity and biotechnology. Where do cytochromes P450 come from, what do they do and what can they do for us? Philos. Trans. R. Soc. London Ser. B Biol. Sci. 2013, 368, 20120476. [Google Scholar] [CrossRef] [PubMed]
- Lepesheva, G.I.; Friggeri, L.; Waterman, M.R. CYP51 as drug targets for fungi and protozoan parasites: Past, present and future. Parasitology 2018, 145, 1820–1836. [Google Scholar] [CrossRef]
- Jawallapersand, P.; Mashele, S.S.; Kovacic, L.; Stojan, J.; Komel, R.; Pakala, S.B.; Krasevec, N.; Syed, K. Cytochrome P450 monooxygenase CYP53 family in fungi: Comparative structural and evolutionary analysis and its role as a common alternative anti-fungal drug target. PLoS ONE 2014, 9, e107209. [Google Scholar] [CrossRef]
- Lamb, D.C.; Follmer, A.H.; Goldstone, J.V.; Nelson, D.R.; Warrilow, A.G.; Price, C.L.; True, M.Y.; Kelly, S.L.; Poulos, T.L.; Stegeman, J.J. On the occurrence of cytochrome P450 in viruses. Proc. Natl. Acad. Sci. USA 2019, 116, 12343–12352. [Google Scholar] [CrossRef]
- Nelson, D.R. Cytochrome P450 nomenclature. Methods Mol. Biol. 1998, 107, 15–24. [Google Scholar]
- Nelson, D.R. Cytochrome P450 nomenclature, 2004. Methods Mol. Biol. 2006, 320, 1–10. [Google Scholar]
- Nelson, D.R. The cytochrome p450 homepage. Hum. Genom. 2009, 4, 59–65. [Google Scholar] [CrossRef]
- Nelson, D.R.; Kamataki, T.; Waxman, D.J.; Guengerich, F.P.; Estabrook, R.W.; Feyereisen, R.; Gonzalez, F.J.; Coon, M.J.; Gunsalus, I.C.; Gotoh, O.; et al. The P450 superfamily: Update on new sequences, gene mapping, accession numbers, early trivial names of enzymes, and nomenclature. DNA Cell Biol. 1993, 12, 1–51. [Google Scholar] [CrossRef]
- Rudolf, J.D.; Chang, C.Y.; Ma, M.; Shen, B. Cytochromes P450 for natural product biosynthesis in Streptomyces: Sequence, structure, and function. Nat. Prod. Rep. 2017, 34, 1141–1172. [Google Scholar] [CrossRef] [PubMed]
- Podust, L.M.; Sherman, D.H. Diversity of P450 enzymes in the biosynthesis of natural products. Nat. Prod. Rep. 2012, 29, 1251–1266. [Google Scholar] [CrossRef] [PubMed]
- Greule, A.; Stok, J.E.; De Voss, J.J.; Cryle, M.J. Unrivalled diversity: The many roles and reactions of bacterial cytochromes P450 in secondary metabolism. Nat. Prod. Rep. 2018, 35, 757–791. [Google Scholar] [CrossRef]
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; de Bruijn, I.; Chooi, Y.H.; Claesen, J.; Coates, R.C.; et al. Minimum Information about a Biosynthetic Gene cluster. Nat. Chem. Biol. 2015, 11, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Khumalo, M.J.; Nzuza, N.; Padayachee, T.; Chen, W.; Yu, J.H.; Nelson, D.R.; Syed, K. Comprehensive Analyses of Cytochrome P450 Monooxygenases and Secondary Metabolite Biosynthetic Gene Clusters in Cyanobacteria. Int. J. Mol. Sci. 2020, 21, 656. [Google Scholar] [CrossRef]
- Malinga, N.A.; Nzuza, N.; Padayachee, T.; Syed, P.R.; Karpoormath, R.; Gront, D.; Nelson, D.R.; Syed, K. An Unprecedented Number of Cytochrome P450s Are Involved in Secondary Metabolism in Salinispora Species. Microorganisms 2022, 10, 871. [Google Scholar] [CrossRef]
- Msomi, N.N.; Padayachee, T.; Nzuza, N.; Syed, P.R.; Kryś, J.D.; Chen, W.; Gront, D.; Nelson, D.R.; Syed, K. In silico analysis of P450s and their role in secondary metabolism in the bacterial class Gammaproteobacteria. Molecules 2021, 26, 1538. [Google Scholar] [CrossRef]
- Msweli, S.; Chonco, A.; Msweli, L.; Syed, P.R.; Karpoormath, R.; Chen, W.; Gront, D.; Nkosi, B.V.Z.; Nelson, D.R.; Syed, K. Lifestyles Shape the Cytochrome P450 Repertoire of the Bacterial Phylum Proteobacteria. Int. J. Mol. Sci. 2022, 23, 5821. [Google Scholar] [CrossRef]
- Mthethwa, B.C.; Chen, W.; Ngwenya, M.L.; Kappo, A.P.; Syed, P.R.; Karpoormath, R.; Yu, J.H.; Nelson, D.R.; Syed, K. Comparative analyses of cytochrome P450s and those associated with secondary metabolism in Bacillus species. Int. J. Mol. Sci. 2018, 19, 3623. [Google Scholar] [CrossRef]
- Nkosi, B.V.Z.; Padayachee, T.; Gront, D.; Nelson, D.R.; Syed, K. Contrasting Health Effects of Bacteroidetes and Firmicutes Lies in Their Genomes: Analysis of P450s, Ferredoxins, and Secondary Metabolite Clusters. Int. J. Mol. Sci. 2022, 23, 5057. [Google Scholar] [CrossRef]
- Nzuza, N.; Padayachee, T.; Syed, P.R.; Kryś, J.D.; Chen, W.; Gront, D.; Nelson, D.R.; Syed, K. Ancient Bacterial Class Alphaproteobacteria Cytochrome P450 Monooxygenases Can Be Found in Other Bacterial Species. Int. J. Mol. Sci. 2021, 22, 5542. [Google Scholar] [CrossRef] [PubMed]
- Padayachee, T.; Nzuza, N.; Chen, W.; Nelson, D.R.; Syed, K. Impact of lifestyle on cytochrome P450 monooxygenase repertoire is clearly evident in the bacterial phylum Firmicutes. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Parvez, M.; Qhanya, L.B.; Mthakathi, N.T.; Kgosiemang, I.K.; Bamal, H.D.; Pagadala, N.S.; Xie, T.; Yang, H.; Chen, H.; Theron, C.W.; et al. Molecular evolutionary dynamics of cytochrome P450 monooxygenases across kingdoms: Special focus on mycobacterial P450s. Sci. Rep. 2016, 6, 33099. [Google Scholar] [CrossRef] [PubMed]
- Senate, L.M.; Tjatji, M.P.; Pillay, K.; Chen, W.; Zondo, N.M.; Syed, P.R.; Mnguni, F.C.; Chiliza, Z.E.; Bamal, H.D.; Karpoormath, R.; et al. Similarities, variations, and evolution of cytochrome P450s in Streptomyces versus Mycobacterium. Sci. Rep. 2019, 9, 3962. [Google Scholar] [CrossRef]
- Mnguni, F.C.; Padayachee, T.; Chen, W.; Gront, D.; Yu, J.-H.; Nelson, D.R.; Syed, K. More P450s are involved in secondary metabolite biosynthesis in Streptomyces compared to Bacillus, Cyanobacteria and Mycobacterium. Int. J. Mol. Sci. 2020, 21, 4814. [Google Scholar] [CrossRef] [PubMed]
- Ortiz de Montellano, P.R. Potential drug targets in the Mycobacterium tuberculosis cytochrome P450 system. J. Inorg. Biochem. 2018, 180, 235–245. [Google Scholar] [CrossRef]
- Van Beilen, J.B.; Holtackers, R.; Lüscher, D.; Bauer, U.; Witholt, B.; Duetz, W.A. Biocatalytic production of perillyl alcohol from limonene by using a novel Mycobacterium sp. cytochrome P450 alkane hydroxylase expressed in Pseudomonas putida. Appl. Environ. Microbiol. 2005, 71, 1737–1744. [Google Scholar] [CrossRef]
- Sogi, K.M.; Holsclaw, C.M.; Fragiadakis, G.K.; Nomura, D.K.; Leary, J.A.; Bertozzi, C.R. Biosynthesis and Regulation of Sulfomenaquinone, a Metabolite Associated with Virulence in Mycobacterium tuberculosis. ACS Infect. Dis. 2016, 2, 800–806. [Google Scholar] [CrossRef]
- Syed, P.R.; Chen, W.; Nelson, D.R.; Kappo, A.P.; Yu, J.H.; Karpoormath, R.; Syed, K. Cytochrome P450 Monooxygenase CYP139 Family Involved in the Synthesis of Secondary Metabolites in 824 Mycobacterial Species. Int. J. Mol. Sci. 2019, 20, 2690. [Google Scholar] [CrossRef]
- Quadri, L.E. Biosynthesis of mycobacterial lipids by polyketide synthases and beyond. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 179–211. [Google Scholar] [CrossRef]
- Ghazaei, C. Mycobacterium tuberculosis and lipids: Insights into molecular mechanisms from persistence to virulence. J. Res. Med. Sci. 2018, 23, 63. [Google Scholar] [CrossRef] [PubMed]
- Bansal-Mutalik, R.; Nikaido, H. Mycobacterial outer membrane is a lipid bilayer and the inner membrane is unusually rich in diacyl phosphatidylinositol dimannosides. Proc. Natl. Acad. Sci. USA 2014, 111, 4958–4963. [Google Scholar] [CrossRef] [PubMed]
- Van Wyk, R.; van Wyk, M.; Mashele, S.S.; Nelson, D.R.; Syed, K. Comprehensive comparative analysis of cholesterol catabolic genes/proteins in mycobacterial species. Int. J. Mol. Sci. 2019, 20, 1032. [Google Scholar] [CrossRef]
- Ngcobo, N.S.; Chiliza, Z.E.; Chen, W.; Yu, J.-H.; Nelson, D.R.; Tuszynski, J.A.; Preto, J.; Syed, K. Comparative Analysis, Structural Insights, and Substrate/Drug Interaction of CYP128A1 in Mycobacterium tuberculosis. Int. J. Mol. Sci. 2020, 21, 4816. [Google Scholar] [CrossRef] [PubMed]
- Belin, P.; Le Du, M.H.; Fielding, A.; Lequin, O.; Jacquet, M.; Charbonnier, J.B.; Lecoq, A.; Thai, R.; Courcon, M.; Masson, C.; et al. Identification and structural basis of the reaction catalyzed by CYP121, an essential cytochrome P450 in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2009, 106, 7426–7431. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, R.C.; Yang, Y.; Wang, Y.; Davis, I.; Liu, A. Substrate-assisted hydroxylation and O-demethylation in the peroxidase-like cytochrome P450 enzyme CYP121. ACS Catal. 2019, 10, 1628–1639. [Google Scholar] [CrossRef]
- Rajput, S.; McLean, K.J.; Poddar, H.; Selvam, I.R.; Nagalingam, G.; Triccas, J.A.; Levy, C.W.; Munro, A.W.; Hutton, C.A. Structure–activity relationships of cyclo (L-tyrosyl-L-tyrosine) derivatives binding to Mycobacterium tuberculosis CYP121: Iodinated analogues promote shift to high-spin adduct. J. Med. Chem. 2019, 62, 9792–9805. [Google Scholar] [CrossRef]
- Johnston, J.B.; Kells, P.M.; Podust, L.M.; Ortiz de Montellano, P.R. Biochemical and structural characterization of CYP124: A methyl-branched lipid omega-hydroxylase from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2009, 106, 20687–20692. [Google Scholar] [CrossRef]
- Johnston, J.B.; Ouellet, H.; Ortiz de Montellano, P.R. Functional redundancy of steroid C26-monooxygenase activity in Mycobacterium tuberculosis revealed by biochemical and genetic analyses. J. Biol. Chem. 2010, 285, 36352–36360. [Google Scholar] [CrossRef]
- Chen, I.-M.A.; Chu, K.; Palaniappan, K.; Ratner, A.; Huang, J.; Huntemann, M.; Hajek, P.; Ritter, S.; Varghese, N.; Seshadri, R. The IMG/M data management and analysis system v. 6.0: New tools and advanced capabilities. Nucleic Acids Res. 2021, 49, D751–D763. [Google Scholar] [CrossRef]
- Ventura, M.; Canchaya, C.; Tauch, A.; Chandra, G.; Fitzgerald, G.F.; Chater, K.F.; van Sinderen, D. Genomics of Actinobacteria: Tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 2007, 71, 495–548. [Google Scholar] [CrossRef] [PubMed]
- Tortoli, E. Phylogeny of the genus Mycobacterium: Many doubts, few certainties. Infect. Genet. Evol. 2012, 12, 827–831. [Google Scholar] [CrossRef] [PubMed]
- Syed, K.; Mashele, S.S. Comparative analysis of P450 signature motifs EXXR and CXG in the large and diverse kingdom of fungi: Identification of evolutionarily conserved amino acid patterns characteristic of P450 family. PLoS ONE 2014, 9, e95616. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J. Biol. Chem. 1992, 267, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Pascal Andreu, V.; de Los Santos, E.L.C.; Del Carratore, F.; Lee, S.Y.; Medema, M.H.; Weber, T. The antiSMASH database version 2: A comprehensive resource on secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2019, 47, D625–D630. [Google Scholar] [CrossRef] [PubMed]

| Category | Salinispora species | Streptomyces species | Mycobacterial species | Cyanobacterial species | Bacteroidetes species | Firmicutes species | Alphaproteobacterial species | Betaproteobacterial species | Gammaproteobacterial species | Deltaproteobacterial species | Epsilonproteobacterial species |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Species analyzed | 126 | 203 | 2666 | 114 | 334 | 972 | 599 | 513 | 1261 | 107 | 216 |
| Species with P450s | 126 | 203 | 2666 | 114 | 77 | 229 | 229 | 290 | 169 | 23 | 53 |
| Percentage of species with P450s | 100 | 100 | 100 | 100 | 23 | 24 | 38 | 57 | 13 | 21 | 25 |
| No. of P450s | 2643 | 5460 | 62,815 | 341 | 98 | 712 | 873 | 603 | 277 | 333 | 53 |
| No. of families | 45 | 253 | 182 | 36 | 21 | 14 | 143 | 79 | 81 | 74 | 2 |
| No. of subfamilies | 103 | 698 | 345 | 79 | 28 | 53 | 214 | 119 | 102 | 171 | 2 |
| Dominant P450 family | CYP105 | CYP107 | CYP135 | CYP110 | CYP1103 | CYP107 | CYP202 | CYP116 | CYP133 & CYP107 | CYP107 | CYP172 |
| Average No. of P450s | 21 | 27 | 24 | 3 | 1 | 3 | 4 | 2 | 2 | 14 | 1 |
| No. of P450s part of BGCs | 1236 | 1231 | 9399 | 27 | 8 | 126 | 21 | 107 | 49 | 69 | 0 |
| No. of P450 families part of BGCs | 35 | 135 | 68 | 6 | 5 | 10 | 16 | 18 | 22 | 37 | 0 |
| Percentage of P450s part of BGCs | 47 | 23 | 15 | 8 | 8 | 18 | 2 | 18 | 18 | 21 | 0 |
| Reference(s) | [28] | [36,37] | This work | [27] | [32] | [34] | [33] | [30] | [29] | [30] | [30] |
| Category | MTBC | MCAC | NTM | MAC | SAP | MCL |
|---|---|---|---|---|---|---|
| Total No. of species analysed | 2128 | 255 | 163 | 106 | 10 | 4 |
| Total No. of P450s | 42,917 | 6519 | 7760 | 5093 | 505 | 21 |
| P450 fragments/pseudo | 2 | 10 | 69 | 3 | 6 | 0 |
| Average No. of P450s | 20 | 26 | 48 | 48 | 51 | 5 |
| Min No. of P450s | 5 | 22 | 9 | 28 | 35 | 3 |
| Maximum No. of P450s | 74 | 52 | 89 | 69 | 95 | 7 |
| No. of P450 families | 66 | 37 | 145 | 59 | 66 | 9 |
| No of P450 subfamilies | 95 | 48 | 261 | 88 | 101 | 9 |
| Dominant P450 family | CYP135 | CYP125 | CYP125 | CYP150 | CYP189 | CYP136 & CYP184 |
| No of the P450s part of BGCs | 8153 | 438 | 450 | 328 | 30 | 0 |
| No. of P450 families part of BGCs | 19 | 13 | 56 | 11 | 15 | 0 |
| Percentage of P450s part of BGCs | 19.0 | 6.7 | 5.8 | 6.4 | 5.9 | 0 |
| Percentage of P450 families part of BGCs | 28.8 | 35.1 | 38.6 | 18.6 | 22.7 | 0 |
| P450 Family | Count | Subfamily | Count | Nature of the Subfamily |
|---|---|---|---|---|
| CYP123 | 3019 | A | 2908 | Bloomed |
| CYP125 | 4085 | A | 3978 | Bloomed |
| CYP136 | 3017 | A | 2687 | Bloomed |
| CYP105 | 674 | Q | 377 | Expanded |
| CYP108 | 714 | B | 703 | Expanded |
| CYP121 | 2029 | A | 2029 | Expanded |
| CYP124 | 2484 | A | 2425 | Expanded |
| CYP126 | 2434 | A | 2429 | Expanded |
| CYP128 | 2243 | A | 2143 | Expanded |
| CYP130 | 2640 | A | 2640 | Expanded |
| CYP132 | 2160 | A | 2160 | Expanded |
| CYP135 | 4702 | A | 2149 | Expanded |
| B | 2543 | Expanded | ||
| CYP137 | 2167 | A | 2167 | Expanded |
| CYP138 | 2810 | A | 2733 | Expanded |
| CYP139 | 2286 | A | 2286 | Expanded |
| CYP140 | 2810 | A | 2401 | Expanded |
| CYP141 | 2105 | A | 2105 | Expanded |
| CYP142 | 2540 | A | 2507 | Expanded |
| CYP143 | 2470 | A | 2387 | Expanded |
| CYP144 | 2700 | A | 2421 | Expanded |
| CYP150 | 1056 | A | 932 | Expanded |
| CYP164 | 514 | A | 505 | Expanded |
| CYP189 | 1002 | A | 987 | Expanded |
| CYP51 | 2680 | B | 2680 | Expanded |
| Category | MTBC | MCAC | NTM | MAC | SAP | MCL |
|---|---|---|---|---|---|---|
| MTBC | 11 | 22 (CYP125) | 49 (CYP189) | 36 (CYP150) | 34 (CYP189) | 8 (CYP136 & CYP164) |
| MCAC | 22 (CYP125) | 6 | 28 (CYP125) | 20 (CYP105) | 19 (CYP125) | 5 (CYP136 & CYP164) |
| NTM | 49 (CYP189) | 28 (CYP125) | 71 | 34 (CYP189) | 29 (CYP189) | 7 (CYP164) |
| MAC | 36 (CYP150) | 20 (CYP105) | 34 (CYP189) | 15 | 25 (CYP189) | 9 (CYP136 & CYP164) |
| SAP | 34 (CYP189) | 19 (CYP125) | 29 (CYP189) | 27 (CYP189) | 23 | 5 (CYP136) |
| MCL | 8 (CYP136 & CYP164) | 5 (CYP136 & CYP164) | 7 (CYP164) | 9 (CYP136 & CYP164) | 5 (CYP136) | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zondo, N.M.; Padayachee, T.; Nelson, D.R.; Syed, K. Saprophytic to Pathogenic Mycobacteria: Loss of Cytochrome P450s Vis a Vis Their Prominent Involvement in Natural Metabolite Biosynthesis. Int. J. Mol. Sci. 2023, 24, 149. https://doi.org/10.3390/ijms24010149
Zondo NM, Padayachee T, Nelson DR, Syed K. Saprophytic to Pathogenic Mycobacteria: Loss of Cytochrome P450s Vis a Vis Their Prominent Involvement in Natural Metabolite Biosynthesis. International Journal of Molecular Sciences. 2023; 24(1):149. https://doi.org/10.3390/ijms24010149
Chicago/Turabian StyleZondo, Ntokozo Minenhle, Tiara Padayachee, David R. Nelson, and Khajamohiddin Syed. 2023. "Saprophytic to Pathogenic Mycobacteria: Loss of Cytochrome P450s Vis a Vis Their Prominent Involvement in Natural Metabolite Biosynthesis" International Journal of Molecular Sciences 24, no. 1: 149. https://doi.org/10.3390/ijms24010149
APA StyleZondo, N. M., Padayachee, T., Nelson, D. R., & Syed, K. (2023). Saprophytic to Pathogenic Mycobacteria: Loss of Cytochrome P450s Vis a Vis Their Prominent Involvement in Natural Metabolite Biosynthesis. International Journal of Molecular Sciences, 24(1), 149. https://doi.org/10.3390/ijms24010149







