The Greatwall–Endosulfine Switch Accelerates Autophagic Flux during the Cell Divisions Leading to G1 Arrest and Entry into Quiescence in Fission Yeast
Abstract
1. Introduction
2. Results
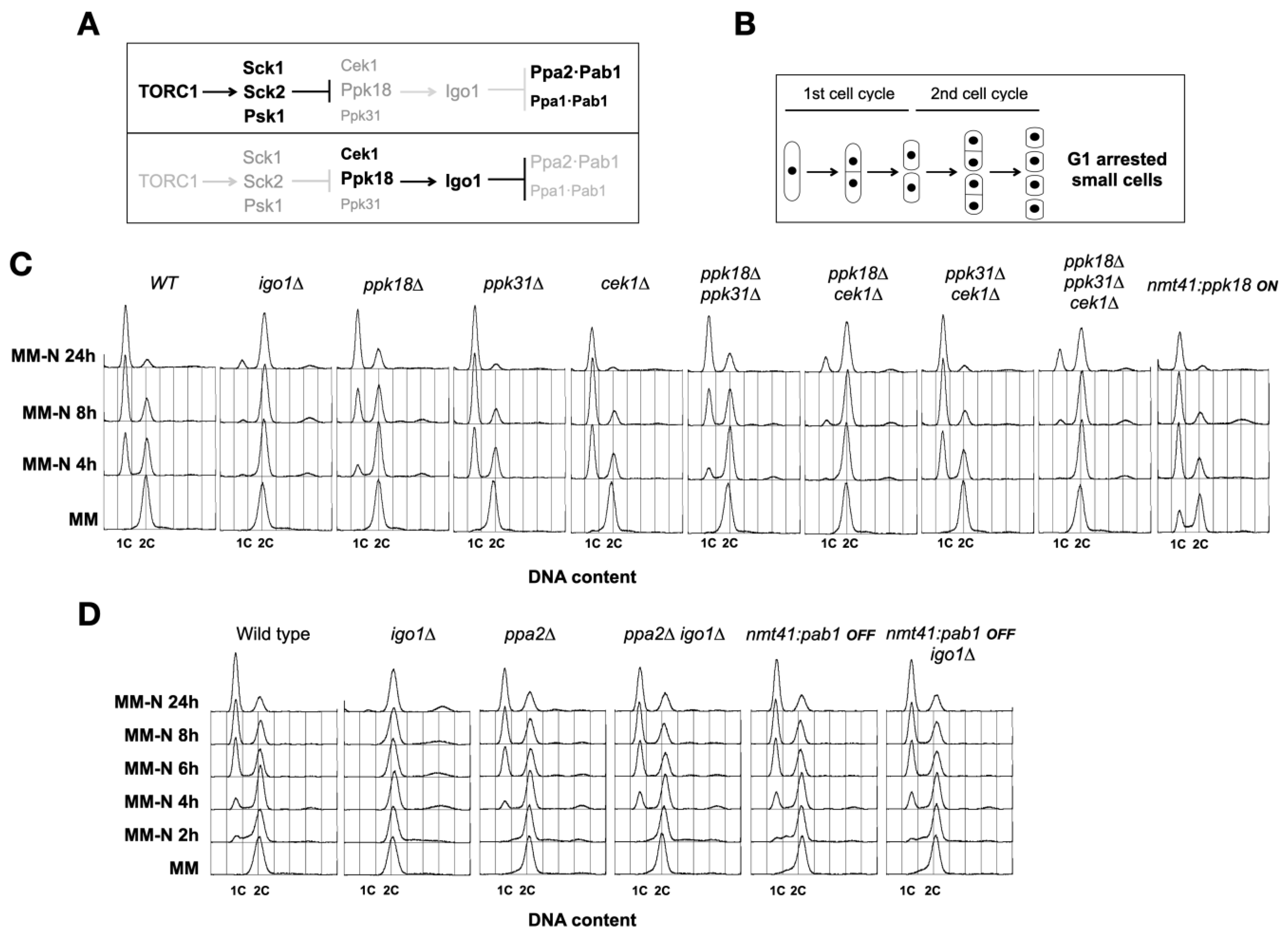
2.1. The Greatwall–Endosulfine-PP2A/B55 Pathway Regulates Proper G1 Arrest under Nitrogen Starvation
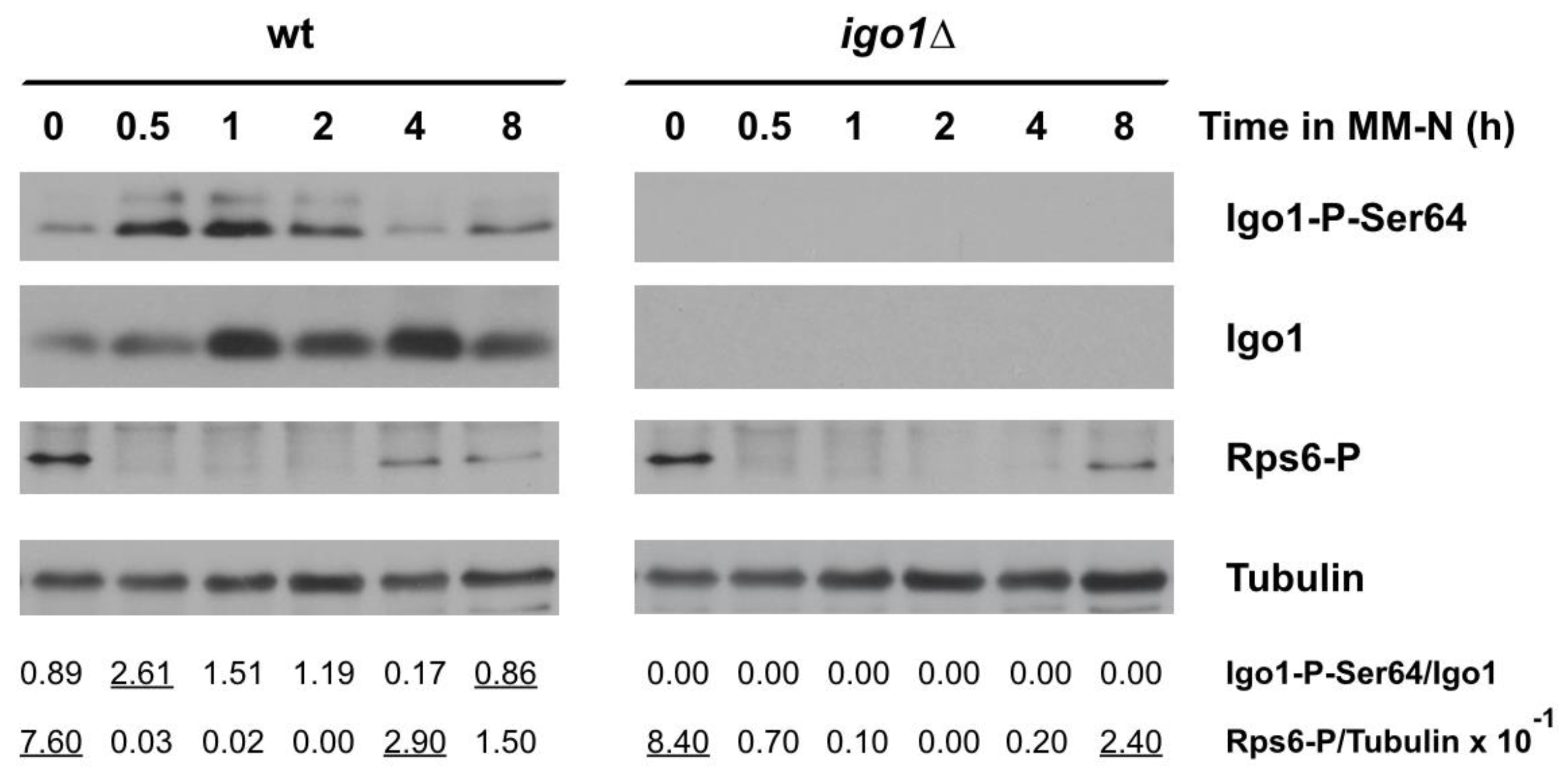
2.2. TORC1 and Greatwall Protein Kinase Activity Oscillates during Nitrogen Starvation
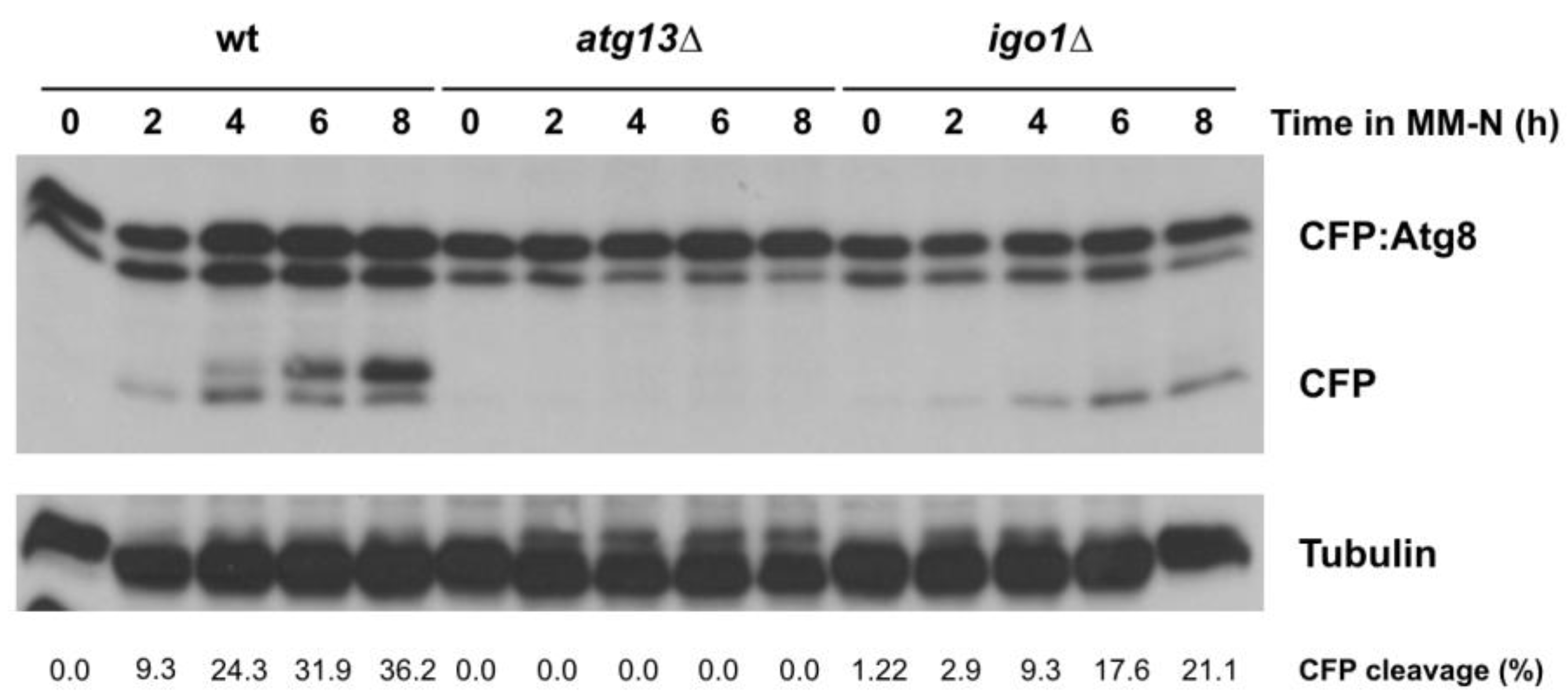
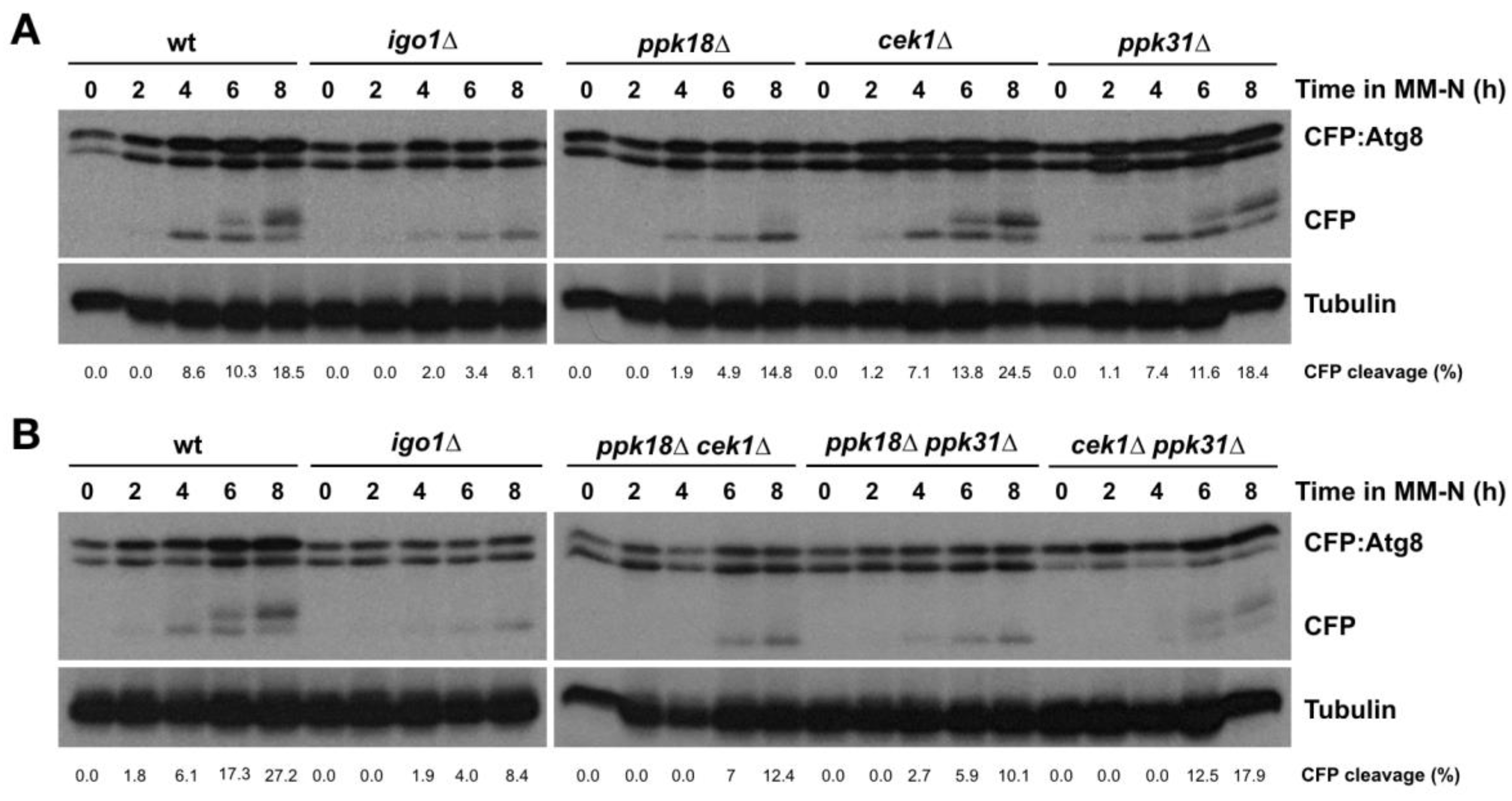
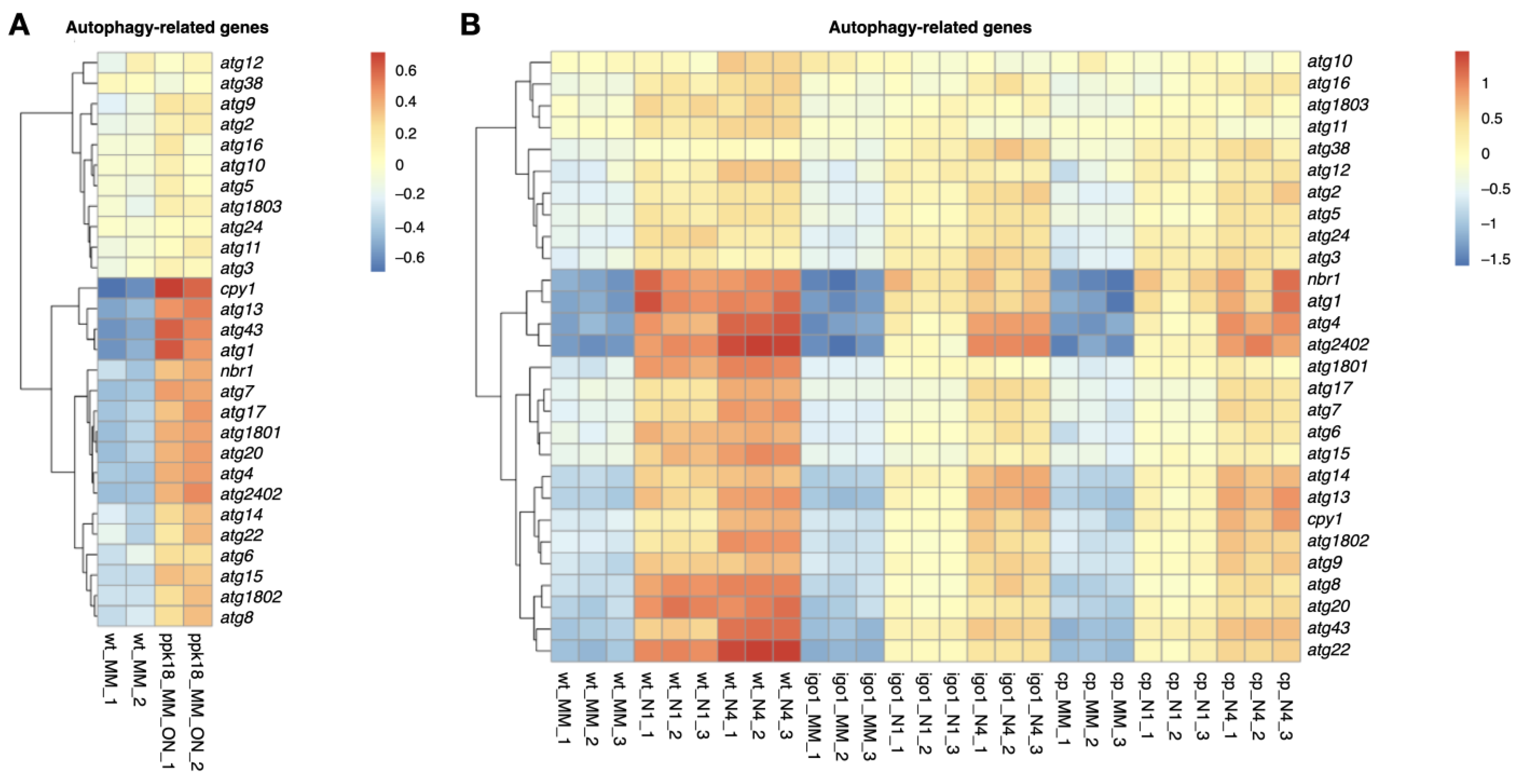
2.3. The Greatwall (Ppk18, Cek1 and Ppk31)–Endosulfine (Igo1) Switch Accelerates Autophagic Flux
2.4. S6 Kinases Are Negative Regulators of Autophagy
2.5. PP2A/Pab1 Negatively Regulates Nitrogen Starvation-Induced Autophagy
3. Discussion
4. Materials and Methods
4.1. Fission Yeast Strains and Methods
4.2. Strain Construction
4.3. Flow Cytometry
4.4. Microscopy
4.5. Protein Extracts & WB
4.6. RNA Extraction, RNA Purification, Library Preparation, and RNAseq
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tsukada, M.; Ohsumi, Y. Isolation and Characterization of Autophagy-Defective Mutants of Saccharomyces cerevisiae. FEBS Lett. 1993, 333, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Klionsky, D.J. Protein Turnover Via Autophagy: Implications for Metabolism. Annu. Rev. Nutr. 2007, 27, 19–40. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.-D.; Du, L.-L. Fission Yeast Autophagy Machinery. Cells 2022, 11, 1086. [Google Scholar] [CrossRef]
- Reggiori, F.; Klionsky, D.J. Autophagic Processes in Yeast: Mechanism, Machinery and Regulation. Genetics 2013, 194, 341–361. [Google Scholar] [CrossRef]
- Mizushima, N. A Brief History of Autophagy from Cell Biology to Physiology and Disease. Nat. Cell Biol. 2018, 20, 521–527. [Google Scholar] [CrossRef]
- Kabeya, Y.; Kamada, Y.; Baba, M.; Takikawa, H.; Sasaki, M.; Ohsumi, Y. Atg17 Functions in Cooperation with Atg1 and Atg13 in Yeast Autophagy. Mol. Biol. Cell 2005, 16, 2544–2553. [Google Scholar] [CrossRef]
- Nanji, T.; Liu, X.; Chew, L.H.; Li, F.K.; Biswas, M.; Yu, Z.Q.; Lu, S.; Dong, M.Q.; Du, L.L.; Klionsky, D.J.; et al. Conserved and Unique Features of the Fission Yeast Core Atg1 Complex. Autophagy 2017, 13, 2018–2027. [Google Scholar] [CrossRef]
- Ganley, I.G.; Lam, D.H.; Wang, J.; Ding, X.; Chen, S.; Jiang, X. ULK1·ATG13·FIP200 Complex Mediates MTOR Signaling and Is Essential for Autophagy. J. Biol. Chem. 2009, 284, 12297–12305. [Google Scholar] [CrossRef]
- Hosokawa, N.; Hara, T.; Kaizuka, T.; Kishi, C.; Takamura, A.; Miura, Y.; Iemura, S.I.; Natsume, T.; Takehana, K.; Yamada, N.; et al. Nutrient-Dependent MTORCl Association with the ULK1-Atg13-FIP200 Complex Required for Autophagy. Mol. Biol. Cell 2009, 20, 1981–1991. [Google Scholar] [CrossRef]
- Jung, C.H.; Jun, C.B.; Ro, S.H.; Kim, Y.M.; Otto, N.M.; Cao, J.; Kundu, M.; Kim, D.H. ULK-Atg13-FIP200 Complexes Mediate MTOR Signaling to the Autophagy Machinery. Mol. Biol. Cell 2009, 20, 1992–2003. [Google Scholar] [CrossRef]
- Kamada, Y.; Yoshino, K.-I.; Kondo, C.; Kawamata, T.; Oshiro, N.; Yonezawa, K.; Ohsumi, Y. Tor Directly Controls the Atg1 Kinase Complex To Regulate Autophagy. Mol. Cell Biol. 2010, 30, 1049–1058. [Google Scholar] [CrossRef] [PubMed]
- Otsubo, Y.; Nakashima, A.; Yamamoto, M.; Yamashita, A. TORC1-Dependent Phosphorylation Targets in Fission Yeast. Biomolecules 2017, 7, 50. [Google Scholar] [CrossRef]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.-L. AMPK and MTOR Regulate Autophagy through Direct Phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef]
- Shang, L.; Chen, S.; Du, F.; Li, S.; Zhao, L.; Wang, X. Nutrient Starvation Elicits an Acute Autophagic Response Mediated by Ulk1 Dephosphorylation and Its Subsequent Dissociation from AMPK. Proc. Natl. Acad. Sci. USA 2011, 108, 4788–4793. [Google Scholar] [CrossRef] [PubMed]
- Chica, N.; Rozalén, A.E.; Pérez-Hidalgo, L.; Rubio, A.; Novak, B.; Moreno, S. Nutritional Control of Cell Size by the Greatwall-Endosulfine-PP2A·B55 Pathway. Curr. Biol. 2016, 26, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Aono, S.; Haruna, Y.; Watanabe, Y.H.; Mochida, S.; Takeda, K. The Fission Yeast Greatwall–Endosulfine Pathway Is Required for Proper Quiescence/G 0 Phase Entry and Maintenance. Genes Cells 2019, 24, 172–186. [Google Scholar] [CrossRef]
- Martín, R.; Portantier, M.; Chica, N.; Nyquist-Andersen, M.; Mata, J.; Lopez-Aviles, S. A PP2A-B55-Mediated Crosstalk between TORC1 and TORC2 Regulates the Differentiation Response in Fission Yeast. Curr. Biol. 2017, 27, 175–188. [Google Scholar] [CrossRef]
- Laboucarié, T.; Detilleux, D.; Rodriguez-Mias, R.A.; Faux, C.; Romeo, Y.; Franz-Wachtel, M.; Krug, K.; Maček, B.; Villén, J.; Petersen, J.; et al. TORC1 and TORC2 Converge to Regulate the SAGA Co-activator in Response to Nutrient Availability. EMBO Rep. 2017, 18, 2197–2218. [Google Scholar] [CrossRef]
- Kohda, T.A.; Tanaka, K.; Konomi, M.; Sato, M.; Osumi, M.; Yamamoto, M. Fission Yeast Autophagy Induced by Nitrogen Starvation Generates a Nitrogen Source That Drives Adaptation Processes. Genes Cells 2007, 12, 155–170. [Google Scholar] [CrossRef]
- Yorimitsu, T.; Zaman, S.; Broach, J.R.; Klionsky, D.J. Protein Kinase A and Sch9 Cooperatively Regulate Induction of Autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell 2007, 18, 4180–4189. [Google Scholar] [CrossRef]
- Mukaiyama, H.; Kajiwara, S.; Hosomi, A.; Giga-Hama, Y.; Tanaka, N.; Nakamura, T.; Takegawa, K. Autophagy-Deficient Schizosaccharomyces pombe Mutants Undergo Partial Sporulation during Nitrogen Starvation. Microbiology 2009, 155, 3816–3826. [Google Scholar] [CrossRef]
- Sun, L.L.; Li, M.; Suo, F.; Liu, X.M.; Shen, E.Z.; Yang, B.; Dong, M.Q.; He, W.Z.; Du, L.L. Global Analysis of Fission Yeast Mating Genes Reveals New Autophagy Factors. PLoS Genet. 2013, 9, e1003715. [Google Scholar] [CrossRef]
- Corral-Ramos, C.; Barrios, R.; Ayté, J.; Hidalgo, E. TOR and MAP Kinase Pathways Synergistically Regulate Autophagy in Response to Nutrient Depletion in Fission Yeast. Autophagy 2022, 18, 375–390. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. MTOR Signaling at a Glance. J. Cell Sci. 2009, 122, 3589–3594. [Google Scholar] [CrossRef]
- Nakashima, A.; Otsubo, Y.; Yamashita, A.; Sato, T.; Yamamoto, M.; Tamanoi, F. Psk1, an AGC Kinase Family Member in Fission Yeast, Is Directly Phosphorylated and Controlled by TORC1 and Functions as S6 Kinase. J. Cell Sci. 2012, 125, 5840–5849. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, N.; Ohkura, H.; Yanagida, M. Distinct, Essential Roles of Type 1 and 2A Protein Phosphatases in the Control of the Fission Yeast Cell Division Cycle. Cell 1990, 63, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Mukaiyama, H.; Nakase, M.; Nakamura, T.; Kakinuma, Y.; Takegawa, K. Autophagy in the Fission Yeast Schizosaccharomyces pombe. FEBS Lett. 2010, 584, 1327–1334. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; McPhee, C.K.; Zheng, L.; Mardones, G.A.; Rong, Y.; Peng, J.; Mi, N.; Zhao, Y.; Liu, Z.; Wan, F.; et al. Termination of Autophagy and Reformation of Lysosomes Regulated by MTOR. Nature 2010, 465, 942–946. [Google Scholar] [CrossRef] [PubMed]
- Shin, C.S.; Huh, W.K. Bidirectional Regulation between TORC1 and Autophagy in Saccharomyces cerevisiae. Autophagy 2011, 7, 854–862. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Dalgaard, J.Z.; Millar, J.B.A.; Arumugam, P. The Rim15-Endosulfine-PP2ACdc55 Signalling Module Regulates Entry into Gametogenesis and Quiescence via Distinct Mechanisms in Budding Yeast. PLoS Genet. 2014, 10, e1004456. [Google Scholar] [CrossRef]
- Otsubo, Y.; Matsuo, T.; Nishimura, A.; Yamamoto, M.; Yamashita, A. TRNA Production Links Nutrient Conditions to the Onset of Sexual Differentiation through the TORC 1 Pathway. EMBO Rep. 2018, 19, e44867. [Google Scholar] [CrossRef]
- Kamada, Y.; Funakoshi, T.; Shintani, T.; Nagano, K.; Ohsumi, M.; Ohsumi, Y. Tor-Mediated Induction of Autophagy via an Apg1 Protein Kinase Complex. J. Cell Biol. 2000, 150, 1507–1513. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, Y.; Suzuki, S.W.; Yamamoto, H.; Kondo-Kakuta, C.; Kimura, Y.; Hirano, H.; Akada, R.; Inagaki, F.; Ohsumi, Y.; Noda, N.N. Structural Basis of Starvation-Induced Assembly of the Autophagy Initiation Complex. Nat. Struct. Mol. Biol. 2014, 21, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Fujioka, Y.; Suzuki, S.W.; Noshiro, D.; Suzuki, H.; Kondo-Kakuta, C.; Kimura, Y.; Hirano, H.; Ando, T.; Noda, N.N.; et al. The Intrinsically Disordered Protein Atg13 Mediates Supramolecular Assembly of Autophagy Initiation Complexes. Dev. Cell 2016, 38, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Bernard, A.; Jin, M.; González-Rodríguez, P.; Füllgrabe, J.; Delorme-Axford, E.; Backues, S.K.; Joseph, B.; Klionsky, D.J. Rph1/KDM4 Mediates Nutrient-Limitation Signaling That Leads to the Transcriptional Induction of Autophagy. Curr. Biol. 2015, 25, 546–555. [Google Scholar] [CrossRef] [PubMed]
- Mata, J.; Lyne, R.; Burns, G.; Bähler, J. The Transcriptional Program of Meiosis and Sporulation in Fission Yeast. Nat. Genet. 2002, 32, 143–147. [Google Scholar] [CrossRef]
- Mata, J.; Bähler, J. Global Roles of Ste11p, Cell Type, and Pheromone in the Control of Gene Expression during Early Sexual Differentiation in Fission Yeast. Proc. Natl. Acad. Sci. USA 2006, 103, 15517–15522. [Google Scholar] [CrossRef]
- Jin, M.; He, D.; Backues, S.K.; Freeberg, M.A.; Liu, X.; Kim, J.K.; Klionsky, D.J. Transcriptional Regulation by Pho23 Modulates the Frequency of Autophagosome Formation. Curr. Biol. 2014, 24, 1314–1322. [Google Scholar] [CrossRef]
- Sun, J.; Mu, Y.; Jiang, Y.; Song, R.; Yi, J.; Zhou, J.; Sun, J.; Jiao, X.; Prinz, R.A.; Li, Y.; et al. Inhibition of P70 S6 Kinase Activity by A77 1726 Induces Autophagy and Enhances the Degradation of Superoxide Dismutase 1 (SOD1) Protein Aggregates. Cell Death Dis. 2018, 9, 407. [Google Scholar] [CrossRef]
- Hać, A.; Domachowska, A.; Narajczyk, M.; Cyske, K.; Pawlik, A.; Herman-Antosiewicz, A. S6K1 Controls Autophagosome Maturation in Autophagy Induced by Sulforaphane or Serum Deprivation. Eur. J. Cell Biol. 2015, 94, 470–481. [Google Scholar] [CrossRef] [PubMed]
- Moreno, S.; Klar, A.; Nurse, P. Molecular Genetic Analysis of Fission Yeast Schizosaccharomyces pombe. Methods Enzymol. 1991, 194, 795–823. [Google Scholar] [CrossRef] [PubMed]
- Foiani, M.; Marini, F.; Gamba, D.; Lucchini, G.; Plevani, P. The B Subunit of the DNA Polymerase Alpha-Primase Complex in Saccharomyces cerevisiae Executes an Essential Function at the Initial Stage of DNA Replication. Mol. Cell. Biol. 1994, 14, 923–933. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vázquez-Bolado, A.; López-San Segundo, R.; García-Blanco, N.; Rozalén, A.E.; González-Álvarez, D.; Suárez, M.B.; Pérez-Hidalgo, L.; Moreno, S. The Greatwall–Endosulfine Switch Accelerates Autophagic Flux during the Cell Divisions Leading to G1 Arrest and Entry into Quiescence in Fission Yeast. Int. J. Mol. Sci. 2023, 24, 148. https://doi.org/10.3390/ijms24010148
Vázquez-Bolado A, López-San Segundo R, García-Blanco N, Rozalén AE, González-Álvarez D, Suárez MB, Pérez-Hidalgo L, Moreno S. The Greatwall–Endosulfine Switch Accelerates Autophagic Flux during the Cell Divisions Leading to G1 Arrest and Entry into Quiescence in Fission Yeast. International Journal of Molecular Sciences. 2023; 24(1):148. https://doi.org/10.3390/ijms24010148
Chicago/Turabian StyleVázquez-Bolado, Alicia, Rafael López-San Segundo, Natalia García-Blanco, Ana Elisa Rozalén, Daniel González-Álvarez, M. Belén Suárez, Livia Pérez-Hidalgo, and Sergio Moreno. 2023. "The Greatwall–Endosulfine Switch Accelerates Autophagic Flux during the Cell Divisions Leading to G1 Arrest and Entry into Quiescence in Fission Yeast" International Journal of Molecular Sciences 24, no. 1: 148. https://doi.org/10.3390/ijms24010148
APA StyleVázquez-Bolado, A., López-San Segundo, R., García-Blanco, N., Rozalén, A. E., González-Álvarez, D., Suárez, M. B., Pérez-Hidalgo, L., & Moreno, S. (2023). The Greatwall–Endosulfine Switch Accelerates Autophagic Flux during the Cell Divisions Leading to G1 Arrest and Entry into Quiescence in Fission Yeast. International Journal of Molecular Sciences, 24(1), 148. https://doi.org/10.3390/ijms24010148




