Abstract
Left Ventricular Non-Compaction (LVNC) is defined by the triad prominent myocardial trabecular meshwork, thin compacted layer, and deep intertrabecular recesses. LVNC associated with dilation is characterized by the coexistence of left ventricular dilation and systolic dysfunction. Pediatric cases with dilated-LVNC have worse outcomes than those with isolated dilated cardiomyopathy and adult patients. Herein, we report a clinical and genetic investigation using trio-based whole-exome sequencing of a pediatric case with early-onset dilated-LVNC. Compound heterozygous mutations were identified in the Striated Muscle Enriched Protein Kinase (SPEG) gene, a key regulator of cardiac calcium homeostasis. A paternally inherited mutation: SPEG; p.(Arg2470Ser) and the second variant, SPEG; p.(Pro2687Thr), is common and occurred de novo. Subsequently, Sanger sequencing was performed for the family in order to segregate the variants. Thus, the index case, his father, and both sisters carried the SPEG: p.(Arg2470Ser) variant. Only the index patient carried both SPEG variants. Both sisters, as well as the patient’s father, showed LVNC without cardiac dysfunction. The unaffected mother did not harbor any of the variants. The in silico analysis of the identified variants (rare and common) showed a decrease in protein stability with alterations of the physical properties as well as high conservation scores for the mutated residues. Interestingly, using the Project HOPE tool, the SPEG; p.(Pro2687Thr) variant is predicted to disturb the second fibronectin type III domain of the protein and may abolish its function. To our knowledge, the present case is the first description of compound heterozygous SPEG mutations involving a de novo variant and causing dilated-LVNC without neuropathy or centronuclear myopathy.
1. Introduction
Left Ventricular Non-Compaction (LVNC) is defined by the triad: prominent myocardial trabecular meshwork, thin compacted layer, and deep intertrabecular recesses [1,2]. It is characterized by incomplete penetrance and variable expressivity. Of note, the anatomy of the trabeculae is different among individuals and can be observed in healthy subjects [3]. Thus, the characterization of pathological LVNC remains challenging.
The clinical diagnosis of LVNC is based mostly on imaging by transthoracic echocardiography. According to diagnostic criteria by Petersen et al., LVNC can be diagnosed if the ratio of non-compacted to compacted myocardium is >2.3 at end-diastole [4]. However, a consensual standardization is lacking for LVNC diagnosis criteria, especially in children.
Although LVNC mainly affects the left ventricle, the right ventricle can be affected as well. Moreover, cases with LVNC in conjunction with a wide range of cardiac disorders have been observed. Therefore, several subtypes of LVNC have been described including dilated-LVNC, hypertrophic-LVNC, mixed dilated-hypertrophic LVNC phenotype, and isolated LVNC [2].
In the pediatric population, LVNC accounts for nearly 10% of all primary cardiomyopathies [5,6]. Moreover, pediatric cases with dilated-LVNC had the worst outcomes followed by those with hypertrophic-LVNC [7,8]. The clinical manifestation in patients with LVNC such as heart failure, arrhythmias, and embolic events are comparable in the adult and pediatric populations [8,9].
The genetic diagnostic yield of LVNC is overall low and highly variable, ranging from 9 to 41% depending on the size of the patient cohort and gene testing strategy (gene panels, WES…) [1,10,11]. Moreover, owing to the small cohort sizes, little is known about LVNC genotype-phenotype correlations.
Emerging data reporting genetic causes of LVNC support an overlap in the etiological basis between this entity and the other types of cardiomyopathies [2,3].
In the present study, we report clinical and genetic investigation of a pediatric case with early-onset dilated-LVNC and asymptomatic family members with the LVNC phenotype. Our study revealed two missense variants in the SPEG gene with a high prediction of pathogenicity, which when acting in concert seem to cause a severe phenotype of the disease, as observed here.
2. Results
2.1. Clinical Presentation and WES Findings
The index patient, a 7-year-old boy (weight = 26.6 Kg (+0.5 SD); height = 125 cm (+0.5 SD)), was born at full term to ostensibly healthy, non-consanguineous parents. Family history was normal with no sudden death nor miscarriages. The neonatal period was uneventful without feeding or growth difficulties. At the age of six months, he was referred to the pediatric cardiology department for unexplained tachypnea. Cardiac ultrasound revealed dilated-LVNC (LV end-diastolic z-value +2.2) with significantly altered LV systolic function (ejection fraction estimated at 32% (Simpson analysis) and non-compaction of the LV. The right ventricle was unaffected. Cardiac MRI performed at the age of 8 months confirmed the diagnosis (Figure 1a. He was started on diuretics, ACE inhibitors, and oral anti-aggregation therapy (aspirin). His clinical course was favorable, with progressive clinical and echocardiographic improvement. The latest echocardiography, performed at the age of 7 years (Figure 1b), showed a non-dilated LV with isolated LVNC and subnormal systolic function (EF 50–55%). The patient remained solely on ACE inhibitors. Of note, the patient presented mild hyperlordosis with fluctuating CPK levels; however, his muscular biopsy was normal without objective clinical muscle deficit. No developmental delay was noted. Ophthalmic examination was normal.
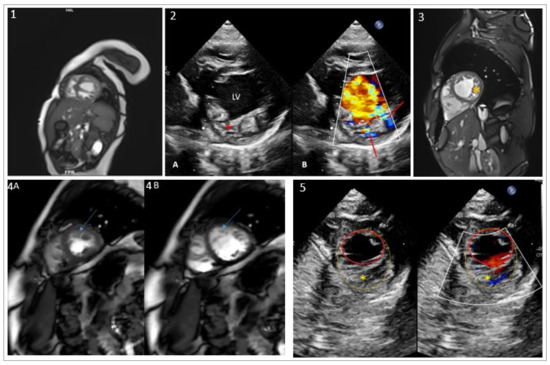
Figure 1.
Echocardiograms, MRI images of the index patient and his relatives, family pedigree and variant conservation score. (1) MRI image of the index patient at the age of 8 months shows dilated-LVNC with altered LV function. (2) Transthoracic echocardiography of the index patient: (2A) Parasternal short axis view showing the left ventricle (LV) with increased trabeculations (red asterisk). (2B) Color Doppler analysis, the red arrows show the blood flow in the intratrabecular recesses. (3) MRI image of the index patient’s father showing LVNC (yellow asterisk) (subject I-1). (4) MRI images of subject II-1 showing LV short axis in systolic (4A) and diastolic (4B) phases with LVNC (blue arrow). (5) Echocardiography of subject II-3 showing trabeculated LV (yellow asterisk). An endocardial border (red circle) and non-compacted layer border to include the trabeculated area (yellow circle) are drawn.
Whole-exome sequencing analysis allowed us to prioritize two missense heterozygous variants in the SPEG gene, one which was a paternally inherited variant: NM_005876: exon30: c.7408C>A; p.(Arg2470Ser) (MAF = 8.422e−5 in the gnomAD database). This variant is located between the first protein kinase domain and the Ig-like 9 domain of the gene. The second variant, NM_005876: exon34: c.8059C>A; p.(Pro2687Thr) (MAF = 0.12), occurred de novo. This variant is located in the second fibronectin type-III domain of the gene (Figure 2A). Subsequently, Sanger sequencing was performed for the family members in order to segregate the variants. Thus, the index-case (II-2), his father (I-1), and both sisters (II-1; II-3) carried the SPEG: p.(Arg2470Ser) variant. Only the index patient carried both SPEG variants. The unaffected mother did not harbor any of the variants (Figure 2B).
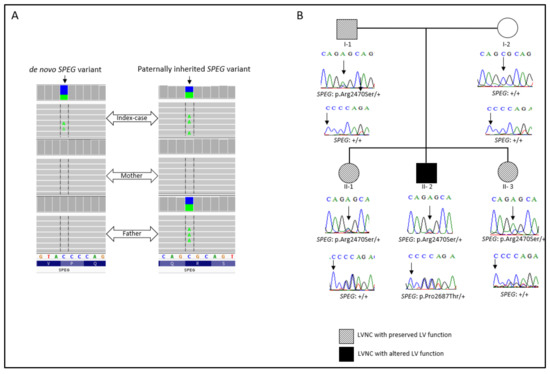
Figure 2.
(A) Integrative genomics viewer visualization showing the de novo and the inherited SPEG variants. (B) Family pedigree. Sanger electropherograms and genotypes are shown below symbols: (+) indicates the wild-type allele, and the arrow indicates the position of the mutation.
Of note, no other clinically relevant variants that may explain the phenotype in this family were found.
Family echocardiographic screening detected LVNC with preserved LV function in the index patient’s father (44 years old) and his older sister (10 years old). The diagnosis was confirmed in both with cardiac MRI (Figure 1-3,4). Both patients were asymptomatic.
His younger sister, 4 years old, was shown to have a trabeculated left ventricle but without criteria of non-compaction and with preserved systolic function (Figure 1-5). MRI assessment has been scheduled at an older age. No other clinical abnormalities were observed in those three first-degree relatives. The mother’s echocardiogram was completely normal.
2.2. In Silico Analysis
Both variants are predicted deleterious by at least three in silico prediction tools. More specifically, the SPEG; p.(Arg2470Ser) variant has been predicted deleterious by Mutation taster, SIFT, and UMD predictor, and the p.(Pro2687Thr) variant has been predicted deleterious by Provean, Polyphen2_HDIV, and _HVAR. Interestingly, the common SPEG; p.(Pro2687Thr) variant had a higher Combined Annotation Dependent Depletion Phred score (score v1.3 = 27) compared to the rare SPEG; p.(Arg2470Ser) variant (score v1.3 = 25.8). The conservation analysis by ConSurf (https://consurf.tau.ac.il/; accessed date 10 December 2021) showed high conservation scores for the SPEG; Arg2470 and Pro2687 residues of 9 and 8, respectively.
The I-Mutant 2.0 software was used to predict protein stability [12]. Both variants showed decreased protein stability with a reliability index of 6 for the SPEG; p.(Arg2470Ser) variant and 9 for the SPEG; p.(Pro2687Thr) variant. Protein stability changes were calculated at pH = 7 and 25 °C.
To assess the physical properties and interactions of the protein, the Project HOPE tool was acquired [8]. The results of this analysis for the SPEG; p.(Arg2470Ser) rare variant showed that the mutant residue (Serine) is less smaller than the wild-type residue (Arginine). Moreover, the charge of the Arginine residue at position 2470 was positive, and the mutant residue charge was neutral. Thus, the mutant residue is more hydrophobic than the wild-type residue, which can result in loss of hydrogen bonds and/or disturb correct folding. Similarly, the SPEG; p.(Pro2687Thr) variant, located in the fibronectin III domain, resulted in a substitution with a residue (Threonine) with a different size and different hydrophobicity properties. The wild-type residue (Proline) is more hydrophobic than the mutant residue. Project HOPE predicted that the p.(Pro2687Thr) variant can disturb this domain and abolish its function. Furthermore, Prolines are known to be very rigid and therefore induce a special backbone conformation which might be required at this position. The variant can disturb this special conformation.
Considering the segregation of the variants with the disease in the family and the favor pathogenic prediction, the SPEG; p.(Pro2687Thr) de novo variant seems to be a key contributor to the early onset and the severity of the disease in the presence of the rare inherited variant SPEG; p.(Arg2470Ser).
3. Discussion
SPEG (Striated Muscle Enriched Protein Kinase) is a protein-coding gene. The protein belongs to the myosin light chain kinase family, which is required for myocyte cytoskeletal development [13]. Due to extensive alternative splicing, the SPEG complex locus encodes several isoforms, including: Aortic Preferentially Expressed Protein 1 (Apeg), Brain Preferentially Expressed Protein (Bpeg), Speg-α, and Speg-β. Contrary to Apeg1 and Bpeg, Speg-α and Speg-β contain two kinase domains and are predominantly expressed in skeletal and cardiac muscle [13]. In vivo studies in mice reported a drastic shift in isoform predominance in the heart from Apeg1 during the embryonic stage to Speg-α and -β in the postnatal period, which can be correlated to the cardiomyocyte maturation occurring in this period [13]. Of note, Liu X and colleagues reported the presence of the four isoforms in fetal hearts [14]. In the same study, the authors demonstrated the implication of SPEG locus in dilated cardiomyopathy. In fact, homozygous SPEG mutant mice led to dilation of right and left atria and ventricles with increased neonatal mortality [14].
Interestingly, along with the Desmin gene (DES), the expression of SPEG gene may be controlled by the desmin locus control region, and both genes shared similar tissues expression profiles [15]. Of note, inherited and de novo mutations in the DES gene have been linked to myofibrillar myopathy and dilated and arrhythmogenic cardiomyopathies [16,17,18,19]. More recently, a de novo missense mutation in the DES gene has been associated to LVNC cardiomyopathy [20].
Firstly, mutations in the SPEG gene have been identified in patients with centronuclear myopathy type 5 (phenotype MIM number 615959) with an autosomal recessive inheritance pattern [21]. Centronuclear myopathies are a group of rare inherited neuromuscular disorders characterized by a progressive weakness defined mainly by numerous centrally placed nuclei on a muscle biopsy and a highly variable clinical presentation including ophthalmoplegia, respiratory failure, and scoliosis [21,22]. Strikingly, Lornage et al. reported a patient affected by myopathy without central nuclei [23].
In 2014, Agrawal et al. identified SPEG mutations in three patients with centronuclear myopathy, two of whom presented dilated cardiomyopathy [21]. Thereafter, patients with non-syndromic dilated cardiomyopathy and no myopathy features or skeletal muscle involvement were reported [24,25]. Although several cases of dilated cardiomyopathy linked to SPEG mutations were reported either associated to myopathy or not, only one case of non-compaction and neuropathy was reported [26]. His cardiac evaluation showed a disclosed enlarged atria, abnormal trabeculation of the left ventricle, and intratrabeculation recesses [26]. Taking together, the SPEG gene seems to be a pleiotropic gene giving rise to different diseases and cardiac phenotypes with variable expressivity.
Recently, different studies showed the role of SPEG in calcium reuptake into the sarcoendoplasmic reticulum through regulating SERCA2a (Sarcoplasmic/Endoplasmic Reticulum Calcium ATPase2) in cardiomyocytes, which is a crucial process in the excitation-contraction coupling [27,28]. Genes implicated in Ca2+ handling such as TTN, RYR2, CASQ2, and RBM20 have been reported in patients with LVNC [29,30]. Similarly, the SPEG gene was defined as a key regulator of cardiac calcium handling owing to its role in Ca2+ regulation through maintenance of transverse tubule formation and phosphorylation of junctional membrane complex formation [27,28]. Inducible deletion of SPEG inhibited SERCA2a calcium transporting activity and impaired Ca2+ reuptake into the sarcoplasmic reticulum through phosphorylation at the Thr484 SERCA2a phosphorylation site [27]. Aberrant calcium handling leads mainly to altered contractility and arrhythmogenesis. Furthermore, Quick et al. showed that the Speg protein binds to both Jph2 (Junctophilin2) and RyR2 proteins. Given that Speg binds to both RyR2 and SERCA2a, it plays a role in the structural organization of sarcoplasmic reticulum complexes which may explain its implication in cardiomyopathies [31].
To date, 15 mutations in the SPEG gene have been linked to cardiac phenotypes in humans through either an autosomal recessive pattern of inheritance or a heterozygous compound model [21,22,23,24,25,26,32,33]. Furthermore, the majority of protein-truncating variants have been found in patients with myopathy associated with dilated cardiomyopathy (Table 1). However, a clear-cut correlation between the phenotype and the variant type, location, or genotype is lacking. Further studies involving a large cohort of patients harboring SPEG mutations are necessary to evaluate a potential genotype–phenotype correlation.

Table 1.
SPEG mutations reported in patients with cardiac phenotypes.
In the present study, we report an early-onset dilated-LVNC case without neuropathy or myopathy features. Furthermore, the cardiac evaluation of both sisters and the father carrying the rare SPEG variant p.(Arg2470Ser) showed an LVNC phenotype without an altered cardiac function. These findings support the fact that the common SPEG; p.(Pro2687Thr) variant is functionally relevant and may act as a risk allele in the presence of the rare SPEG variant.
Further characterization of SPEG variants is needed to better understand their physiopathological mechanism and cumulative effect.
4. Materials and Methods
4.1. Whole-Exome Sequencing (WES)
Peripheral blood sample was collected after obtaining the written informed consent. Genomic DNA was extracted by standard techniques.
Whole-exome sequencing was performed for the patient and his parents using the Agilent SureSelect Human All Exon kit V4 (Agilent Technologies, Santa Clara, CA, USA). The captured libraries were sequenced on Illumina NextSeq500 sequencing platform (Illumina, San Diego, CA, USA). Raw fastQ files were aligned to the hg19 reference human genome (University of California Santa Cruz, UCSC) using BWA software. Variant calling workflow was performed according to the GATK best practices. The output files were annotated using ANNOVAR software.
4.2. In Silico Analysis Tools
4.2.1. Variant Prioritization
Variant prioritization was carried out using Variant Annotation and Filtering Tool (VarAFT http://varaft.eu/) version 2.17; accessed date 10 December 2021. To pinpoint candidate causative variants, we adopted the following filtering strategy: firstly, a minimum variant allele frequency of 2% was applied using gnomAD database (http://gnomad.broadinstitute.org/; accessed date 10 December 2021). Then, we removed non-coding and synonymous variants. The remaining variants including missense and protein-truncating variants were filtered based on their in silico pathogenicity prediction to assess their likely causality.
A pedigree-based analysis was performed considering autosomal dominance and recessiveness as well as X-linked dominance and recessiveness patterns of inheritance.
4.2.2. Residue Conservation Analysis
Residue conservation was determined using the ConSurf server (https://consurf.tau.ac.il/; accessed date: 4 January 2022). ConSurf is a bioinformatics tool for estimating the evolutionary conservation of amino and nucleic acid positions in a protein/DNA/RNA molecule based on the phylogenetic relations between homologous sequences. The degree to which an amino or nucleic acid position is evolutionarily conserved is strongly dependent on its structural and functional importance [34].
4.2.3. Protein Physical Properties, Stability, and Interaction Assessment
The prediction of variants’ effects on the protein was performed using Project HOPE and I-Mutant 2.0 software [12,35].
A flowchart explaining the study methodology is summarized in Figure 3.
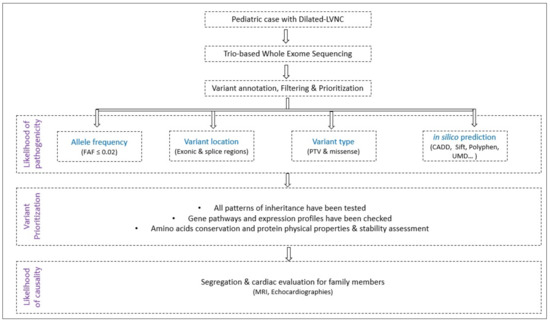
Figure 3.
Study design flowchart. (FAF: Filtering Allele Frequency; PTV: Protein-Truncating Variants).
Author Contributions
Conceptualization, S.Z. and C.O.; Methodology, H.J.; Validation, S.Z. and C.O.; Clinical investigation of the patients and family members, F.E.L., C.W., A.C. and C.O.; Analysis and interpretation of data, H.J; Molecular investigation and in silico analysis, H.J.; writing—original draft preparation, H.J., F.E.L. and C.O.; Writing—review and editing, H.J., S.Z. and C.O. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no specific grant from any funding agency, commercial, or not-for-profit sectors.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of “Assistance Publique Hôpitaux de Marseille” (Registration number: 2016-A00958-53; date of approval: 13/08/2021; ref [16] 57 MS 3).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article.
Acknowledgments
We would like to thank the family for their collaboration. This work was supported by the “Association Française contre les Myopathies-AFM Telethon” (MoThARD-Project), and the “Institut National de la Santé et de la Recherche Médicale” to S.Z. H.J. received Postdoctoral fellowships from the “Association Française contre les Myopathies-AFM Telethon”.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Lorca, R.; Martín, M.; Pascual, I.; Astudillo, A.; Molina, B.D.; Cigarrán, H.; Cuesta-Llavona, E.; Avanzas, P.; Reguero, J.R.; Coto, E.; et al. Characterization of Left Ventricular Non-Compaction Cardiomyopathy. J. Clin. Med. 2020, 9, 2524. [Google Scholar] [CrossRef] [PubMed]
- Towbin, J.A.; Lorts, A.; Jefferies, J.L. Left ventricular non-compaction cardiomyopathy. Lancet 2015, 386, 813–825. [Google Scholar] [CrossRef]
- Di Toro, A.; Giuliani, L.; Smirnova, A.; Favalli, V.; Serio, A.; Urtis, M.; Grasso, M.; Arbustini, E. Myths to debunk: The non-compacted myocardium. Eur. Heart J. Suppl. 2020, 22 (Suppl. L), L6–L10. [Google Scholar] [CrossRef] [PubMed]
- Petersen, S.E.; Selvanayagam, J.B.; Wiesmann, F.; Robson, M.D.; Francis, J.M.; Anderson, R.H.; Watkins, H.; Neubauer, S. Left ven-tricular non-compaction: Insights from cardiovascular magnetic resonance imaging. J. Am. Coll. Cardiol. 2005, 46, 101–105. [Google Scholar]
- Nugent, A.W.; Daubeney, P.E.; Chondros, P.; Carlin, J.; Cheung, M.; Wilkinson, L.C.; Davis, A.; Kahler, S.G.; Chow, C.; Wilkinson, J.L.; et al. The Epidemiology of Childhood Cardiomyopathy in Australia. N. Engl. J. Med. 2003, 348, 1639–1646. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.Y.; Moreno-Betancur, M.; Nugent, A.W.; Cheung, M.; Colan, S.; Turner, C.; Sholler, G.F.; Robertson, T.; Justo, R.; Bullock, A.; et al. Long-Term Outcomes of Childhood Left Ventricular Noncompaction Cardiomyopathy: Results from a National Population-Based Study. Circulation 2018, 138, 367–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jefferies, J.L.; Wilkinson, J.D.; Sleeper, L.A.; Colan, S.D.; Lu, M.; Pahl, E.; Kantor, P.F.; Everitt, M.D.; Webber, S.A.; Kaufman, B.D.; et al. Cardiomyopathy phenotypes and outcomes for children with left ventricular myocardial noncompaction: Results from the pediatric cardiomyopathy registry. J. Card. Fail. 2015, 21, 877–884. [Google Scholar] [CrossRef] [Green Version]
- Ichida, F. Left ventricular noncompaction—Risk stratification and genetic consideration. J. Cardiol. 2020, 75, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Takasaki, A.; Ozawa, S.W.; Nakaoka, H.; Okabe, M.; Miyao, N.; Saito, K.; Ibuki, K.; Hirono, K.; Yoshimura, N.; et al. Long-Term Prognosis of Patients with Left Ventricular Noncompaction—Comparison Between Infantile and Juvenile Types. Circ. J. 2017, 81, 694–700. [Google Scholar] [CrossRef] [Green Version]
- Ross, S.B.; Singer, E.S.; Driscoll, E.; Nowak, N.; Yeates, L.; Puranik, R.; Sy, R.W.; Rajagopalan, S.; Barratt, A.; Ingles, J.; et al. Genetic architecture of left ventricular noncompaction in adults. Hum. Genome Var. 2020, 7, 33. [Google Scholar] [CrossRef]
- Hershberger, R.E.; Lindenfeld, J.; Mestroni, L.; Seidman, C.E.; Taylor, M.R.; Towbin, J.A. Genetic Evaluation of Cardiomyopathy—A Heart Failure Society of America Practice Guideline. J. Card. Fail. 2009, 15, 83–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capriotti, E.; Fariselli, P.; Casadio, R. I-Mutant2.0: Predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005, 33, W306–W310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, S.; Rosen, S.; Li, Q.; Agrawal, P. Striated Preferentially Expressed Protein Kinase (SPEG) in Muscle Development, Function, and Disease. Int. J. Mol. Sci. 2021, 22, 5732. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ramjiganesh, T.; Chen, Y.-H.; Chung, S.W.; Hall, S.R.; Schissel, S.L.; Padera, R.F., Jr.; Liao, R.; Ackerman, K.G.; Kajstura, J.; et al. Disruption of Striated Preferentially Expressed Gene Locus Leads to Dilated Cardiomyopathy in Mice. Circulation 2009, 119, 261–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tam, J.L.; Triantaphyllopoulos, K.; Todd, H.; Raguz, S.; de Wit, T.; Morgan, J.E.; Partridge, T.A.; Makrinou, E.; Grosveld, F.; Antoniou, M. The human desmin locus: Gene organization and LCR-mediated transcriptional control. Genomics 2006, 87, 733–746. [Google Scholar] [CrossRef] [Green Version]
- Goebel, H.H. Desmin-related myopathies. Curr. Opin. Neurol. 1997, 10, 426–429. [Google Scholar]
- Huang, Y.-S.; Xing, Y.-L.; Li, H.-W. Heterozygous desmin gene (DES) mutation contributes to familial dilated cardiomyopathy. J. Int. Med. Res. 2021, 49, 03000605211006598. [Google Scholar] [CrossRef]
- Klauke, B.; Kossmann, S.; Gaertner, A.; Brand, K.; Stork, I.; Brodehl, A.; Dieding, M.; Walhorn, V.; Anselmetti, D.; Gerdes, D.; et al. De novo desmin-mutation N116S is associated with arrhythmogenic right ventricular cardiomyopathy. Hum. Mol. Genet. 2010, 19, 4595–4607. [Google Scholar] [CrossRef] [Green Version]
- Lorenzon, A.; Beffagna, G.; Bauce, B.; De Bortoli, M.; Mura, I.E.L.; Calore, M.; Dazzo, E.; Basso, C.; Nava, A.; Thiene, G.; et al. Desmin Mutations and Arrhythmogenic Right Ventricular Cardiomyopathy. Am. J. Cardiol. 2013, 111, 400–405. [Google Scholar] [CrossRef]
- Tamiya, R.; Saito, Y.; Fukamachi, D.; Nagashima, K.; Aizawa, Y.; Ohkubo, K.; Hatta, T.; Sezai, A.; Tanaka, M.; Ishikawa, T.; et al. Desmin-related myopathy characterized by non-compaction cardiomyopathy, cardiac conduction defect, and coronary artery dissection. ESC Heart Fail. 2020, 7, 1338–1343. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, P.B.; Pierson, C.R.; Joshi, M.; Liu, X.; Ravenscroft, G.; Moghadaszadeh, B.; Talabere, T.; Viola, M.; Swanson, L.C.; Haliloğlu, G.; et al. SPEG Interacts with Myotubularin, and Its Deficiency Causes Centronuclear Myopathy with Dilated Cardiomyopathy. Am. J. Hum. Genet. 2014, 95, 218–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, J.; Ma, W.; Chen, Y.; Jiang, R.; Zeng, Q.; Tan, J.; Jiang, H.; Li, Q.; Zhang, V.W.; Wang, J.; et al. Novel SPEG variant cause centronuclear myopathy in China. J. Clin. Lab. Anal. 2020, 34, e23054. [Google Scholar] [CrossRef] [PubMed]
- Lornage, X.; Sabouraud, P.; Lannes, B.; Gaillard, D.; Schneider, R.; Deleuze, J.-F.; Boland, A.; Thompson, J.; Böhm, J.; Biancalana, V.; et al. Novel SPEG Mutations in Congenital Myopathy without Centralized Nuclei. J. Neuromuscul. Dis. 2018, 5, 257–260. [Google Scholar] [CrossRef] [PubMed]
- Almannai, M.; Luo, S.; Faqeih, E.; Al Mutairi, F.; Li, Q.; Agrawal, P.B. Homozygous SPEG Mutation Is Associated with Isolated Dilated Cardiomyopathy. Circ. Genom. Precis. Med. 2021, 14, e003310. [Google Scholar] [CrossRef]
- Levitas, A.; Muhammad, E.; Zhang, Y.; Perea Gil, I.; Serrano, R.; Diaz, N.; Arafat, M.; Gavidia, A.A.; Kapiloff, M.S.; Mercola, M.; et al. A novel recessive mutation in SPEG causes early onset dilated cardiomyopathy. PLoS Genet. 2020, 16, e1009000. [Google Scholar] [CrossRef]
- Wang, H.; Schänzer, A.; Kampschulte, B.; Daimagüler, H.-S.; Logeswaran, T.; Schlierbach, H.; Petzinger, J.; Ehrhardt, H.; Hahn, A.; Cirak, S. A novel SPEG mutation causes non-compaction cardiomyopathy and neuropathy in a floppy infant with centronuclear myopathy. Acta Neuropathol. Commun. 2018, 6, 83. [Google Scholar] [CrossRef]
- Quan, C.; Li, M.; Du, Q.; Chen, Q.; Wang, H.; Campbell, D.; Fang, L.; Xue, B.; MacKintosh, C.; Gao, X.; et al. SPEG Controls Calcium Reuptake into the Sarcoplasmic Reticulum Through Regulating SERCA2a by Its Second Kinase-Domain. Circ. Res. 2019, 124, 712–726. [Google Scholar] [CrossRef] [Green Version]
- Campbell, H.; Aguilar-Sanchez, Y.; Quick, A.P.; Dobrev, D.; Wehrens, X.H. SPEG: A key regulator of cardiac calcium homeostasis. Cardiovasc. Res. 2020, 117, 2175–2185. [Google Scholar] [CrossRef] [PubMed]
- Sedaghat-Hamedani, F.; Haas, J.; Zhu, F.; Geier, C.; Kayvanpour, E.; Liss, M.; Lai, A.; Frese, K.; Pribe-Wolferts, R.; Amr, A.; et al. Clinical genetics and outcome of left ventricular non-compaction cardiomyopathy. Eur. Heart J. 2017, 38, 3449–3460. [Google Scholar]
- Nozaki, Y.; Kato, Y.; Uike, K.; Yamamura, K.; Kikuchi, M.; Yasuda, M.; Ohno, S.; Horie, M.; Murayama, T.; Kurebayashi, N.; et al. Co-Phenotype of Left Ventricular Non-Compaction Cardiomyopathy and Atypical Catecholaminergic Polymorphic Ventricular Tachycardia in Association with R169Q, a Ryanodine Receptor Type 2 Missense Mutation. Circ. J. 2020, 84, 226–234. [Google Scholar] [CrossRef] [Green Version]
- Quick, A.P.; Wang, Q.; Philippen, L.E.; Barreto-Torres, G.; Chiang, D.; Beavers, D.; Wang, G.; Khalid, M.; Reynolds, J.O.; Campbell, H.M.; et al. SPEG (Striated Muscle Preferentially Expressed Protein Kinase) Is Essential for Cardiac Function by Regulating Junctional Membrane Complex Activity. Circ. Res. 2017, 120, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Qualls, A.; Donkervoort, S.; Herkert, J.C.; D’Gama, A.M.; Bharucha-Goebel, D.; Collins, J.; Bs, K.R.C.; Foley, A.R.; Schoots, M.H.; Jongbloed, J.D.; et al. Novel SPEG mutations in congenital myopathies: Genotype–phenotype correlations. Muscle Nerve 2019, 59, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Castiglioni, C.; Bayram, A.K.; Fattori, F.; Pekuz, S.; Araneda, D.; Per, H.; Erazo, R.; Gümüş, H.; Zorludemir, S.; et al. Insights from genotype–phenotype correlations by novel SPEG mutations causing centronuclear myopathy. Neuromuscul. Disord. 2017, 27, 836–842. [Google Scholar] [CrossRef] [PubMed]
- Ashkenazy, H.; Erez, E.; Martz, E.; Pupko, T.; Ben-Tal, N. ConSurf 2010: Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010, 38, W529–W533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venselaar, H.; Te Beek, T.A.; Kuipers, R.K.; Hekkelman, M.L.; Vriend, G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinform. 2010, 11, 1–10. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).