DOT1L Methyltransferase Regulates Calcium Influx in Erythroid Progenitor Cells in Response to Erythropoietin
Abstract
1. Introduction
2. Results
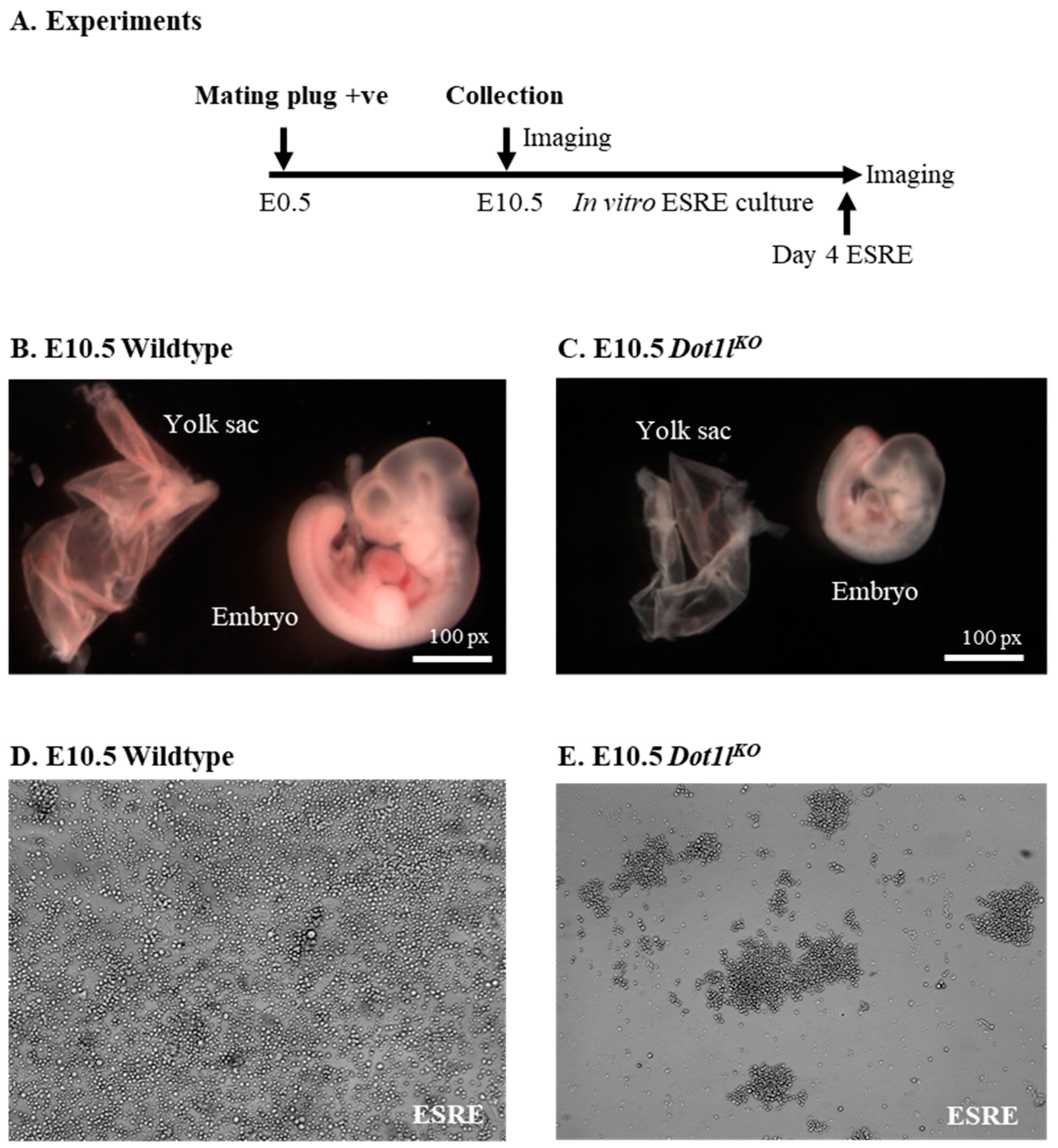
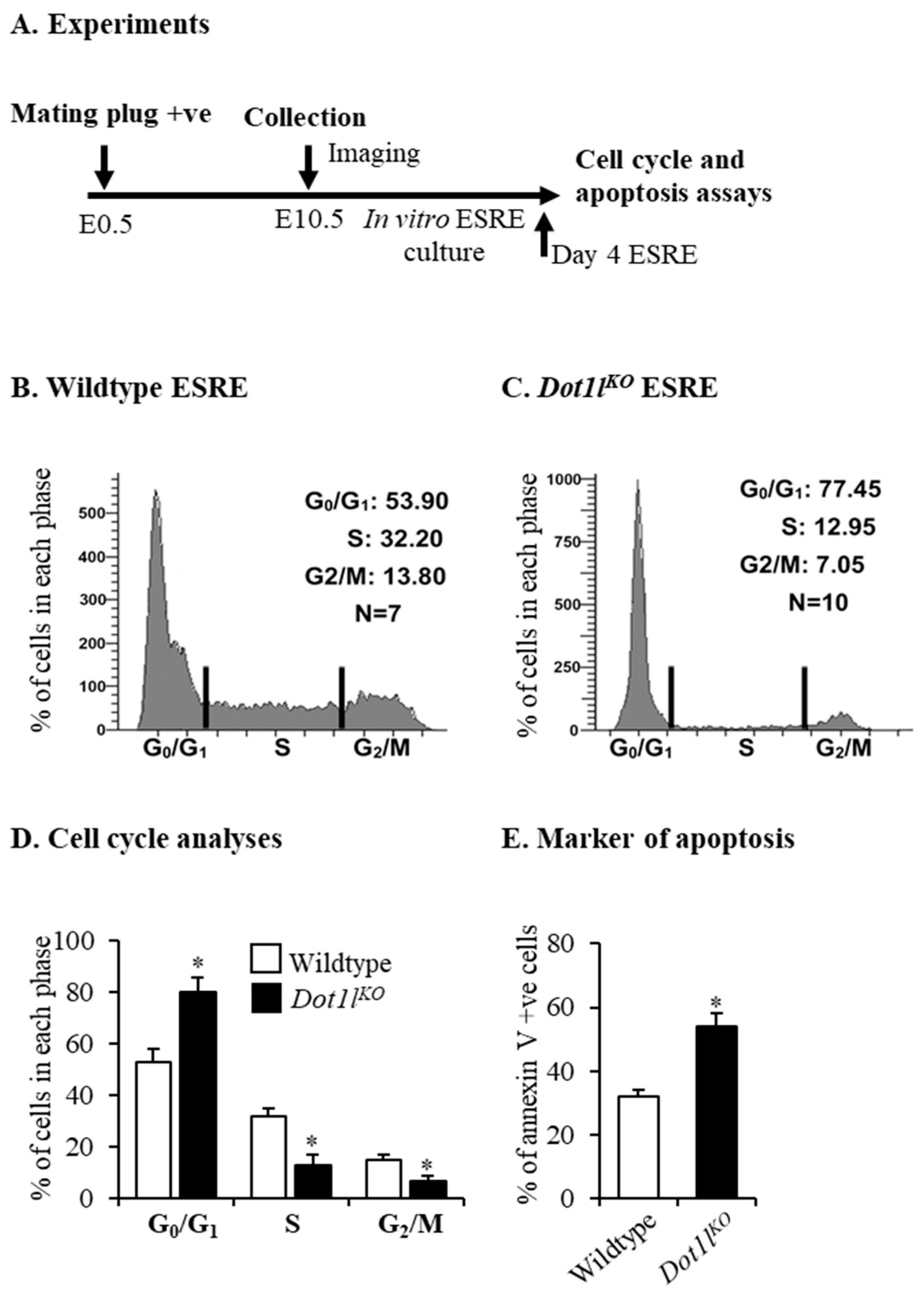
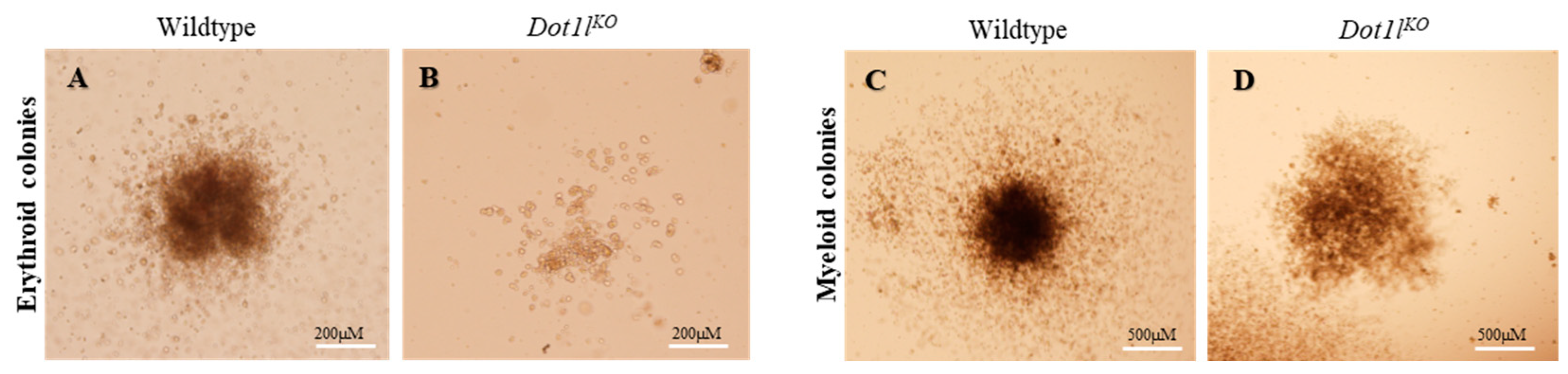
2.1. Dot1lKO Erythroblasts Displayed Decreased Proliferation, Cell Cycle Arrest, and Increased Apoptosis
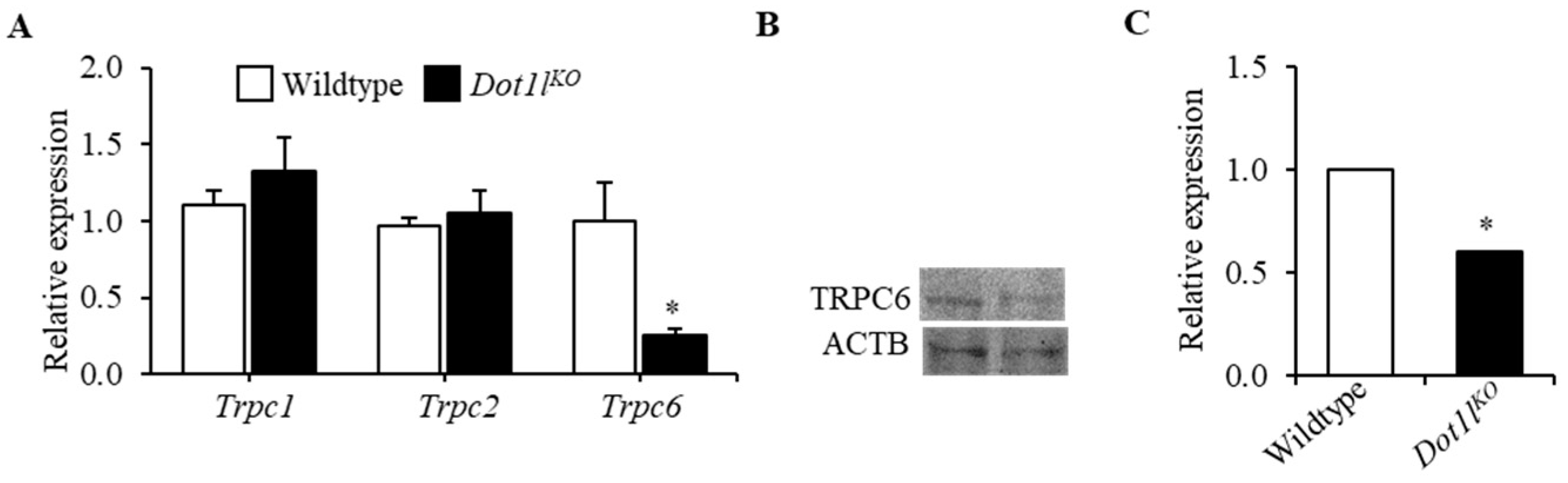
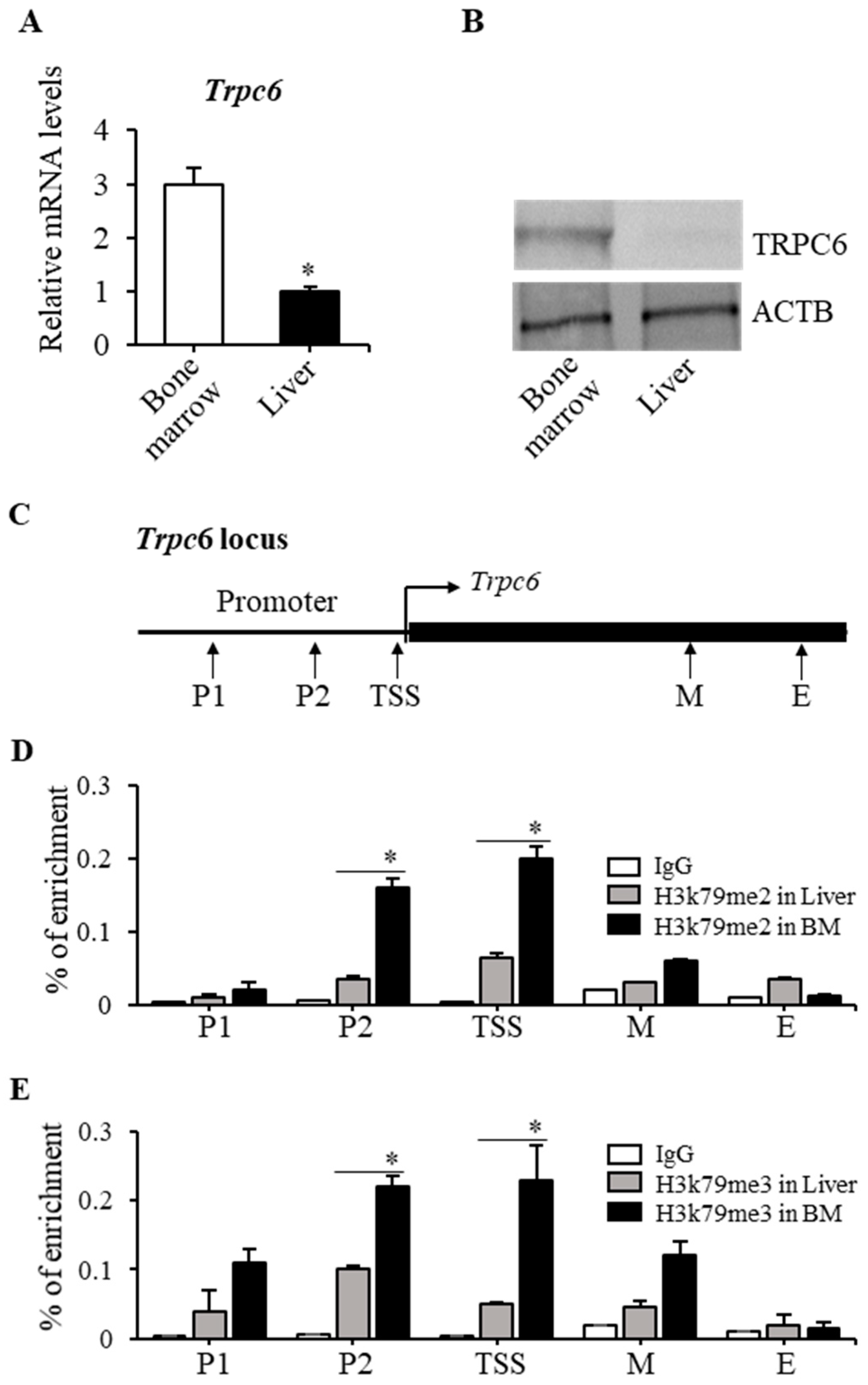
2.2. TRPC6 Expression Was Downregulated in Dot1lKO Hematopoietic Progenitor Cells
2.3. H3K79 Methylation Was Closely Associated with the Trpc6 Expression Level
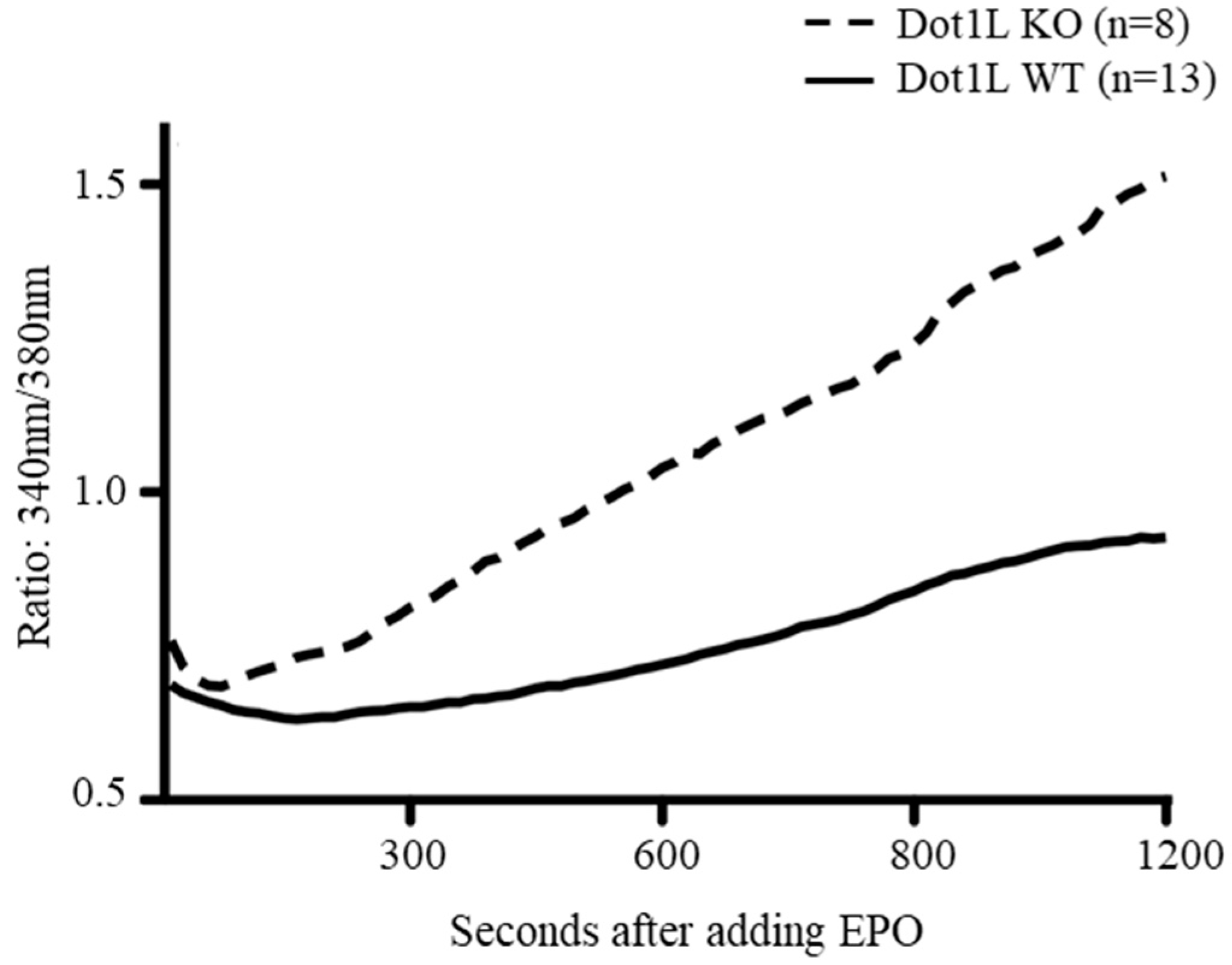
2.4. Abnormal Calcium Influx in Dot1lKO KIT-Positive HPCs in Response to EPO
3. Discussion
4. Materials and Methods
4.1. Mouse Lines and Isolation of Cells from Yolk Sac
4.2. Assessment of Cell Proliferation, Cell Cycle Analyses, and Apoptosis Assays
4.3. Analysis of Definitive Erythropoiesis from Yolk Sac Cells
4.4. Isolation of KIT-Positive Yolk Sac Cells
4.5. RNA Extraction, cDNA Preparation, and RT-qPCR
4.6. Western Blot Analyses
4.7. Extraction of Bone Marrow and Liver Cells
4.8. ChIP Assay for H3K79 Di- and Tri-Methylation in Trpc6 Locus
4.9. Measuring Calcium Influx of Yolk Sac Cells in Response to EPO
4.10. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tsiftsoglou, A.S.; Vizirianakis, I.S.; Strouboulis, J. Erythropoiesis: Model systems, molecular regulators, and developmental programs. IUBMB Life 2009, 61, 800–830. [Google Scholar] [CrossRef] [PubMed]
- Ingley, E.; Tilbrook, P.A.; Klinken, S.P. New insights into the regulation of erythroid cells. IUBMB Life 2004, 56, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.S.; Lim, S.K.; D’Agati, V.; Costantini, F. Differential effects of an erythropoietin receptor gene disruption on primitive and definitive erythropoiesis. Genes Dev. 1996, 10, 154–164. [Google Scholar] [CrossRef]
- Wu, H.; Liu, X.; Jaenisch, R.; Lodish, H.F. Generation of committed erythroid BFU-E and CFU-E progenitors does not require erythropoietin or the erythropoietin receptor. Cell 1995, 83, 59–67. [Google Scholar] [CrossRef]
- Miller, B.A.; Bell, L.L.; Lynch, C.J.; Cheung, J.Y. Erythropoietin modulation of intracellular calcium: A role for tyrosine phosphorylation. Cell Calcium 1994, 16, 481–490. [Google Scholar] [CrossRef]
- Fisher, J.W. Erythropoietin: Physiology and pharmacology update. Exp. Biol. Med. 2003, 228, 1–14. [Google Scholar] [CrossRef]
- Tong, Q.; Chu, X.; Cheung, J.Y.; Conrad, K.; Stahl, R.; Barber, D.L.; Mignery, G.; Miller, B.A. Erythropoietin-modulated calcium influx through TRPC2 is mediated by phospholipase Cgamma and IP3R. Am. J. Physiol. Cell Physiol. 2004, 287, C1667–C1678. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tong, Q.; Hirschler-Laszkiewicz, I.; Zhang, W.; Conrad, K.; Neagley, D.W.; Barber, D.L.; Cheung, J.Y.; Miller, B.A. TRPC3 is the erythropoietin-regulated calcium channel in human erythroid cells. J. Biol. Chem. 2008, 283, 10385–10395. [Google Scholar] [CrossRef]
- Miller, B.A.; Cheung, J.Y.; Tillotson, D.L.; Hope, S.M.; Scaduto, R.C., Jr. Erythropoietin stimulates a rise in intracellular-free calcium concentration in single BFU-E derived erythroblasts at specific stages of differentiation. Blood 1989, 73, 1188–1194. [Google Scholar] [CrossRef]
- Hensold, J.O.; Dubyak, G.; Housman, D.E. Calcium ionophore, A23187, induces commitment to differentiation but inhibits the subsequent expression of erythroid genes in murine erythroleukemia cells. Blood 1991, 77, 1362–1370. [Google Scholar] [CrossRef]
- Chu, X.; Tong, Q.; Cheung, J.Y.; Wozney, J.; Conrad, K.; Mazack, V.; Zhang, W.; Stahl, R.; Barber, D.L.; Miller, B.A. Interaction of TRPC2 and TRPC6 in erythropoietin modulation of calcium influx. J. Biol. Chem. 2004, 279, 10514–10522. [Google Scholar] [CrossRef] [PubMed]
- Lintschinger, B.; Balzer-Geldsetzer, M.; Baskaran, T.; Graier, W.F.; Romanin, C.; Zhu, M.X.; Groschner, K. Coassembly of Trp1 and Trp3 proteins generates diacylglycerol- and Ca2+-sensitive cation channels. J. Biol. Chem. 2000, 275, 27799–27805. [Google Scholar] [CrossRef] [PubMed]
- Strübing, C.; Krapivinsky, G.; Krapivinsky, L.; Clapham, D.E. TRPC1 and TRPC5 form a novel cation channel in mammalian brain. Neuron 2001, 29, 645–655. [Google Scholar] [CrossRef]
- Hoenderop, J.G.; Voets, T.; Hoefs, S.; Weidema, F.; Prenen, J.; Nilius, B.; Bindels, R.J. Homo- and heterotetrameric architecture of the epithelial Ca2+ channels TRPV5 and TRPV6. EMBO J. 2003, 22, 776–785. [Google Scholar] [CrossRef]
- Hofmann, T.; Schaefer, M.; Schultz, G.; Gudermann, T. Subunit composition of mammalian transient receptor potential channels in living cells. Proc. Natl. Acad. Sci. USA 2002, 99, 7461–7466. [Google Scholar] [CrossRef]
- Goel, M.; Sinkins, W.G.; Schilling, W.P. Selective association of TRPC channel subunits in rat brain synaptosomes. J. Biol. Chem. 2002, 277, 48303–48310. [Google Scholar] [CrossRef] [PubMed]
- Mariño-Ramírez, L.; Kann, M.G.; Shoemaker, B.A.; Landsman, D. Histone structure and nucleosome stability. Expert Rev. Proteom. 2005, 2, 719–729. [Google Scholar] [CrossRef]
- Gil, M.; Ramil, F.; AgÍs, J.A. Hydroids (Cnidaria, Hydrozoa) from Mauritanian Coral Mounds. Zootaxa 2020, 4878, 4872–4878. [Google Scholar] [CrossRef]
- Jones, B.; Su, H.; Bhat, A.; Lei, H.; Bajko, J.; Hevi, S.; Baltus, G.A.; Kadam, S.; Zhai, H.; Valdez, R.; et al. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet. 2008, 4, e1000190. [Google Scholar] [CrossRef]
- Amorim, C.A.; Rondina, D.; Rodrigues, A.P.; Goncalves, P.B.; de Figueiredo, J.R.; Giorgetti, A. Cryopreservation of isolated ovine primordial follicles with propylene glycol and glycerol. Fertil. Steril. 2004, 81 (Suppl. 1), 735–740. [Google Scholar] [CrossRef]
- Feng, Q.; Wang, H.; Ng, H.H.; Erdjument-Bromage, H.; Tempst, P.; Struhl, K.; Zhang, Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. CB 2002, 12, 1052–1058. [Google Scholar] [CrossRef]
- Steger, D.J.; Lefterova, M.I.; Ying, L.; Stonestrom, A.J.; Schupp, M.; Zhuo, D.; Vakoc, A.L.; Kim, J.E.; Chen, J.; Lazar, M.A.; et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol. Cell. Biol. 2008, 28, 2825–2839. [Google Scholar] [CrossRef]
- van Leeuwen, F.; Gafken, P.R.; Gottschling, D.E. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 2002, 109, 745–756. [Google Scholar] [CrossRef]
- Huyen, Y.; Zgheib, O.; Ditullio, R.A., Jr.; Gorgoulis, V.G.; Zacharatos, P.; Petty, T.J.; Sheston, E.A.; Mellert, H.S.; Stavridi, E.S.; Halazonetis, T.D. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 2004, 432, 406–411. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Yang, Y.; Ortega, M.M.; Copeland, J.N.; Zhang, M.; Jacob, J.B.; Fields, T.A.; Vivian, J.L.; Fields, P.E. Early mammalian erythropoiesis requires the Dot1L methyltransferase. Blood 2010, 116, 4483–4491. [Google Scholar] [CrossRef]
- Malcom, C.A.; Piaseka-Srader, J.; Chakravarthi, V.P.; Borosha, S.; Ratri, A.; Alvarez, N.; Vivian, J.L.; Fields, T.A.; Rumi, M.A.; Fields, P.E. The Role of DOT1L Methyltransferase Activity in Fetal Hematopoiesis. bioRxiv 2020, 2020, 319889. [Google Scholar] [CrossRef]
- Edvardsson, K.; Nguyen-Vu, T.; Kalasekar, S.M.; Pontén, F.; Gustafsson, J.; Williams, C. Estrogen receptor β expression induces changes in the microRNA pool in human colon cancer cells. Carcinogenesis 2013, 34, 1431–1441. [Google Scholar] [CrossRef] [PubMed]
- Access to fertility services by transgender persons: An Ethics Committee opinion. Fertil. Steril. 2015, 104, 1111–1115. [CrossRef]
- England, S.J.; McGrath, K.E.; Frame, J.M.; Palis, J. Immature erythroblasts with extensive ex vivo self-renewal capacity emerge from the early mammalian fetus. Blood 2011, 117, 2708–2717. [Google Scholar] [CrossRef]
- Malcom, C.A.; Ratri, A.; Piasecka-Srader, J.; Borosha, S.; Chakravarthi, V.P.; Alvarez, N.S.; Vivian, J.L.; Fields, T.A.; Karim Rumi, M.A.; Fields, P.E. Primitive Erythropoiesis in the Mouse is Independent of DOT1L Methyltransferase Activity. Front. Cell Dev. Biol. 2022, 9, 813503. [Google Scholar] [CrossRef]
- Borosha, S.; Ratri, A.; Ghosh, S.; Malcom, C.A.; Chakravarthi, V.P.; Vivian, J.L.; Fields, T.A.; Rumi, M.A.K.; Fields, P.E. DOT1L Mediated Gene Repression in Extensively Self-Renewing Erythroblasts. Front. Genet. 2022, 13, 828806. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.; Grillot, D.; Benito, A.; Richard, C.; Nuñez, G.; Fernández-Luna, J.L. Erythropoietin can promote erythroid progenitor survival by repressing apoptosis through Bcl-XL and Bcl-2. Blood 1996, 88, 1576–1582. [Google Scholar] [CrossRef]
- Koury, M.J.; Bondurant, M.C. Erythropoietin retards DNA breakdown and prevents programmed death in erythroid progenitor cells. Science 1990, 248, 378–381. [Google Scholar] [CrossRef] [PubMed]
- Hirschler-Laszkiewicz, I.; Zhang, W.; Keefer, K.; Conrad, K.; Tong, Q.; Chen, S.J.; Bronson, S.; Cheung, J.Y.; Miller, B.A. Trpc2 depletion protects red blood cells from oxidative stress-induced hemolysis. Exp. Hematol. 2012, 40, 71–83. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Numaga-Tomita, T.; Nishida, M. TRPC Channels in Cardiac Plasticity. Cells 2020, 9, 454. [Google Scholar] [CrossRef] [PubMed]
- Föller, M.; Lang, F. Ion Transport in Eryptosis, the Suicidal Death of Erythrocytes. Front. Cell Dev. Biol. 2020, 8, 597. [Google Scholar] [CrossRef]
- Greka, A.; Mundel, P. Balancing calcium signals through TRPC5 and TRPC6 in podocytes. J. Am. Soc. Nephrol. 2011, 22, 1969–1980. [Google Scholar] [CrossRef]
- Gees, M.; Colsoul, B.; Nilius, B. The role of transient receptor potential cation channels in Ca2+ signaling. Cold Spring Harb. Perspect. Biol. 2010, 2, a003962. [Google Scholar] [CrossRef]
- Imagawa, S.; Smith, B.R.; Palmer-Crocker, R.; Bunn, H.F. The effect of recombinant erythropoietin on intracellular free calcium in erythropoietin-responsive cells. Blood 1989, 73, 1452–1457. [Google Scholar] [CrossRef]
- Khristi, V.; Chakravarthi, V.P.; Singh, P.; Ghosh, S.; Pramanik, A.; Ratri, A.; Borosha, S.; Roby, K.F.; Wolfe, M.W.; Rumi, M.A.K. ESR2 regulates granulosa cell genes essential for follicle maturation and ovulation. Mol. Cell. Endocrinol. 2018, 474, 214–226. [Google Scholar] [CrossRef]
- Khristi, V.; Ratri, A.; Ghosh, S.; Pathak, D.; Borosha, S.; Dai, E.; Roy, R.; Chakravarthi, V.P.; Wolfe, M.W.; Karim Rumi, M.A. Disruption of ESR1 alters the expression of genes regulating hepatic lipid and carbohydrate metabolism in male rats. Mol. Cell. Endocrinol. 2019, 490, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Chakravarthi, V.P.; Ghosh, S.; Roby, K.F.; Wolfe, M.W.; Rumi, M.A.K. A Gatekeeping Role of ESR2 to Maintain the Primordial Follicle Reserve. Endocrinology 2020, 161, 37. [Google Scholar] [CrossRef] [PubMed]
- Chuong, E.B.; Rumi, M.A.; Soares, M.J.; Baker, J.C. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat. Genet. 2013, 45, 325–329. [Google Scholar] [CrossRef] [PubMed]
- Chakravarthi, V.P.; Khristi, V.; Ghosh, S.; Yerrathota, S.; Dai, E.; Roby, K.F.; Wolfe, M.W.; Rumi, M.A.K. ESR2 Is Essential for Gonadotropin-Induced Kiss1 Expression in Granulosa Cells. Endocrinology 2018, 159, 3860–3873. [Google Scholar] [CrossRef] [PubMed]

| Gene | Reference mRNA | Forward Primer | Reverse Primer |
|---|---|---|---|
| Trpc1 | NM_0011643.4 | 366F: cggttgtcagtccgcagat | 456R: tcgttttggccgatgattaagta |
| Trpc2 | NM_011644.3_ | 631F: ctcaagggtatgttgaagcagt | 741R: gttgtttgggcttaccacact |
| Trpc6 | NM_004621.6 | 164F: gcttccggggtaatgaaaaca | 255R: gtatgctggtcctcgattagc |
| Target | Chromosome 9 Locus | Forward Primer | Reverse Primer |
|---|---|---|---|
| Promoter-1 | 8543340-680 | aagcagggctcactgaatctgg | ggcattttccgatggtgtctg |
| Promoter-2 | 8543915-4113 | cccaaataaagaatgtgcctgg | cgctgaagagttactatgtcaaccg |
| Trpc6 TSS | 8544511-618 | gagagccaggactatttgctgatg | tgccctcgcccatacttacaag |
| Trpc6 Middle | 8549504-630 | tctacctcctgatgctgggcttac | ggggtttgaagagatgagagtgc |
| Trpc6 End | 8679716-902 | tgccctacaaagcaatgaaagg | aaagagagcgtgagcccaacac |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, Y.; Borosha, S.; Ratri, A.; Lee, E.B.; Wang, H.; Fields, T.A.; Kinsey, W.H.; Vivian, J.L.; Rumi, M.A.K.; Fields, P.E. DOT1L Methyltransferase Regulates Calcium Influx in Erythroid Progenitor Cells in Response to Erythropoietin. Int. J. Mol. Sci. 2022, 23, 5137. https://doi.org/10.3390/ijms23095137
Feng Y, Borosha S, Ratri A, Lee EB, Wang H, Fields TA, Kinsey WH, Vivian JL, Rumi MAK, Fields PE. DOT1L Methyltransferase Regulates Calcium Influx in Erythroid Progenitor Cells in Response to Erythropoietin. International Journal of Molecular Sciences. 2022; 23(9):5137. https://doi.org/10.3390/ijms23095137
Chicago/Turabian StyleFeng, Yi, Shaon Borosha, Anamika Ratri, Eun Bee Lee, Huizhen Wang, Timothy A. Fields, William H. Kinsey, Jay L. Vivian, M. A. Karim Rumi, and Patrick E. Fields. 2022. "DOT1L Methyltransferase Regulates Calcium Influx in Erythroid Progenitor Cells in Response to Erythropoietin" International Journal of Molecular Sciences 23, no. 9: 5137. https://doi.org/10.3390/ijms23095137
APA StyleFeng, Y., Borosha, S., Ratri, A., Lee, E. B., Wang, H., Fields, T. A., Kinsey, W. H., Vivian, J. L., Rumi, M. A. K., & Fields, P. E. (2022). DOT1L Methyltransferase Regulates Calcium Influx in Erythroid Progenitor Cells in Response to Erythropoietin. International Journal of Molecular Sciences, 23(9), 5137. https://doi.org/10.3390/ijms23095137






