HKT1;1 and HKT1;2 Na+ Transporters from Solanum galapagense Play Different Roles in the Plant Na+ Distribution under Salinity
Abstract
:1. Introduction
2. Results
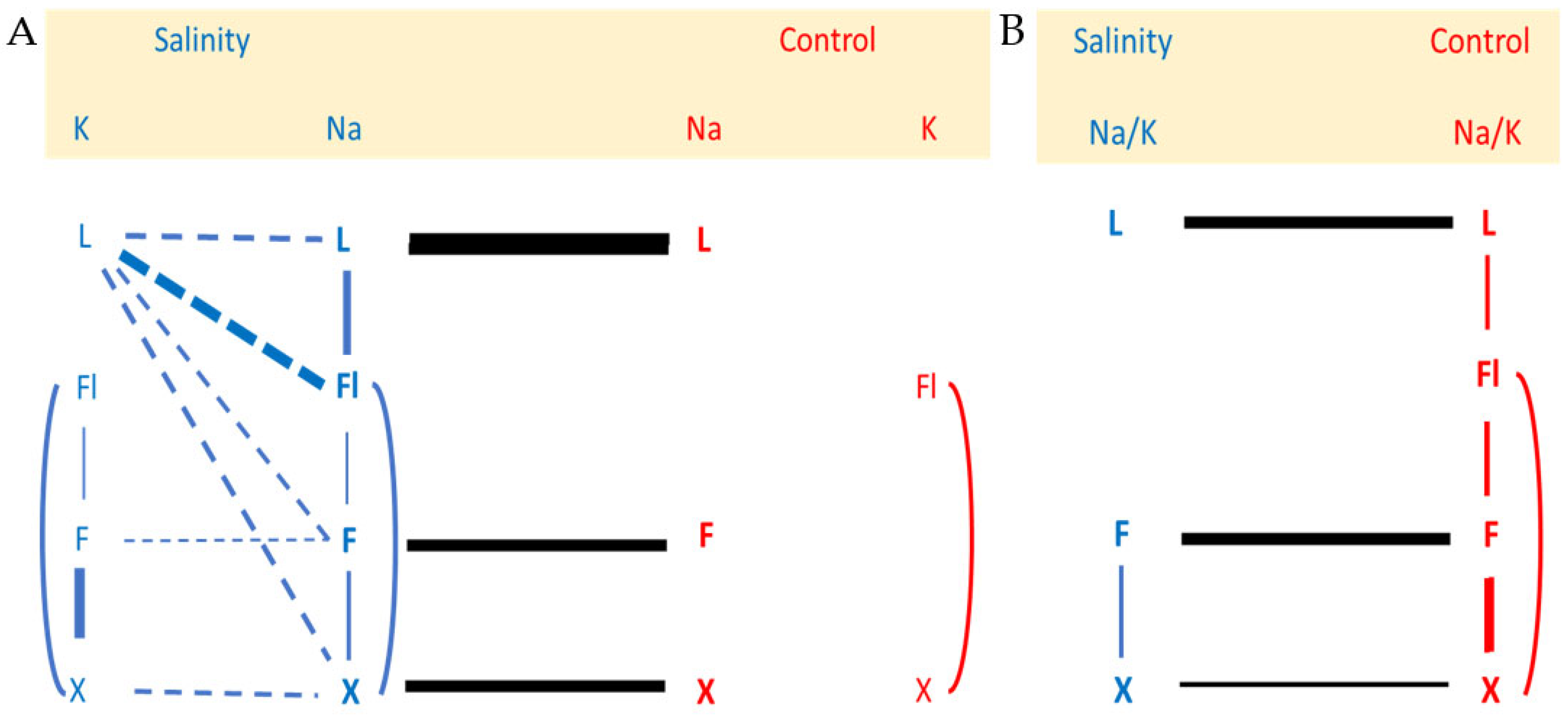
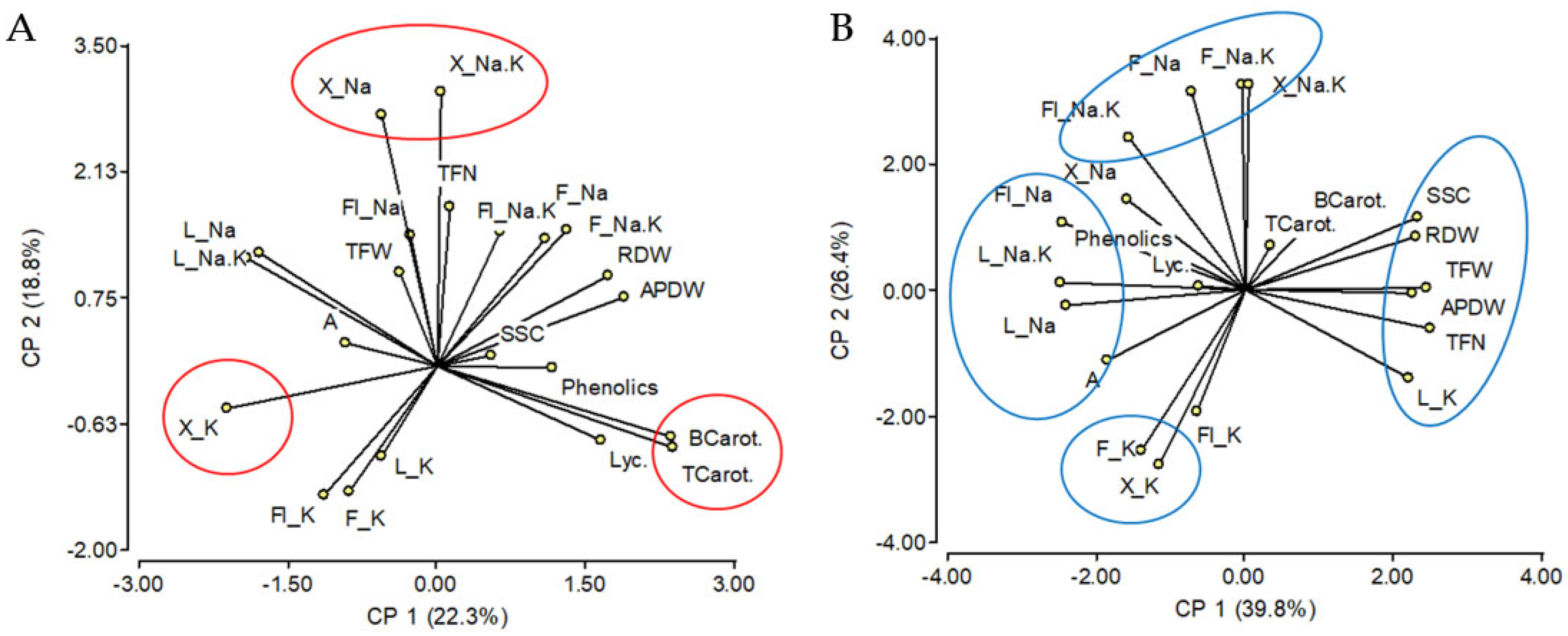
2.1. Effects of Salinity Treatment on Na and K Contents of the Different Tissues and Their Relationships
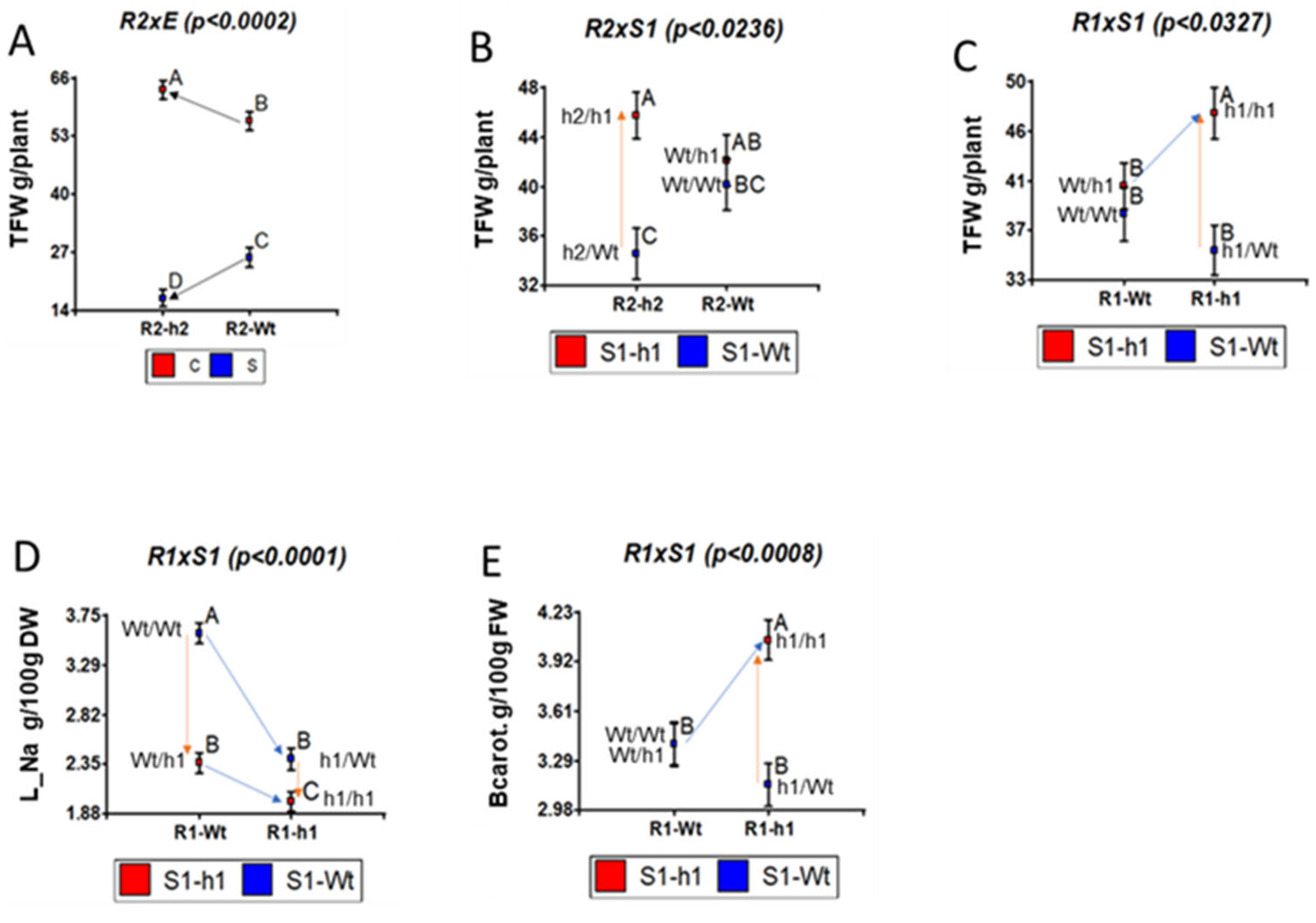
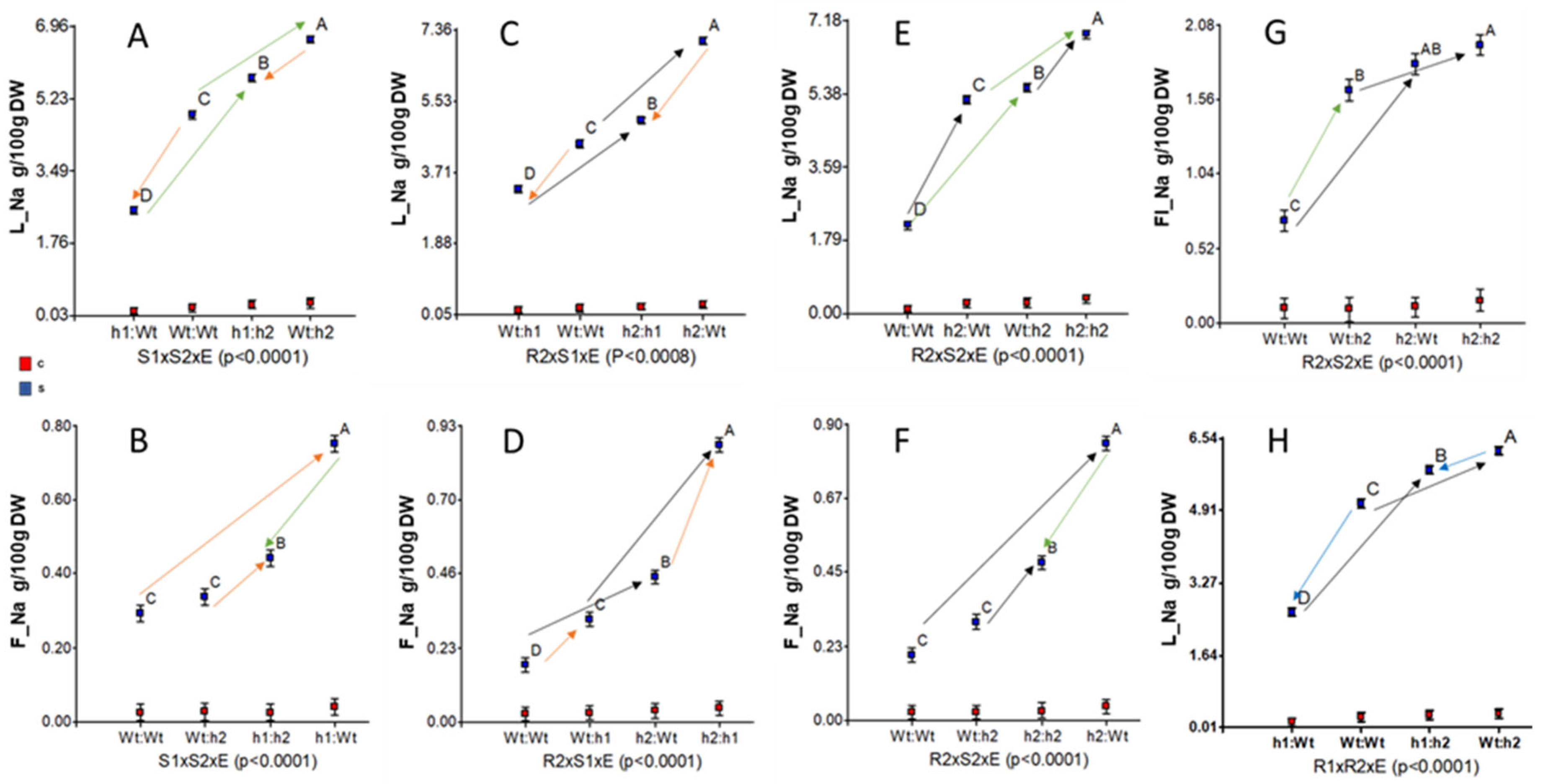
2.2. Rootstock and Scion Contributions of S. galapagense Alleles at HKT1;1 and HKT1;2 to Na Distribution, Salt Tolerance and Fruit Quality
2.3. Silencing Rootstock and Scion S. galapagense Alleles at HKT1;1 and HKT1;2 Affect K+ Tissular Distribution under Both Control and Salinity Conditions in a Complex Way
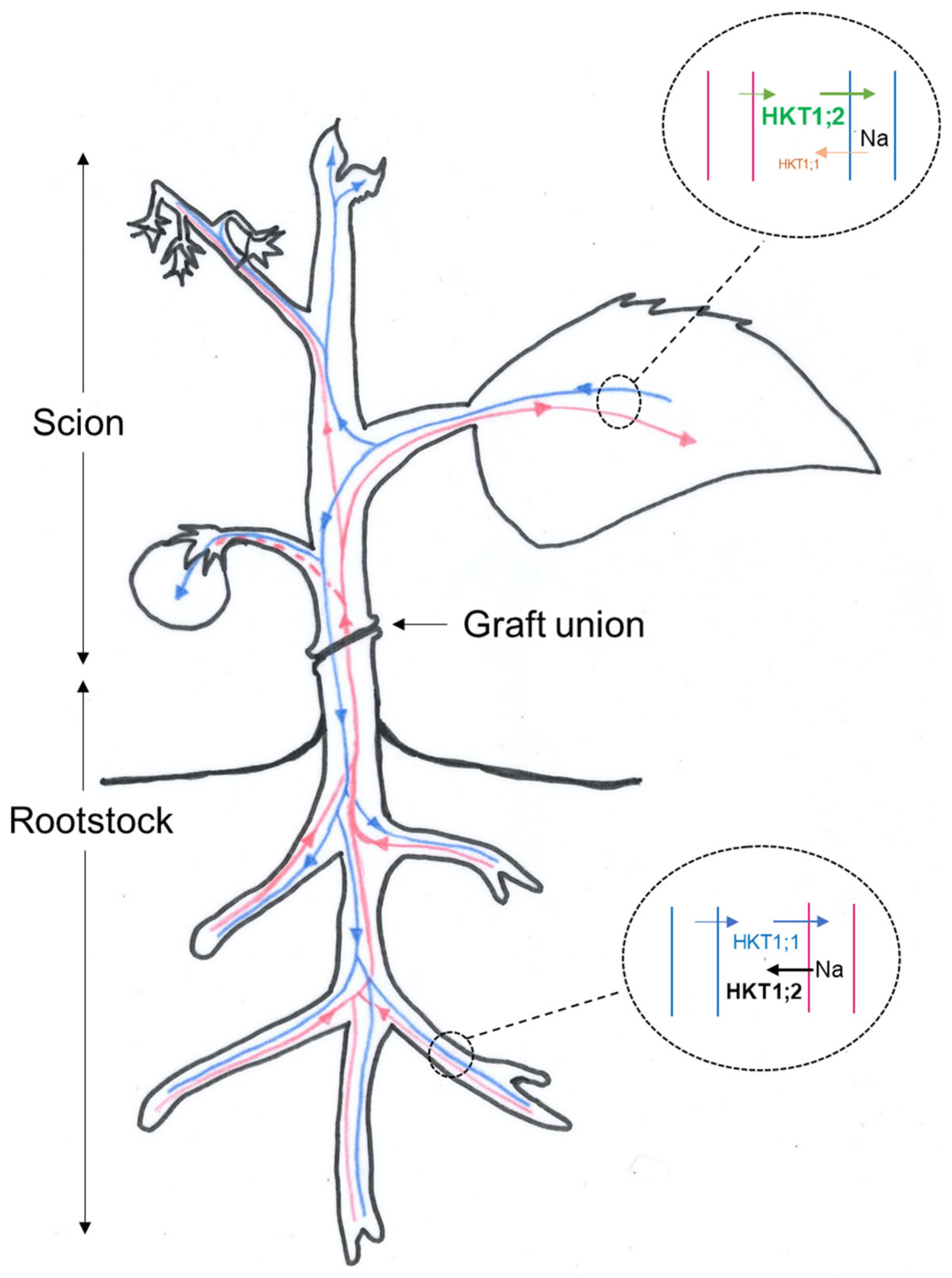
3. Discussion
3.1. A Model for Na Recirculation
3.2. Leaf K Content Might Regulate Fruit Soluble Solids Content under Salinity
4. Materials and Methods
4.1. Plant Materials
4.2. Plant Growth Conditions and Phenotyping
4.3. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Munns, R.; Gilliham, M. Salinity tolerance of crops-what is the cost? New Phytol. 2015, 208, 668–673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero-Aranda, M.R.; Soria, T.; Cuartero, J. Tomato plant-water uptake and plant-water relationships under saline growth conditions. Plant Sci. 2001, 160, 265–272. [Google Scholar] [CrossRef]
- Tal., M.; Shannon, M.C. Salt tolerance in the wild relatives of the cultivated tomato: Responses of Lycopersicon esculentum, L. cheesmaniae, L. peruvianum, S. pennellii and F1 hybrids to high salinity. Aust. J. Plant Physiol. 1983, 10, 109–117. [Google Scholar] [CrossRef]
- Asins, M.J.; Bretó, M.P.; Cambra, M.; Carbonell, E.A. Salt tolerance in Lycopersicon species. I. Character definition and change in gene expression. Theor. Appl. Genet. 1993, 86, 737–743. [Google Scholar] [CrossRef]
- Pailles, Y.; Awlia, M.; Julkoswka, M.; Passone, L.; Zemmouri, K.; Negrao, S.; Schmöckel, S.M.; Tester, M. Diverse traits contribute to salinity tolerance of wild tomato seedlings from the Galapagos Islands. Plant Physiol. 2020, 182, 534–546. [Google Scholar] [CrossRef] [Green Version]
- Bonarota, M.-S.; Kosma, D.K.; Barrios-Masias, F.H. Salt tolerance mechanisms in the Lycopersion clade and their trade-offs. AoB Plants 2022, 14, plab072. [Google Scholar]
- Rick, C.M. Lycopersicon Mill. In Flora of the Galápagos Islands; Wiggins, I.L., Porter, D.M., Eds.; University of Stanford Press: Standford, CA, USA, 1971; pp. 468–471. [Google Scholar]
- Rick, C.M. Genetic and systematic studies on accessions of Lycopersicon from the Galapagos Islands. Amer. J. Bot. 1956, 43, 687–696. [Google Scholar] [CrossRef]
- Rush, D.W.; Epstein, E. Breeding and selecting for crop tolerance by the incorporation of wild germplasm into a domesticated tomato. J. Am. Soc. Hortic. Sci. 1976, 106, 669–670. [Google Scholar]
- Nuez, F.; Prohens, J.; Blanca, J.M. Relationships, origin, and diversity of Galapagos tomatoes: Implications for the conservation of natural populations. Am. J. Bot. 2004, 9, 86–99. [Google Scholar] [CrossRef]
- Asins, M.J. Present and future of QTL analysis in plant breeding. Plant Breed. 2002, 121, 281–291. [Google Scholar] [CrossRef]
- Monforte, A.; Asins, M.J.; Carbonell, E.A. Salt tolerance in Lycopersicon species. V. May genetic variability at quantitative trait loci affect their analysis? Theor. Appl. Genet. 1997, 95, 284–293. [Google Scholar] [CrossRef]
- Villalta, I.; Bernet, G.P.; Carbonell, E.A.; Asins, M.J. Comparative QTL analysis of salinity tolerance in terms of fruit yield using two Solanum populations of F7 lines. Theor. Appl. Genet. 2007, 114, 1001–1017. [Google Scholar] [CrossRef] [PubMed]
- Estañ, M.T.; Villalta, I.; Bolarín, M.C.; Carbonell, E.A.; Asins, M.J. Identification of fruit yield loci controlling the salt tolerance conferred by Solanum rootstocks. Theor. Appl. Genet. 2009, 118, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Frary, A.; Keles, D.; Pinar, H.; Gol, D.; Doganlar, S. NaCl tolerance in Lycopersicon pennellii introgression lines: QTL related to physiological responses. Biol. Plant. 2011, 55, 461–468. [Google Scholar] [CrossRef]
- Asins, M.J.; Bolarín, M.C.; Pérez-Alfocea, F.; Estañ, M.T.; Martinez-Andújar, C.; Albacete, A.; Villalta, I.; Bernet, G.P.; Dodd, I.; Carbonell, E.A. Genetic analysis of physiological components of salt tolerance conferred by Solanum rootstocks. What is the rootstock doing for the scion? Theor. Appl. Genet. 2010, 121, 105–115. [Google Scholar] [CrossRef]
- Asins, M.J.; Villalta, I.; Aly, M.M.; Olías, R.; Álvarez De Morales, P.; Huertas, R.; Li, J.; Jaime-Pérez, N.; Haro, R.; Raga, V.; et al. Two closely linked tomato HKT coding genes are positional candidates for the major tomato QTL involved in Na+/K+ homeostasis. Plant Cell Environ. 2013, 36, 1171–1191. [Google Scholar] [CrossRef]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [Green Version]
- Kashyap, S.P.; Kumari, N.; Mishra, P.; Moharana, D.P.; Aamir, M. Tapping the potential of Solanum lycopersicum L. pertaining to salinity tolerance: Perspectives and challenges. Genet. Resour. Crop Evol. 2021. [Google Scholar] [CrossRef]
- Almeida, P.M.F.; de Boer, G.J.; de Boer, A.H. Assessment of natural variation in the first pore domain of the tomato HKT1;2 transporter and characterization of mutated versions of SlHKT1;2 expressed in Xenopus laevis oocytes and via complementation of the salt sensitive athkt1;1 mutant. Front. Plant Sci. 2014, 5, 600. [Google Scholar] [CrossRef] [Green Version]
- Villalta, I.; Reina-Sánchez, A.; Bolarín, M.C.; Cuartero, J.; Belver, A.; Venema, K.; Carbonell, E.A.; Asins, M.J. Genetic analysis of Na+ and K+ concentrations in leaf and stem as physiological components of salt tolerance in tomato. Theor. Appl. Genet. 2008, 116, 869–880. [Google Scholar] [CrossRef]
- Asins, M.J.; Raga, V.; Roca, D.; Carbonell, E.A. Genetic dissection of tomato rootstock effects on scion traits under moderate salinity. Theor. Appl. Genet. 2015, 128, 667–679. [Google Scholar] [CrossRef] [PubMed]
- Jaime-Pérez, N.; Pineda, B.; García-Sogo, B.; Atares, A.; Athman, A.; Byrt, C.S.; Olias, R.; Asins, M.J.; Gilliham, M.; Moreno, V.; et al. The Na+ transporter encoded by the HKT1;2 gene modulates Na+/K+ homeostasis in tomato shoots under salinity. Plant Cell Environ. 2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero-Aranda, M.R.; González Fernández, P.; Pérez Tienda, J.R.; López Díaz, M.R.; Espinosa, J.; Granum, E.; Traverso, J.A.; Pineda, B.; Garcia-Sogo, B.; Moreno, V.; et al. Na+ transporter HKT1;2 reduces flower Na+ content and considerably mitigates the decline in tomato fruit yields under saline conditions. Plant Physiol. Biochem. 2020, 154, 341–352. [Google Scholar] [CrossRef]
- Ali, A.; Raddatz, N.; Pardo, J.M.; Yun, D.-J. HKT sodium and potassium transporters in Arabidopsis thaliana and related halophyte species. Physiol Plant. 2021, 171, 546–558. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Liang, X.; Yin, P.; Zhang, M.; Jiang, C. A domestication-associated reduction in K+-preferring HKT transporter activity underlies maize shoot K+ accumulation and salt tolerance. New Phytol. 2019, 222, 301–317. [Google Scholar] [CrossRef]
- Huang, L.; Kuang, L.; Wu, L.; Shen, Q.; Han, Y.; Jiang, L.; Wu, D.; Zhang, G. The HKT transporter HvHKT1;5 negatively regulates salt tolerance. Plant Physiol. 2020, 182, 584–596. [Google Scholar] [CrossRef]
- Fisher, D.B. Long distance transport. In Biochemistry and Molecular Biology of Plants, 4th ed.; Buchanan, B.B., Gruissen, W., Russell, L.J., Eds.; American Society of Plant Biologists: Rockville, MD, USA, 2000; pp. 730–784. [Google Scholar]
- Romero-Aranda, M.R.; Espinosa, J.; González-Fernández, P.; Jaime-Fernández, E.; Traverso, J.A.; Asins, M.J.; Belver, A. Role of Na+ transporters HKT1;1 and HKT1;2 in tomato salt tolerance. I. Function loss of cheesmaniae alleles in roots and aerial parts. Plant Physiol. Biochem. 2021, 168, 282–293. [Google Scholar] [CrossRef]
- Windt, C.W.; Gerkema, E.; Van As, H. Most water in the tomato truss is imported through the xylem, not the phloem: A nuclear magnetic resonance flow imaging study. Plant Physiol. 2009, 151, 830–842. [Google Scholar] [CrossRef] [Green Version]
- Diouf, I.A.; Derivot, L.; Pascual, L.; Causse, M. Water deficit and salinity stress reveal many specific QTL for plant growth and fruit quality traits in tomato. Front. Plant Sci. 2018, 9, 279. [Google Scholar] [CrossRef] [Green Version]
- Ragel, P.; Raddatz, N.; Leidi, E.O.; Quintero, F.J.; Pardo, J.M. Regulation of K+ nutrition in plants. Front. Plant Sci. 2019, 10, 281. [Google Scholar] [CrossRef] [Green Version]
- Isayenkov, S.V.; Maathuis, F.J.M. Plant salinity stress: Many unanswered questions remain. Front. Plant Sci. 2019, 10, 80. [Google Scholar] [CrossRef] [Green Version]
- Shabala, S.; Pottosin, I. Regulation of potassium transport in plants under hostile conditions: Implications for abiotic and biotic stress tolerance. Physiol. Plant. 2014, 151, 257–279. [Google Scholar] [CrossRef]
- Ahmad, I.; Maathuis, F.J.M. Cellular and tissue distribution of potassium: Physiological relevance, mechanisms and regulation. J. Plant Physiol. 2014, 171, 708–714. [Google Scholar] [CrossRef] [PubMed]
- Frary, A.; Gol, D.; Keles, D.; Okmen, B.; Pinar, H.; Sigva, H.O.; Yemenicioğlu, A.; Doganlar, S. Salt tolerance in Solanum pennellii: Antioxidant response and related QTL. BMC Plant Biol. 2010, 10, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rouphael, Y.; Petropoulus, S.A.; Cardarelli, M.; Colla, G. Salinity as eustressor for enhancing quality of vegetables. Sci. Hortic. 2018, 234, 361–369. [Google Scholar] [CrossRef]
- Darwin, S.C.; Knapp, S.; Peralta, I.E. Taxonomy of tomatoes in Galapagos Islands: Native and introduced species of Solanum section Lycopersicum (Solanaceae). Syst. Biodivers. 2003, 1, 29–53. [Google Scholar] [CrossRef]
- Kimura, S.; Koenig, D.; Kang, J.; Yoong, F.Y.; Sinha, N. Natural variation in leaf morphology results from mutation of a novel KNOX gene. Curr. Biol. 2008, 18, 672–677. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Hong, Y.; Zhu, G.; Li, Y.; Niu, Q.; Yao, J.; Zhu, J.K. Loss of salt tolerance during tomato domestication conferred by variation in a Na+/K+ transporter. EMBO J. 2020, 39, e103256. [Google Scholar] [CrossRef]
- Di Rienzo, J.A.; Casanoves, F.; Balzarini, M.G.; Gonzalez, L.; Tablada, M.; Robledo, C.W. InfoStat Versión 2020; Centro de Transferencia InfoStat, FCA, Universidad Nacional de Córdoba: Cordova, Argentina, 2020. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asins, M.J.; Romero-Aranda, M.R.; Espinosa, J.; González-Fernández, P.; Jaime-Fernández, E.; Traverso, J.A.; Carbonell, E.A.; Belver, A. HKT1;1 and HKT1;2 Na+ Transporters from Solanum galapagense Play Different Roles in the Plant Na+ Distribution under Salinity. Int. J. Mol. Sci. 2022, 23, 5130. https://doi.org/10.3390/ijms23095130
Asins MJ, Romero-Aranda MR, Espinosa J, González-Fernández P, Jaime-Fernández E, Traverso JA, Carbonell EA, Belver A. HKT1;1 and HKT1;2 Na+ Transporters from Solanum galapagense Play Different Roles in the Plant Na+ Distribution under Salinity. International Journal of Molecular Sciences. 2022; 23(9):5130. https://doi.org/10.3390/ijms23095130
Chicago/Turabian StyleAsins, Maria J., Maria R. Romero-Aranda, Jesus Espinosa, Paloma González-Fernández, Emilio Jaime-Fernández, Jose A. Traverso, Emilio A. Carbonell, and Andres Belver. 2022. "HKT1;1 and HKT1;2 Na+ Transporters from Solanum galapagense Play Different Roles in the Plant Na+ Distribution under Salinity" International Journal of Molecular Sciences 23, no. 9: 5130. https://doi.org/10.3390/ijms23095130
APA StyleAsins, M. J., Romero-Aranda, M. R., Espinosa, J., González-Fernández, P., Jaime-Fernández, E., Traverso, J. A., Carbonell, E. A., & Belver, A. (2022). HKT1;1 and HKT1;2 Na+ Transporters from Solanum galapagense Play Different Roles in the Plant Na+ Distribution under Salinity. International Journal of Molecular Sciences, 23(9), 5130. https://doi.org/10.3390/ijms23095130







