Comparative Analysis of the Metabolome and Transcriptome between the Green and Yellow-Green Regions of Variegated Leaves in a Mutant Variety of the Tree Species Pteroceltis tatarinowii
Abstract
:1. Introduction
2. Results
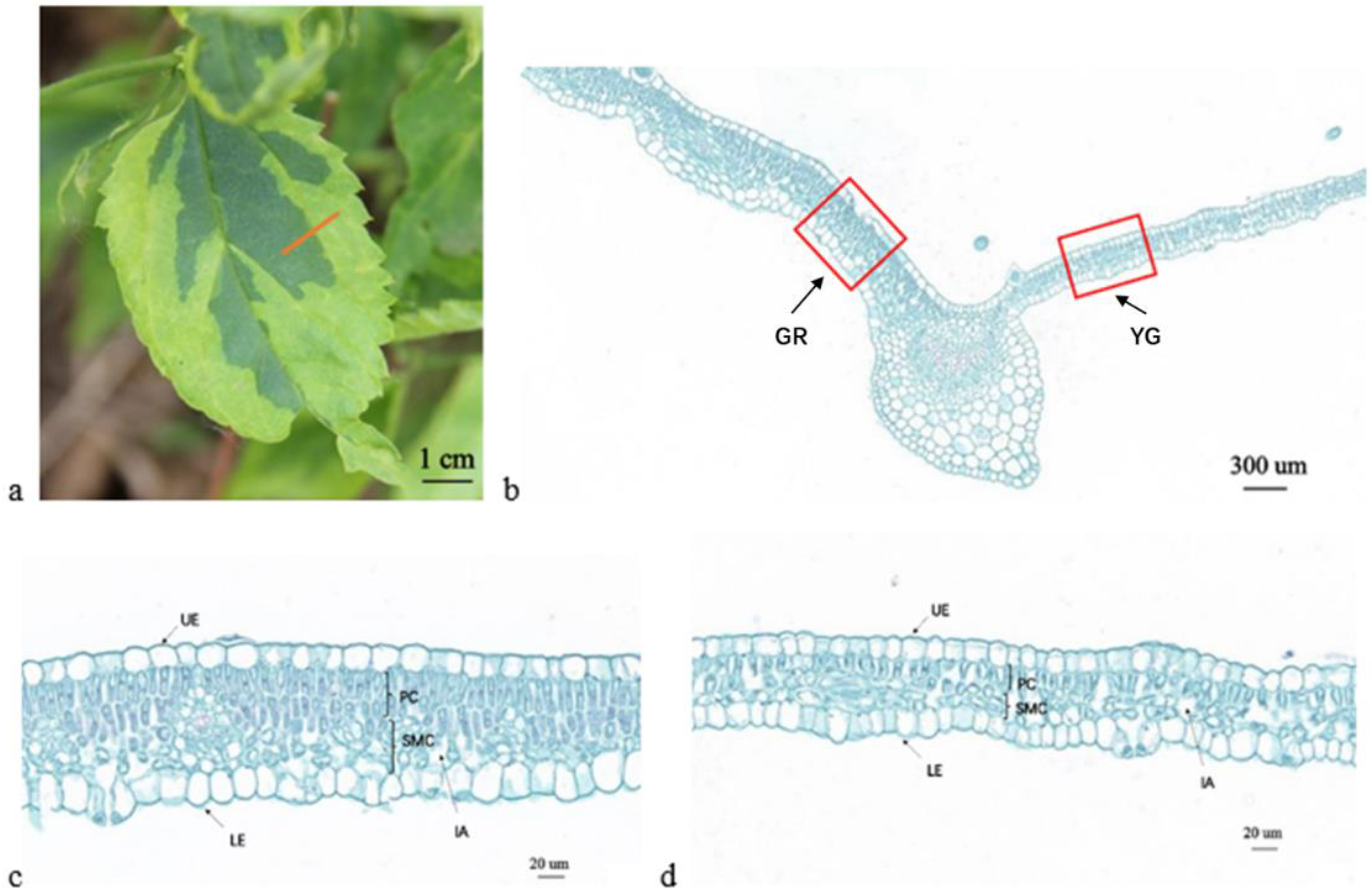
2.1. Morphological Observations
2.2. Microstructure and Ultramicrostructure Observations
2.3. Determination of Pigment Contents
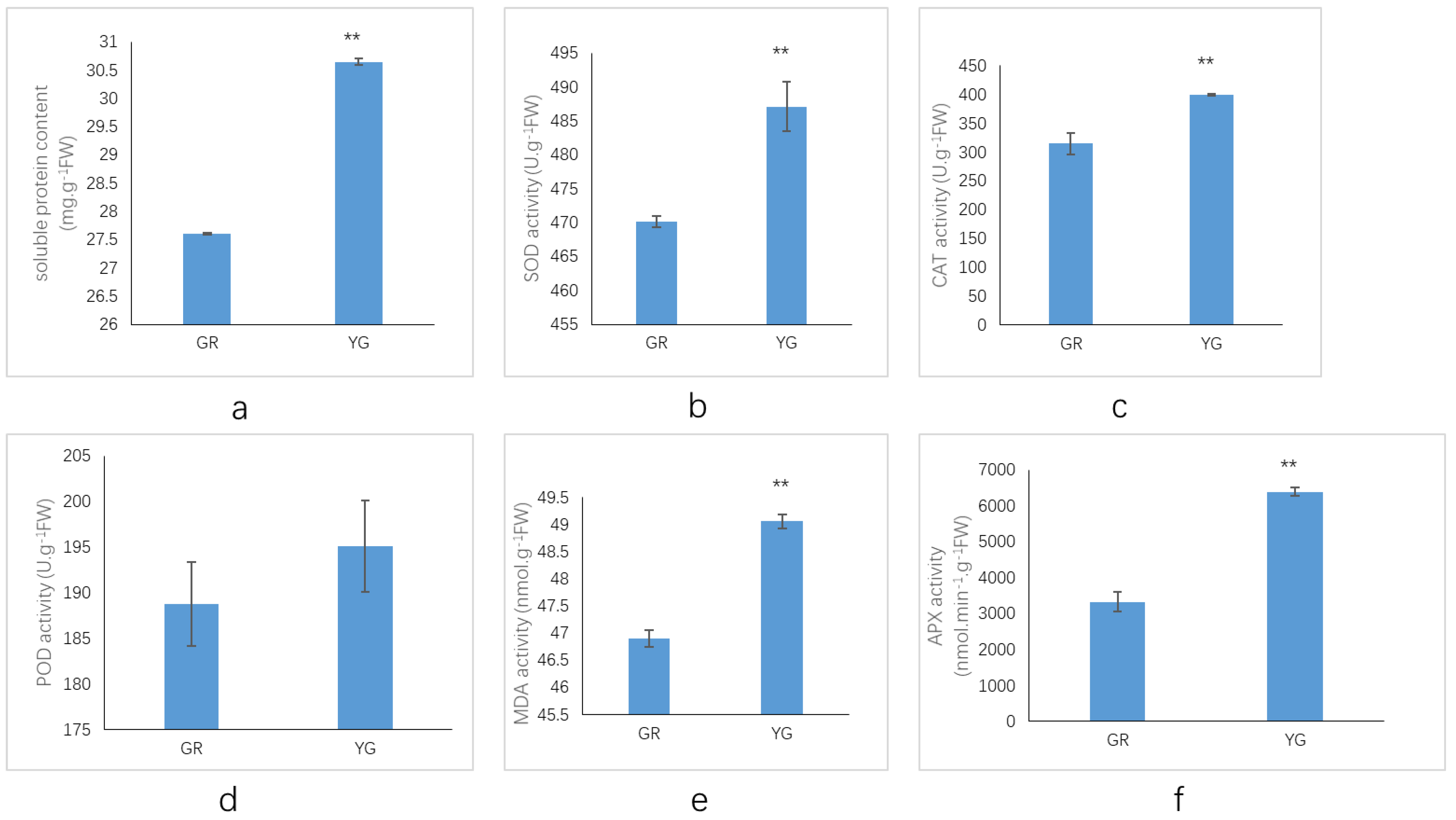
2.4. Assays of Protective Enzyme Activities and MDA Contents
2.5. RNA-Seq and Assembly Analyses
2.5.1. Quality Determination and Analysis of RNA Samples from Transcriptome Sequencing
2.5.2. Differentially Expressed Gene (DEG) Analysis
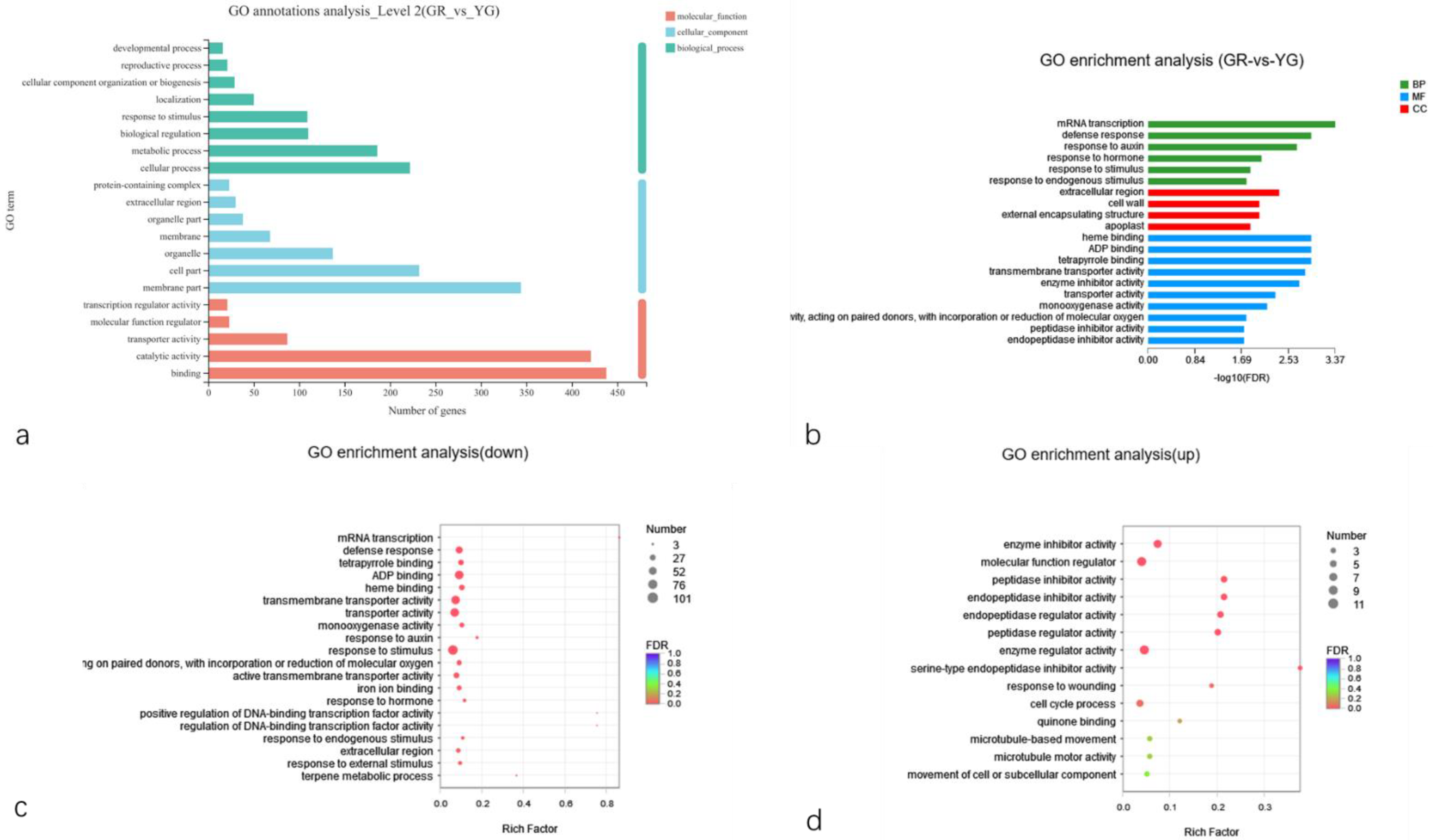
2.5.3. GO Classification and Enrichment Analysis of DEGs
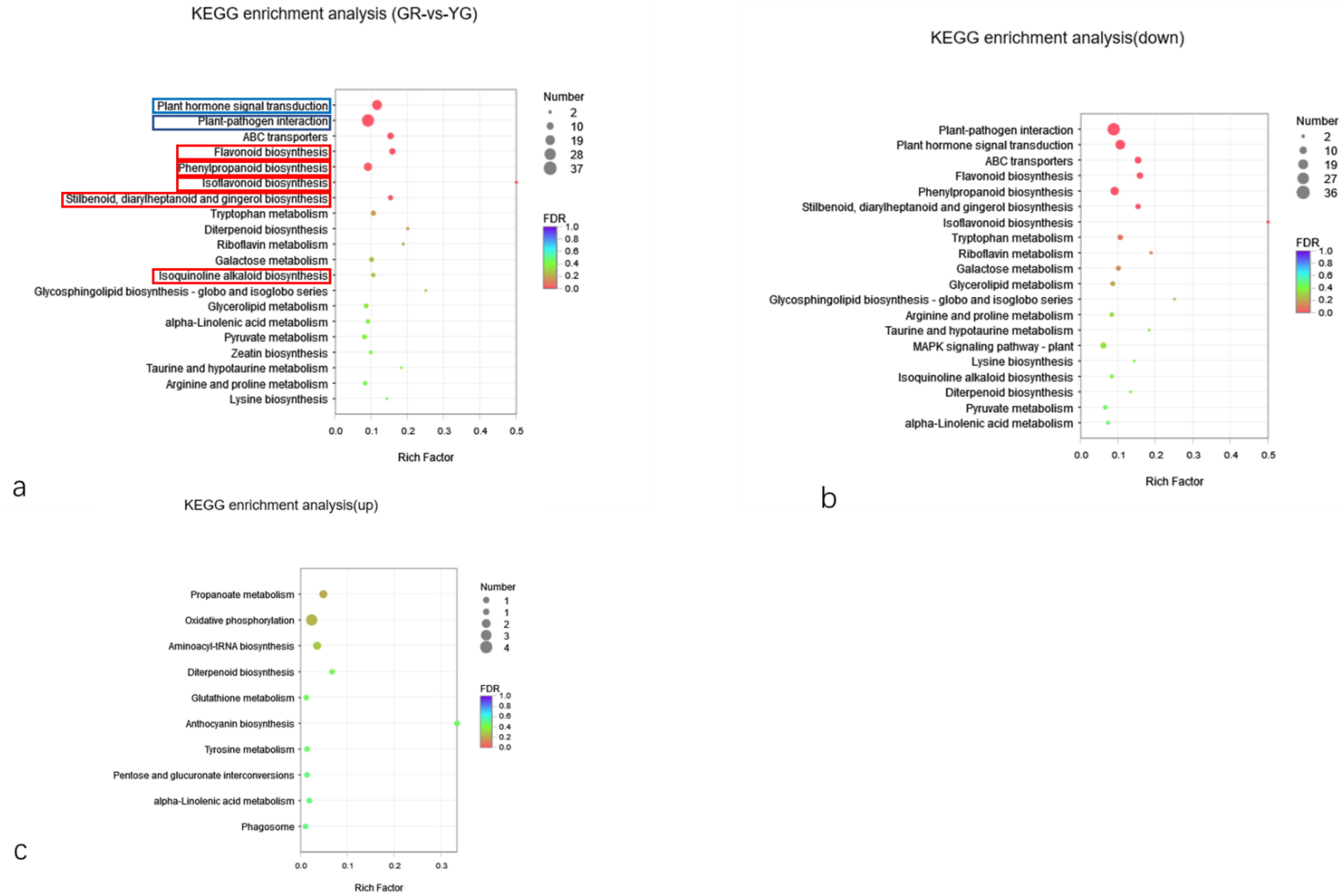
2.5.4. KEGG Classification and Enrichment Analysis of DEGs
2.5.5. Flavonoid Biosynthesis Pathways in Different Leaf Colour Regions
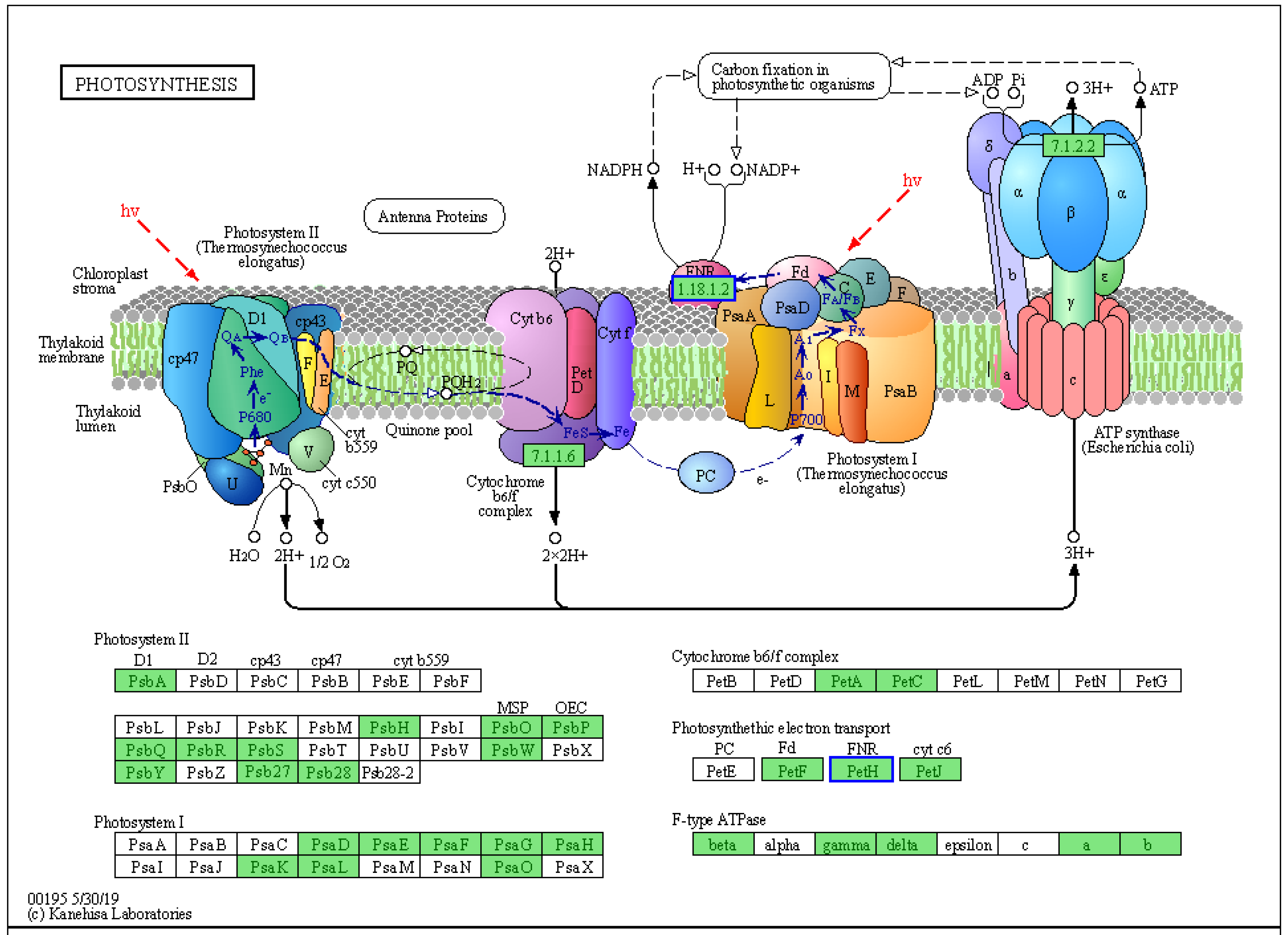
2.5.6. Pigment- and Photosynthesis-Related Pathways
2.5.7. Amino Acid and Lipid Metabolic Pathways
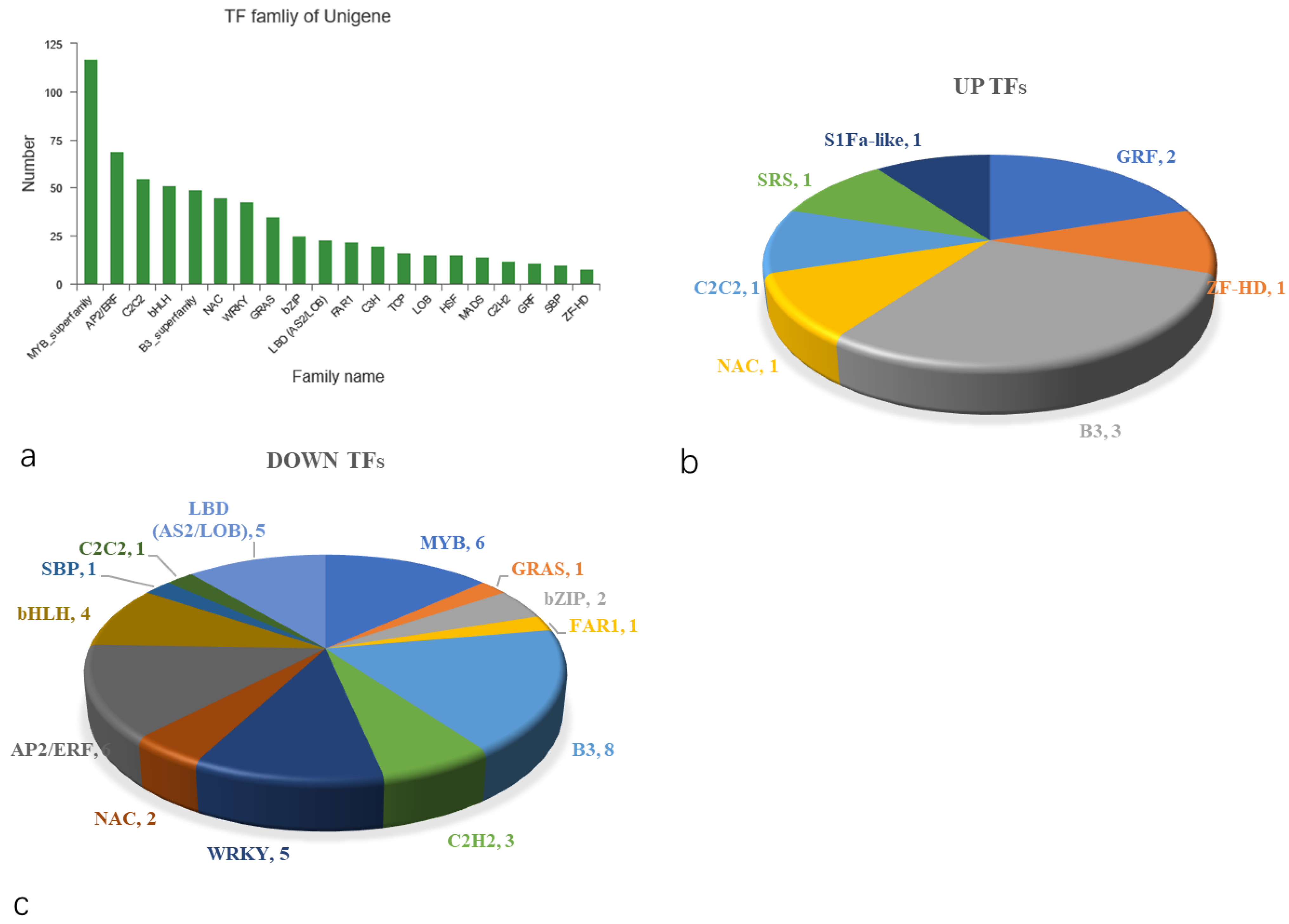
2.5.8. Computer Prediction Analysis of Transcriptome Factors (TFs)
2.6. RT-qPCR Validation
2.7. Analysis of Differentially Accumulated Metabolites
2.7.1. Statistics and Analysis of Differentially Accumulated Metabolites
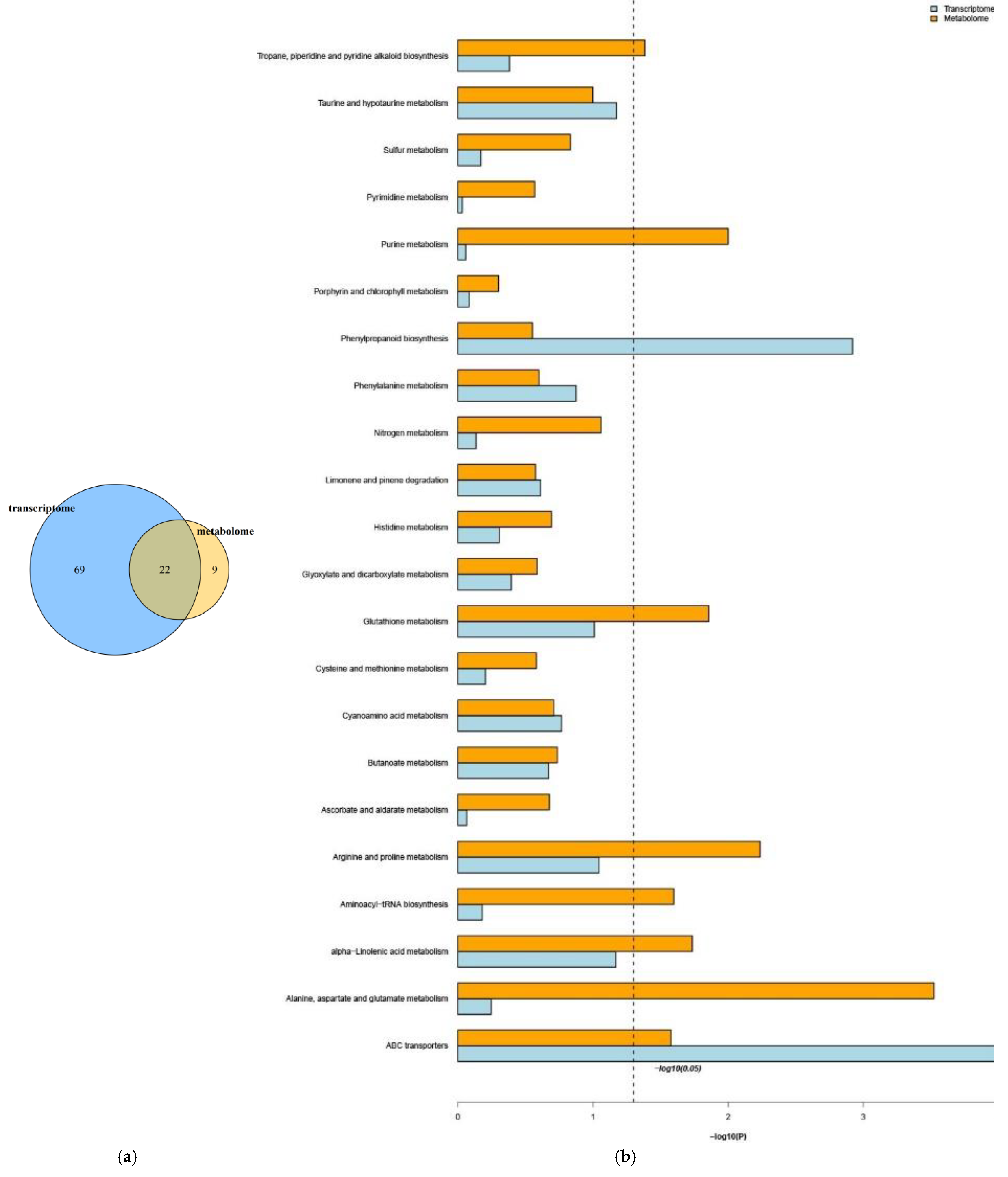
2.7.2. KEGG Annotations of Differentially Accumulated Metabolites
2.8. Correlation Analysis of DEGs and Differentially Accumulated Metabolites
3. Discussion
3.1. Explanation of Morphological Variation in P. tatarinowii ‘Jinyuyuan’ Leaves
3.2. Genes Associated with Variegated Leaf Colour Formation in P. tatarinowii ‘Jinyuyuan’
3.3. Analysis of the Redox Capacity of P. tatarinowii ‘Jinyuyuan’
4. Materials and Methods
4.1. Plant Materials
4.2. Morphological Observations
4.3. Microstructure and Ultramicrostructure Observations
4.4. Determination of Pigment Contents
4.5. Assays of Protective Enzyme Activities and MDA Contents
4.6. RNA Preparation, RNA Sequencing (RNA-seq) and Differential Gene Expression Analysis
4.7. RT-qPCR Validation
4.8. UPLC-MS
4.9. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Konoplyova, A.; Petropoulou, Y.; Yiotis, C.; Psaras, G.K.; Manetas, Y. The fine structure and photosynthetic cost of structural leaf variegation. Flora-Morphol. Distrib. Funct. Ecol. Plants 2008, 203, 653–662. [Google Scholar] [CrossRef]
- Yu, F.; Fu, A.; Aluru, M.; Park, S.; Xu, Y.; Liu, H.; Liu, X.; Foudree, A.; Nambogga, M.; Rodermel, S. Variegation mutants and mechanisms of chloroplast biogenesis. Plant Cell Environ. 2007, 30, 350–365. [Google Scholar] [CrossRef]
- Li, Y.; Li, W.; Hu, D.; Shen, P.; Zhang, G.; Zhu, Y. Comparative analysis of the metabolome and transcriptome between green and albino zones of variegated leaves from Hydrangea macrophylla ‘Maculata’ infected by hydrangea ringspot virus. Plant Physiol. Biochem. 2020, 157, 195–210. [Google Scholar] [CrossRef]
- Caringella, M.A.; Bongers, F.J.; Sacck, L. Leaf hydraulic conductance varies with vein anatomy across Arabidopsis thaliana wild-type and leaf vein mutants. Plant Cell Environ. 2015, 38, 2735–2746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, C.; Wang, L.; Ye, W.; Wu, L.W.; Cui, Y.T.; Chen, P.; Pan, J.J.; Zhang, D.; Hu, J.; Zeng, D.L.; et al. Single-point mutation of an histidine-aspartic domain-containing gene involving in Chloroplast Ribosome Biogenesis Leads to White Fine Stripe Leaf in Rice. Sci. Rep. 2017, 7, 3298. [Google Scholar] [CrossRef]
- Wang, R.; Yang, F.; Zhang, X.Q.; Wu, D.; Xu, Y. Characterization of a thermo-inducible chlorophyll-deficient mutant in barley. Front. Plant Sci. 2017, 8, 1936. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.Y.; Chang, A.X.; Du, C.Y. Genetic, physiological and biochemical analysis of the Formation of Yellow-green Leaf Color of Burley Tobacco (Nicotiana tabacum). Int. J. Agric. Biol. 2015, 17, 767–772. [Google Scholar] [CrossRef]
- Rodermel, S. Arabidopsis Variegation Mutants. Arab. Book/Am. Soc. Plant Biol. 2002, 1, e0079. [Google Scholar] [CrossRef] [Green Version]
- Sakamoto, W.; Tamura, T.; Hanba-Tomita, Y.; Sodmergen Murata, M. The VAR1 locus of Arabidopsis encodes a chloroplsatic FtsH and is responsible for leaf variegation in the mutant alleles. Genes Cells 2002, 7, 769–780. [Google Scholar] [CrossRef]
- Chen, M.; Choi, Y.D.; Voytas, D.; Rodermel, S. Mutations in the Arabidopsis VAR2 locus cause leaf variegation due to the loss of a chloroplastic FtsH protease. Plant J. 2000, 22, 303–313. [Google Scholar] [CrossRef]
- Sakamoto, W.; Zaltsman, A.; Adam, Z.; Takahashi, Y. Coordinated regulation and complex formation of yellow variegated1 and yellow variegated2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 2003, 15, 2843–2855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, F.; Zhu, Y.; Wu, W.; Xie, Y.; Huang, J. Leaf variegation of thylakoid formation1 is suppressed by mutations of specific σ-factors in Arabidopsis. Plant Physiol. 2015, 168, 1066–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Z.; Wu, W.; Huang, W.; Huang, J. Down-regulation of specific plastid ribosomal proteins suppresses thf1 leaf variegation, implying a role of THF1 in plastid gene expression. Photosynth. Res. 2015, 126, 301–310. [Google Scholar] [CrossRef]
- Rosso, D.; Ivanov, A.G.; Fu, A.; Geisler-Lee, J.; Hendrickson, L.; Geisler, M.; Stewart, G.; Krol, M.; Hurry, V.; Rodermel, S.R.; et al. IMMUTANS does not act as a stress-induced safety valve in the protection of the photosynthetic apparatus of Arabidopsis during steady-state photosynthesis. Plant Physiol. 2006, 142, 574–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, R.; Wang, Y.; Wang, P.; Li, C.; Xiao, F.; Chen, N.; Li, N.; Li, C.; Sun, C.; Li, L.; et al. A single nucleotide mutation of IspF gene involved in the MEP pathway for isoprenoid biosynthesis causes yellow-green leaf phenotype in rice. Plant Mol. Biol. 2018, 96, 5–16. [Google Scholar] [CrossRef]
- Liu, X.; Yu, F.; Rodermel, S. Arabidopsis Chloroplast FtsH, var2 and Suppressors of var2 Leaf Variegation: A Review. J. Integr. Plant Biol. 2010, 52, 750–761. [Google Scholar] [CrossRef] [PubMed]
- Miura, E.; Kato, Y.; Matsushima, R.; Albrecht, V.; Laalami, S.; Sakamoto, W. The Balance between Protein Synthesis and Degradation in Chloroplasts Determines Leaf Variegation in Arabidopsis yellow variegated Mutants. Plant Cell 2007, 19, 1313–1328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.-M.; Cui, J.; Dai, H.; Zhou, Y.; Li, N.; Zhang, Z. Comparative transcriptome profiling of genes and pathways involved in leaf-patterning of Clivia miniata var. variegata. Gene 2018, 677, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Zhang, J.; Zhao, Q.; Liu, Y.; Chen, S.; Guo, H.; Shi, L.; Dai, S. Proteomic Analysis Reveals the Leaf Color Regulation Mechanism in Chimera Hosta “Gold Standard” Leaves. Int. J. Mol. Sci. 2016, 17, 346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, G.; Yang, F.; Shi, S.; Li, N.; Wang, Z.; Liu, H.; Huang, D.; Wang, C. Transcriptome Characterization of Cymbidium sinense ‘Dharma’ Using 454 Pyrosequencing and Its Application in the Identification of Genes Associated with Leaf Color Variation. PLoS ONE 2015, 10, e0128592. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, M.; Ding, Y.; Zhou, P.; Fang, Y. Composition of photosynthetic pigments and photosynthetic characteristics in green and yellow sectors of the variegated Aucuba japonica ‘Variegata’ leaves. Flora 2018, 240, 25–33. [Google Scholar] [CrossRef]
- Li, W.X.; Yang, S.B.; Lu, Z.G.; He, Z.C.; Ye, Y.L.; Zhao, B.B.; Wang, L.; Jin, B. Cytological, physiological, and transcriptomic analyses of golden leaf coloration in Ginkgo biloba L. Hortic. Res. 2018, 5, 12. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Cao, J.; Li, J.; Zhou, B.; Tang, J.; Miao, A. Phenotypic, histological and proteomic analyses reveal multiple differences associated with chloroplast development in yellow and variegated variants from Camellia sinensis. Sci. Rep. 2016, 6, 33369. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.-X.; Fan, S.-T.; Chen, L.; Zhao, Y.; Yang, Y.-M.; Ai, J.; Li, X.-Y.; Liu, Y.-X.; Qin, H.-Y. Actinidia kolomikta leaf colour and optical characteristics. Biol. Plant. 2015, 59, 767–772. [Google Scholar] [CrossRef]
- Wang, N.; Wang, N.; Zhu, T.; Zhu, T.; Lu, N.; Lu, N.; Wang, Z.; Wang, Z.; Yang, G.; Yang, G.; et al. Quantitative Phosphoproteomic and Physiological Analyses Provide Insights into the Formation of the Variegated Leaf in Catalpa fargesii. Int. J. Mol. Sci. 2019, 20, 1895. [Google Scholar] [CrossRef] [Green Version]
- Fang, S.; Li, G.; Fu, X. Biomass production and bark yield in the plantations of Pteroceltis tatarinowii. Biomass Bioenergy 2004, 26, 319–328. [Google Scholar] [CrossRef]
- Xu, D. Photosynthetic organisms capable of producing oxygen. In Photosynthesis; Beijing Science Press: Beijing, China, 2013; pp. 115–120. [Google Scholar]
- Darzi, Y.; Letunic, I.; Bork, P.; Yamada, T. iPath3.0: Interactive pathways explorer v3. Nucleic Acids Res. 2018, 46, 510–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Tang, D.; Liu, M.; Ruan, J. Integrated analyses of the transcriptome and metabolome of the leaves of albino tea cultivars reveal coordinated regulation of the carbon and nitrogen metabolism. Sci. Hortic. 2018, 231, 272–281. [Google Scholar] [CrossRef]
- Tsai, C.C.; Wu, Y.J.; Sheue, C.R.; Liao, P.C.; Chen, Y.H.; Li, S.J.; Liu, J.W.; Chang, H.T.; Liu, W.L.; Ko, Y.Z.; et al. Molecular basis underlying leaf variegation of a moth orchid mutant (Phalaenopsis aphrodite subsp. formosana). Front. Plant Sci. 2017, 8, 1333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eini, O.; Ebadzad-Sahraei, G.; Behjatnia, S.A.A. Molecular characterization and construction of an infectious clone of a pepper isolate of Beet curly top Iran virus. Mol. Biol. Res. Commun. 2016, 5, 101–113. [Google Scholar]
- Shilpi, S.; Kumar, A.; Biswas, S.; Roy, A.; Mandal, B. A recombinant Tobacco curly shoot virus causes leaf curl disease in tomato in a north-eastern state of India and has potentiality to trans-replicate a non-cognate betasatellite. Virus Genes 2015, 50, 87–96. [Google Scholar] [CrossRef]
- Guggisberg, A.M.; Park, J.; Edwards, R.L.; Kelly, M.L.; Hodge, D.M.; Tolia, N.H.; Odom, A.R. A sugar phosphatase regulates the methylerythritol phosphate (MEP) pathway in malaria parasites. Nat. Commun. 2014, 5, 4467. [Google Scholar] [CrossRef] [Green Version]
- Lonosky, P.M.; Zhang, X.; Honavar, V.G.; Dobbs, D.L.; Fu, A.; Rodermel, S.R. A Proteomic Analysis of Maize Chloroplast Biogenesis. Plant Physiol. 2004, 134, 560–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.H.; Wang, B.H.; Dai, Z.Y.; Li, A.H.; Liu, G.Q.; Zuo, S.M.; Zhang, H.X.; Pan, X.B. Morphological structure and genetic mapping of new leaf-color mutant gene in rice (Oryza sativa). Chin. J. Rice Sci. 2012, 19, 79–85. [Google Scholar] [CrossRef]
- Maekawa, S.; Takabayashi, A.; Reyes, T.H.; Yamamoto, H.; Tanaka, A.; Sato, T.; Yamaguchi, J. Pale-Green Phenotype of atl31 atl6 Double Mutant Leaves Is Caused by Disruption of 5-Aminolevulinic Acid Biosynthesis in Arabidopsis thaliana. PLoS ONE 2015, 10, e0117662. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.M.; Zhang, X.; Wang, J.L.; Wan, J.M. Leaf chloroplast ultrastructure and photosynthetic properties of a chlorophyll-deficient mutant of rice. Photosynthetica 2014, 52, 217–222. [Google Scholar] [CrossRef]
- Yang, H.Y.; Xia, X.W.; Fang, W.; Fu, Y.; Zhou, M.B. Identification of genes involved in spontaneous leaf color variation in Pseudosasa japonica. Genet. Mol. Res. 2015, 14, 11827–11840. [Google Scholar] [CrossRef] [PubMed]
- Eyang, Y.; Echen, X.; Exu, B.; Eli, Y.; Ema, Y.; Ewang, G. Phenotype and transcriptome analysis reveals chloroplast development and pigment biosynthesis together influenced the leaf color formation in mutants of Anthurium andraeanum ‘Sonate’. Front. Plant Sci. 2015, 6, 139. [Google Scholar] [CrossRef] [Green Version]
- Winkel-Shirley, B. Flavonoid Biosynthesis. A Colorful Model for Genetics, Biochemistry, Cell Biology, and Biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef]
- Chen, L.; Hu, B.; Qin, Y.; Hu, G.; Zhao, J. Advance of the negative regulation of anthocyanin biosynthesis by MYB transcription factors. Plant Physiol. Biochem. 2019, 136, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Springob, K.; Nakajima, J.-I.; Yamazaki, M.; Saito, K. Recent advances in the biosynthesis and accumulation of anthocyanins. Nat. Prod. Rep. 2003, 20, 288–303. [Google Scholar] [CrossRef]
- Shan, X.; Zhang, Y.; Peng, W.; Wang, Z.; Xie, D. Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J. Exp. Bot. 2009, 60, 3849–3860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aluru, M.R.; Zola, J.; Foudree, A.; Rodermel, S.R. Chloroplast Photooxidation-Induced Transcriptome Reprogramming in Arabidopsis immutans White Leaf Sectors. Plant Physiol. 2009, 150, 904–923. [Google Scholar] [CrossRef] [Green Version]
- Bréhélin, C.; Kessler, F.; Van Wijk, K.J. Plastoglobules: Versatile lipoprotein particles in plastids. Trends Plant Sci. 2007, 12, 260–266. [Google Scholar] [CrossRef] [Green Version]
- Velikova, V.; Müller, C.; Ghirardo, A.; Rock, T.M.; Aichler, M.; Walch, A.; Schmitt-Kopplin, P.; Schnitzler, J.-P. Knocking Down of Isoprene Emission Modifies the Lipid Matrix of Thylakoid Membranes and Influences the Chloroplast Ultrastructure in Poplar. Plant Physiol. 2015, 168, 859–870. [Google Scholar] [CrossRef] [Green Version]
- Asada, K. THE WATER-WATER CYCLE IN CHLOROPLASTS: Scavenging of Active Oxygens and Dissipation of Excess Photons. Annu. Rev. Plant Biol. 1999, 50, 601–639. [Google Scholar] [CrossRef]
- Ishikawa, T.; Shigeoka, S. Recent Advances in Ascorbate Biosynthesis and the Physiological Significance of Ascorbate Peroxidase in Photosynthesizing Organisms. Biosci. Biotechnol. Biochem. 2008, 72, 1143–1154. [Google Scholar] [CrossRef]
- Fischer, A.H.; Jacobson, K.A.; Rose, J.; Zeller, R. Hematoxylin and eosin staining of tissue and cell sections. Cold Spring Harb. Protoc. 2008, 5, pdb-prot4986. [Google Scholar] [CrossRef]
- Huang, S.-S.; Chen, J.; Dong, X.-J.; Patton, J.; Pei, Z.-M.; Zheng, H.-L. Calcium and calcium receptor CAS promote Arabidopsis thaliana de-etiolation. Physiol. Plant. 2012, 144, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Esfandiari, E.; Shakiba, M.R.; Mahboob, S.A.; Alyari, H.; Shahabivan, S. The Effect of Water Stress on the Antioxidant Content, Protective Enzyme Activities, Proline Content and Lipid Peroxidation in Wheat Seedling. Pak. J. Biol. Sci. 2008, 11, 1916–1922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, Q.; Cui, G.; Chen, Y.; Zhou, X.; Deng, B.; Huang, P.; Zhu, S.; Chen, J.; Zhao, T. Combined analysis of mRNA and miRNA transcriptomes reveals the regulatory mechanism of PVY resistance in tobacco. Ind. Crop. Prod. 2021, 176, 114322. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| GR | YG | |
|---|---|---|
| L* | 47.70 ± 0.70 B | 56.70 ± 1.17 A |
| a* | −7.68 ± 1.33 | −7.14 ± 0.73 |
| b* | −5.46 ± 0.86 B | 4.80 ± 1.11 A |
| c* | 9.50 ± 0.76 | 8.65 ± 0.82 |
| h | 35.78 ± 8.45 A | −33.72 ± 6.72 B |
| RHSCC | 137B | N144D |
| Type | Upper Epidermis Thickness (μm) | Lower Epidermis Thickness (μm) | Leaf Thickness (μm) | Palisade Cells Thickness (μm) | Spongy Cells Thickness (μm) | Palisade Cells/Spongy Cells |
|---|---|---|---|---|---|---|
| GR | 8.580 ± 2.614 | 7.029 ± 1.668 | 105.585 ± 6.924 A | 44.626 ± 8.310 | 48.660 ± 7.197 a | 0.953 ± 0.316 |
| YG | 8.487 ± 1.569 | 6.218 ± 1.509 | 84.288 ± 7.815 B | 37.644 ± 6.918 | 35.967 ± 7.809 b | 1.073 ± 0.219 |
| GR | YG | |
|---|---|---|
| Chl a (mg g−1 FW) | 0.40 ± 0.0021 A | 0.31 ± 0.0003 B |
| Chl b (mg g−1 FW) | 0.19 ± 0.0026 A | 0.15 ± 0.0024 B |
| Chl a + b (mg g−1 FW) | 0.59 ± 0.0032 A | 0.47 ± 0.0028 B |
| Chl a/b | 2.14 ± 0.0320 a | 2.06 ± 0.0307 b |
| Car (ug g−1 FW) | 118.12 ± 0.0307 A | 110.50 ± 0.0178 B |
| Chl a + b /Car | 4.97 ± 0.0269 A | 4.22 ± 0.0244 B |
| Transcriptome | Metabolomic | ||||||
|---|---|---|---|---|---|---|---|
| Pathway ID | Description | Number | Pathway ID | Description | Number | ||
| Down | Up | Down | Up | ||||
| map00250 | Alanine, aspartate and glutamate metabolism | 2 | 0 | map00220 | Arginine biosynthesis | 3 | |
| map00260 | Glycine, serine and threonine metabolism | 2 | 1 | map00250 | Alanine, aspartate and glutamate metabolism | 3 | |
| map00270 | Cysteine and methionine metabolism | 4 | 0 | map00270 | Cysteine and methionine metabolism | 1 | |
| map00280 | Valine, leucine and isoleucine degradation | 1 | 1 | map00330 | Arginine and proline metabolism | 2 | 1 |
| map00300 | Lysine biosynthesis | 2 | 0 | map00340 | Histidine metabolism | 1 | |
| map00310 | Lysine degradation | 1 | 0 | map00360 | Phenylalanine metabolism | 1 | |
| map00330 | Arginine and proline metabolism | 5 | 0 | map00400 | Phenylalanine, tyrosine and tryptophan biosynthesis | 1 | |
| map00340 | Histidine metabolism | 1 | 0 | map00430 | Taurine and hypotaurine metabolism | 1 | |
| map00350 | Tyrosine metabolism | 4 | 1 | map00460 | Cyanoamino acid metabolism | 1 | |
| map00360 | Phenylalanine metabolism | 4 | 1 | map00472 | D-Arginine and D-ornithine metabolism | 1 | |
| map00380 | Tryptophan metabolism | 7 | 0 | map00480 | Glutathione metabolism | 2 | |
| map00410 | beta-Alanine metabolism | 3 | 1 | ||||
| map00430 | Taurine and hypotaurine metabolism | 2 | 0 | ||||
| map00450 | Selenocompound metabolism | 1 | 0 | ||||
| map00460 | Cyanoamino acid metabolism | 5 | 0 | ||||
| map00480 | Glutathione metabolism | 5 | 1 | ||||
| Pathway ID | Description | Number | ||
|---|---|---|---|---|
| Down | Up | |||
| Transcriptome | map00061 | Fatty acid biosynthesis | 1 | 1 |
| map00062 | Fatty acid elongation | 2 | 0 | |
| map00071 | Fatty acid degradation | 1 | ||
| map00073 | Cutin, suberine and wax biosynthesis | 1 | ||
| map00100 | Steroid biosynthesis | 1 | ||
| map00561 | Glycerolipid metabolism | 6 | 0 | |
| map00564 | Glycerophospholipid metabolism | 3 | ||
| map00590 | Arachidonic acid metabolism | 1 | ||
| map00591 | Linoleic acid metabolism | 2 | 0 | |
| map00592 | alpha-Linolenic acid metabolism | 4 | 1 | |
| map00600 | Sphingolipid metabolism | 2 | 0 | |
| map01040 | Biosynthesis of unsaturated fatty acids | 1 | ||
| Metabolomic | map00592 | alpha-Linolenic acid metabolism | 1 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiao, Q.; Wu, C.; Cheng, T.-T.; Yan, Y.; Zhang, L.; Wan, Y.-L.; Wang, J.-W.; Liu, Q.-Z.; Feng, Z.; Liu, Y. Comparative Analysis of the Metabolome and Transcriptome between the Green and Yellow-Green Regions of Variegated Leaves in a Mutant Variety of the Tree Species Pteroceltis tatarinowii. Int. J. Mol. Sci. 2022, 23, 4950. https://doi.org/10.3390/ijms23094950
Qiao Q, Wu C, Cheng T-T, Yan Y, Zhang L, Wan Y-L, Wang J-W, Liu Q-Z, Feng Z, Liu Y. Comparative Analysis of the Metabolome and Transcriptome between the Green and Yellow-Green Regions of Variegated Leaves in a Mutant Variety of the Tree Species Pteroceltis tatarinowii. International Journal of Molecular Sciences. 2022; 23(9):4950. https://doi.org/10.3390/ijms23094950
Chicago/Turabian StyleQiao, Qian, Chong Wu, Tian-Tian Cheng, Yu Yan, Lin Zhang, Ying-Lin Wan, Jia-Wei Wang, Qing-Zhong Liu, Zhen Feng, and Yan Liu. 2022. "Comparative Analysis of the Metabolome and Transcriptome between the Green and Yellow-Green Regions of Variegated Leaves in a Mutant Variety of the Tree Species Pteroceltis tatarinowii" International Journal of Molecular Sciences 23, no. 9: 4950. https://doi.org/10.3390/ijms23094950
APA StyleQiao, Q., Wu, C., Cheng, T.-T., Yan, Y., Zhang, L., Wan, Y.-L., Wang, J.-W., Liu, Q.-Z., Feng, Z., & Liu, Y. (2022). Comparative Analysis of the Metabolome and Transcriptome between the Green and Yellow-Green Regions of Variegated Leaves in a Mutant Variety of the Tree Species Pteroceltis tatarinowii. International Journal of Molecular Sciences, 23(9), 4950. https://doi.org/10.3390/ijms23094950





