Strawberry FaSnRK1α Regulates Anaerobic Respiratory Metabolism under Waterlogging
Abstract
:1. Introduction
2. Results
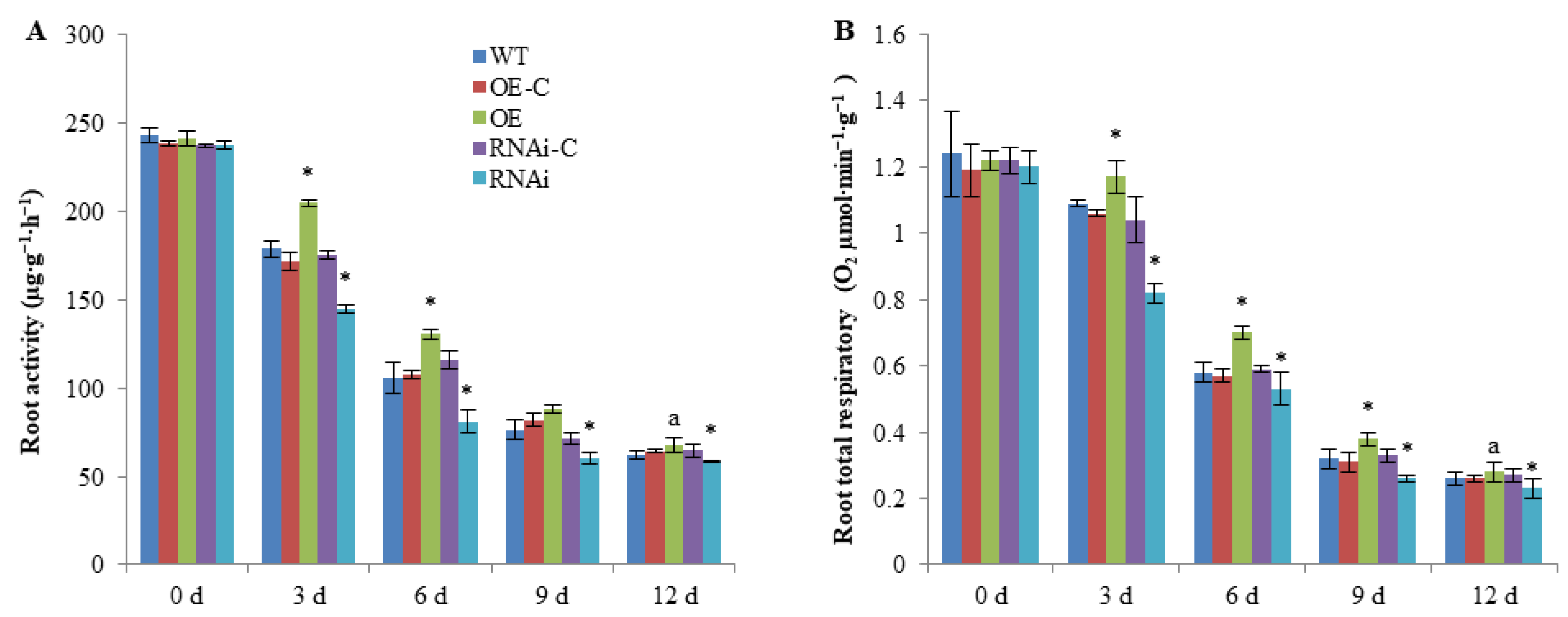
2.1. FaSnRK1α Regulates Root Activity and Respiratory Rate under Waterlogging
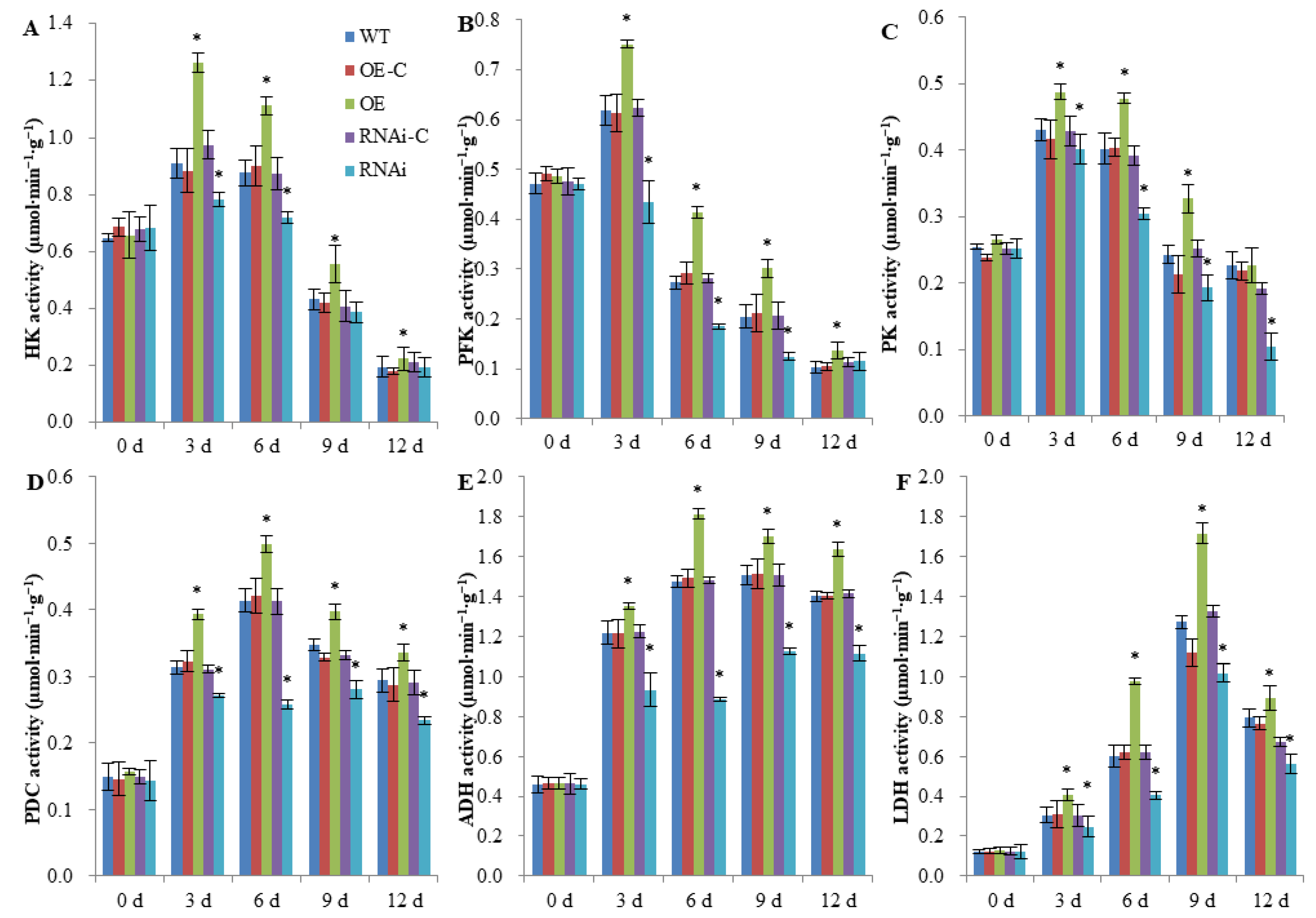
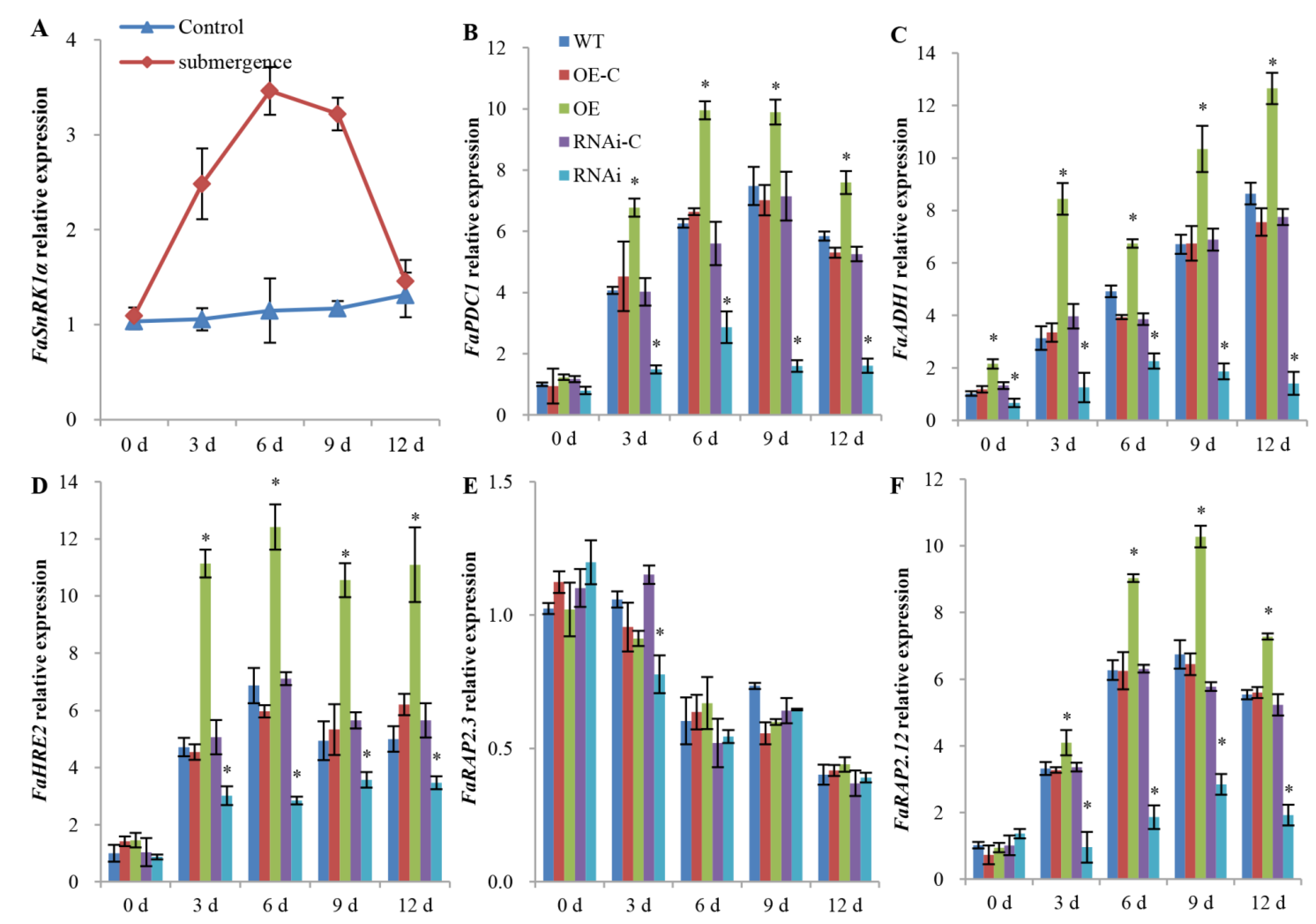
2.2. FaSnRK1α Regulates Anaerobic Respiration Metabolism under Waterlogging
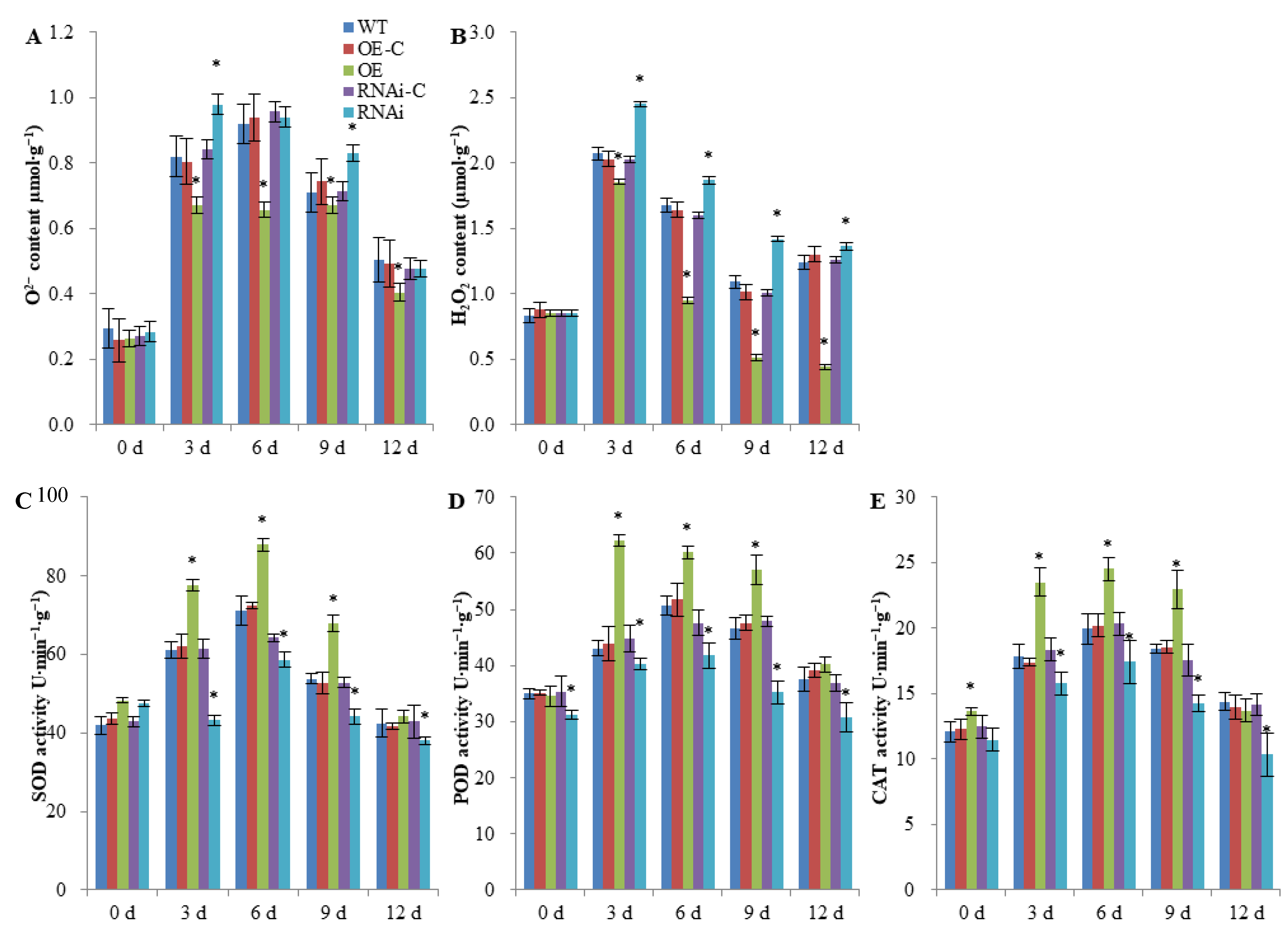
2.3. FaSnRK1α Regulates ROS Accumulation and Antioxidant Enzyme Activities under Waterlogging
2.4. Transcriptomic Analysis of Roots of WT and FaSnRK1α-RNAi Plants
2.5. FaSnRK1α Regulates the Expression of Anoxia-Related Genes and ERFVIIs
3. Discussion
4. Summary
5. Materials and Methods
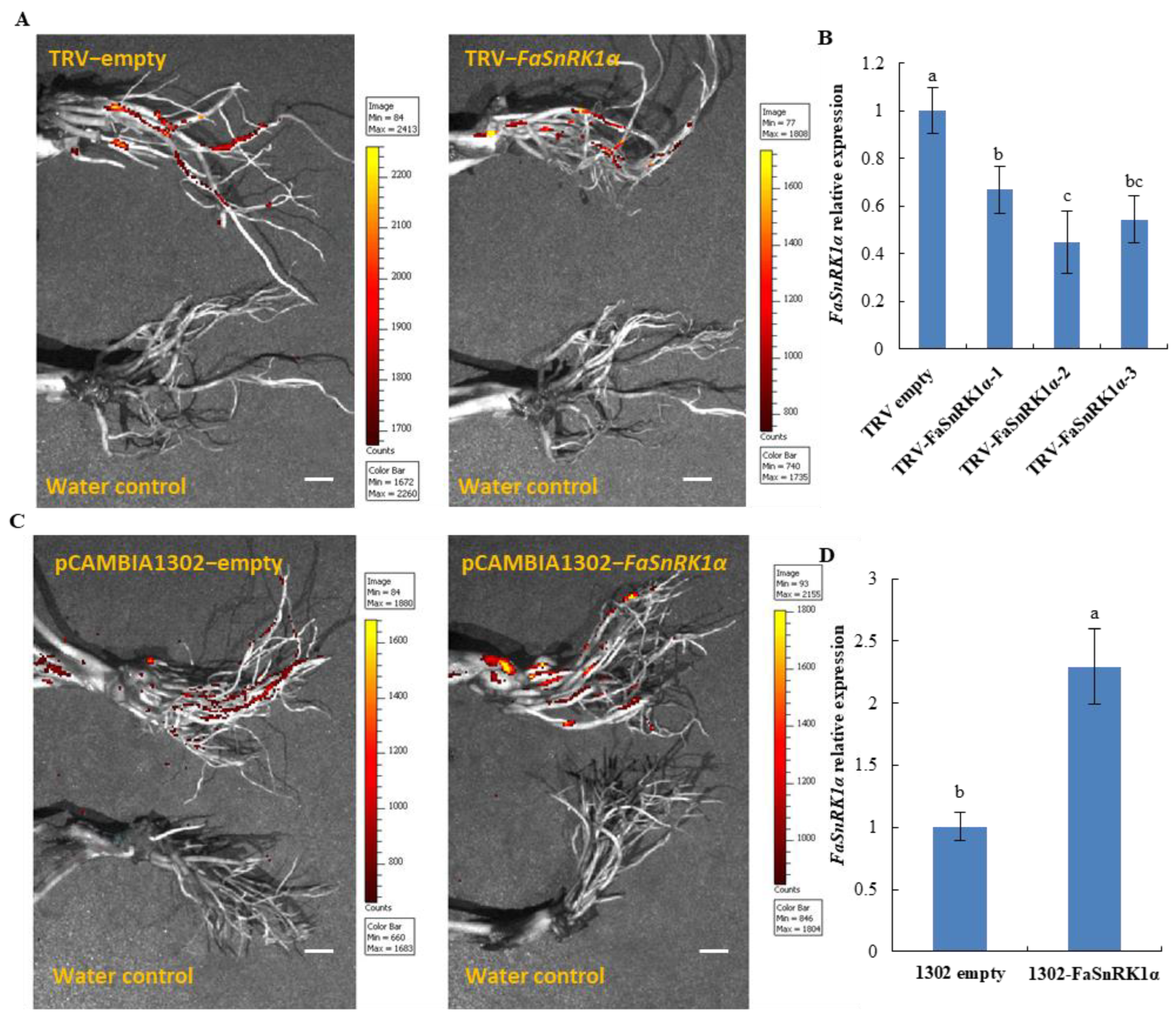
5.1. Vector Construction and Agrobacterium-Mediated Infiltration
5.2. Plant Materials and Treatment
5.3. Determination of Root Activity and Root Respiration Rate
5.4. Determination of Respiration-Related Enzyme Activities
5.5. ROS Accumulation and Antioxidant Capacity
5.6. RNA Isolation and Library Preparation
5.7. RNA Sequencing and Differentially Expressed Gene (DEG) Analysis
5.8. RT-qPCR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Drew, M.C. Plant injury and adaptation to oxygen deficiency in the root environment: A review. Plant Soil 1983, 75, 179–199. [Google Scholar] [CrossRef]
- Jackson, M.B.; Colmer, T.D. Response and adaptation by plants to flooding stress. Ann. Bot. 2005, 96, 501–505. [Google Scholar] [CrossRef] [PubMed]
- Justin, S.H.F.W.; Armstrong, W. The anatomical characteristics of roots and plant response to soil flooding. New Phytol. 1987, 106, 465–495. [Google Scholar] [CrossRef]
- Evans, D.E. Aerenchyma formation. New Phytol. 2003, 161, 35–49. [Google Scholar] [CrossRef]
- Jackson, M.B. Ethylene and responses of plants to soil waterlogging and submergence. Ann. Rev. Plant Physiol. 1985, 36, 145–174. [Google Scholar] [CrossRef]
- Stünzi, J.T.; Kende, H. Gas composition in the internal air spaces of deep water rice in relation to growth induced by submergence. Plant Cell Physiol. 1989, 30, 49–56. [Google Scholar] [CrossRef] [Green Version]
- Sairam, R.K.; Kumutha, D.; Ezhilmathi, K.; Deshmukh, P.S.; Srivastava, G.C. Physiology and biochemistry of waterlogging tolerance in plants. Biol. Plant. 2008, 52, 401–412. [Google Scholar] [CrossRef]
- Visser, E.J.W.; Bögemann, G.; Gerard, M.; Blom, C.W.P.M.; Voesenek, L.A.C.J. Ethylene accumulation in waterlogged Rumex plants promotes formation of adventitious roots. J. Exp. Bot. 1996, 47, 403–410. [Google Scholar] [CrossRef] [Green Version]
- Giuntoli, B.; Perata, P. Group VII Ethylene Response Factors in Arabidopsis: Regulation and Physiological Roles. Plant Physiol. 2018, 176, 1143–1155. [Google Scholar] [CrossRef] [Green Version]
- Gasch, P.; Fundinger, M.; Müller, J.T.; Lee, T.; Bailey-Serres, J.; Mustroph, A. Redundant ERF-VII transcription factors bind to an evolutionarily conserved cis-motif to regulate hypoxia-responsive gene expression in Arabidopsis. Plant Cell 2016, 28, 160–180. [Google Scholar] [CrossRef] [Green Version]
- Licausi, F.; Pucciariello, C.; Perata, P. New role for an old rule: N-end rule-mediated degradation of ethylene responsive factor proteins governs low oxygen response in plants (F). J. Integr. Plant Biol. 2013, 55, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Abbas, M.; Berckhan, S.; Rooney, D.J.; Gibbs, D.J.; Vicente, C.J.; Sousa, C.C.; Bassel, G.W.; Marín-de la, R.N.; León, J.; Alabadí, D.; et al. Oxygen sensing coordinates photomorphogenesis to facilitate seedling survival. Curr. Biol. 2015, 25, 1483–1488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, M.V.; Iyer, S.; Amerhauser, C.; Lehmann, M.; van Dongen, J.T.; Geigenberger, P. Oxygen sensing via the ethylene response transcription factor RAP2.12 affects plant metabolism and performance under both normoxia and hypoxia. Plant Physiol. 2016, 172, 141–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Licausi, F.; van Dongen, J.T.; Giuntoli, B.; Novi, G.; Santaniello, A.; Geigenberger, P.; Perata, P. HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J. 2010, 62, 302–315. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.Y.; Hsu, F.C.; Li, J.P.; Wang, N.N.; Shih, M. The AP2/ERF transcription factor AtERF73/HRE1 modulates ethylene responses during hypoxia in Arabidopsis. Plant Physiol. 2011, 156, 202–212. [Google Scholar] [CrossRef] [Green Version]
- Papdi, C.; Pérez-Salamó, I.; Joseph, M.P.; Giuntoli, B.; Bögre, L.; Koncz, C.; Szabados, L. The low oxygen, oxidative and osmotic stress responses synergistically act through the ethylene response factor VII genes RAP2.12, RAP2.2 and RAP2.3. Plant J. 2015, 82, 772–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hess, N.; Klode, M.; Anders, M.; Sauter, M. The hypoxia responsive transcription factor genes ERF71/HRE2 and ERF73/HRE1 of Arabidopsis are differentially regulated by ethylene. Physiol. Plant. 2011, 143, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.H.; Hong, J.W.; Kim, E.C.; Yoo, S.D. Regulatory functions of SnRK1 in stress-responsive gene expression and in plant growth and development. Plant Physiol. 2012, 158, 1955. [Google Scholar] [CrossRef] [Green Version]
- Peng, H.P.; Chan, C.S.; Shih, M.C.; Yang, S.F. Signaling events in the hypoxic induction of alcohol dehydrogenase gene in Arabidopsis. Plant Physiol. 2001, 126, 742–749. [Google Scholar] [CrossRef] [Green Version]
- Hinz, M.; Wilson, I.W.; Yang, J.; Buerstenbinder, K.; Llewellyn, D.; Dennis, E.S.; Sauter, M.; Dolferus, R. Arabidopsis RAP2.2: An ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 2010, 153, 757–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crozet, P.; Margalha, L.; Confraria, A.; Rodrigues, A.; Martinho, C.; Adamo, M.; Elias, C.A.; Baena-González, E. Mechanisms of regulation of SNF1/AMPK/SnRK1 protein kinases. Front Plant Sci. 2014, 5, 190. [Google Scholar] [CrossRef]
- Emanuelle, S.; Doblin, M.S.; Stapleton, D.I.; Bacic, A.; Gooley, P.R. Molecular insights into the enigmatic metabolic regulator, SnRK1. Trends Plant Sci. 2016, 21, 341–353. [Google Scholar] [CrossRef]
- Kudahettige, N.P.; Pucciariello, C.; Parlanti, S.; Alpi, A.; Perata, P. Regulatory interplay of the Sub1A and CIPK15 pathways in the regulation of α-amylase production in flooded rice plants. Plant Biol. 2011, 13, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.A.; Lin, C.C.; Lee, K.W.; Chen, J.L.; Huang, L.F.; Ho, S.L.; Liu, H.J.; Hsing, Y.I.; Yu, S.M. The SnRK1A protein kinase plays a key role in sugar signaling during germination and seedling growth of rice. Plant Cell 2007, 19, 2484–2499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.W.; Chen, P.W.; Lu, C.A.; Chen, S.; Ho, T.H.D.; Yu, S.M. Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Sci. Signal. 2009, 2, ra61. [Google Scholar] [CrossRef]
- Yu, S.M.; Lee, H.T.; Lo, S.F.; Ho, T.D. How does rice cope with too little oxygen during its early life? New Phytol. 2021, 229, 36–41. [Google Scholar] [CrossRef]
- Lin, C.R.; Lee, K.W.; Chen, C.Y.; Hong, Y.F.; Chen, J.L.; Lu, C.A.; Chen, K.T.; Ho, T.H.D.; Yu, S.M. SnRK1A-interacting negative regulators modulate the nutrient starvation signaling sensor SnRK1 in source-sink communication in cereal seedlings under abiotic stress. Plant Cell 2014, 26, 808–827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, H.Y.; Wen, T.N.; Wang, Y.T.; Shih, M.C. Quantitative phosphoproteomics of protein kinase SnRK1 regulated protein phosphorylation in Arabidopsis under submergence. J. Exp. Bot. 2016, 67, 2745–2760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baena-Gonza’lez, E.; Sheen, J. Convergent energy and stress signaling. Trends Plant Sci. 2008, 13, 474–482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polge, C.; Thomas, M. Snf1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 2007, 12, 20–28. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Chan, Z. ROS regulation during abiotic stress responses incrop plants. Front Plant Sci. 2015, 6, 1092. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.R.; Liang, J.H.; Wang, G.F.; Sun, M.X.; Peng, F.T.; Xiao, Y.S. Overexpression of PpSnRK1α in tomato enhanced salt tolerance by regulating aba signaling pathway and reactive oxygen metabolism. BMC Plant Biol. 2020, 20, 128. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Bjornson, M.; Dandekar, A.; Dehesh, K. Determinants of timing and amplitude in the plant general stress response. J. Integr. Plant Biol. 2016, 58, 119–126. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, R.R.; Weits, D.A.; Feulner, C.; van Dongen, J.T. Oxygen Sensing and Integrative Stress Signaling in Plants. Plant Physiol. 2018, 176, 1131–1142. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.Y. Hydrogen peroxide controls transcriptional responses of ERF73/HRE1 and ADH1 via modulation of ethylene signaling during hypoxic stress. Planta 2014, 239, 877–885. [Google Scholar] [CrossRef]
- Pucciariello, C.; Banti, V.; Perata, P. ROS signaling as common element in low oxygen and heat stresses. Plant Physiol. Biol. 2012, 59, 3–10. [Google Scholar] [CrossRef] [Green Version]
- Pucciariello, C.; Parlanti, S.; Banti, V.; Novi, G.; Perata, P. Reactive oxygen species-driven transcription in Arabidopsis under oxygen deprivation. Plant Physiol. 2012, 159, 184–196. [Google Scholar] [CrossRef] [Green Version]
- Yu, W.; Peng, F.; Xiao, Y.; Wang, G.; Luo, J. Overexpression of PpSnRK1α in Tomato Promotes Fruit Ripening by Enhancing RIPENING INHIBITOR Regulation Pathway. Front. Plant Sci. 2018, 9, 1856. [Google Scholar] [CrossRef] [Green Version]
- Licausi, F.; Kosmacz, M.; Weits, D.A.; Giuntoli, B.; Giorgi, F.M.; Voesenek, L.A.C.J.; Perata, P.; van Dongen, J.T. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 2011, 479, 419–422. [Google Scholar] [CrossRef]
- Zhao, S.J.; Shi, G.A.; Dong, X.C. Techniques of Plant Physiological Experiment, 1st ed.; China Agricultural Science and Technology Press: Beijing, China, 2002; pp. 47–75. [Google Scholar]
- Ma, N.N.; Zuo, Y.Q.; Liang, X.Q.; Yin, B.; Wang, G.D.; Meng, Q.W. The multiple stress-responsive transcription factor SlNAC1 improves the chilling tolerance of tomato. Physiol. Plant. 2013, 149, 474–486. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Peng, F.; Zhang, S.; Xiao, Y.; Zhang, Y. The protein kinase FaSnRK1α regulates sucrose accumulation in strawberry fruits. Plant Physiol. Biochem. 2020, 151, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Qu, G.Z.; Li, H.Y.; Wu, Y.J.; Wang, C.; Liu, G.F.; Yang, C.P. Enhanced salt tolerance of transgenic poplar plants expressing a manganese superoxide dismutase from Tamarix and rossowii. Mol. Biol. Rep. 2009, 37, 1119. [Google Scholar] [CrossRef] [PubMed]
- Aebi, H. Catalase. In Methods of Enzymatic Analysis, 2nd ed.; Bergmeyer, H.U., Ed.; Academic Press: New York, NY, USA, 1974; pp. 673–684. [Google Scholar]
- Pütter, J. Peroxidases. In Methods of Enzymatic Analysis, 2nd ed.; Bergmeyer, H.U., Ed.; Academic Press: New York, NY, USA, 1974; pp. 685–690. [Google Scholar]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, J.; Yu, W.; Xiao, Y.; Zhang, Y.; Peng, F. Strawberry FaSnRK1α Regulates Anaerobic Respiratory Metabolism under Waterlogging. Int. J. Mol. Sci. 2022, 23, 4914. https://doi.org/10.3390/ijms23094914
Luo J, Yu W, Xiao Y, Zhang Y, Peng F. Strawberry FaSnRK1α Regulates Anaerobic Respiratory Metabolism under Waterlogging. International Journal of Molecular Sciences. 2022; 23(9):4914. https://doi.org/10.3390/ijms23094914
Chicago/Turabian StyleLuo, Jingjing, Wenying Yu, Yuansong Xiao, Yafei Zhang, and Futian Peng. 2022. "Strawberry FaSnRK1α Regulates Anaerobic Respiratory Metabolism under Waterlogging" International Journal of Molecular Sciences 23, no. 9: 4914. https://doi.org/10.3390/ijms23094914
APA StyleLuo, J., Yu, W., Xiao, Y., Zhang, Y., & Peng, F. (2022). Strawberry FaSnRK1α Regulates Anaerobic Respiratory Metabolism under Waterlogging. International Journal of Molecular Sciences, 23(9), 4914. https://doi.org/10.3390/ijms23094914





