Effects of Module Truncation of a New Alginate Lyase VxAly7C from Marine Vibrio xiamenensis QY104 on Biochemical Characteristics and Product Distribution
Abstract
1. Introduction
2. Results
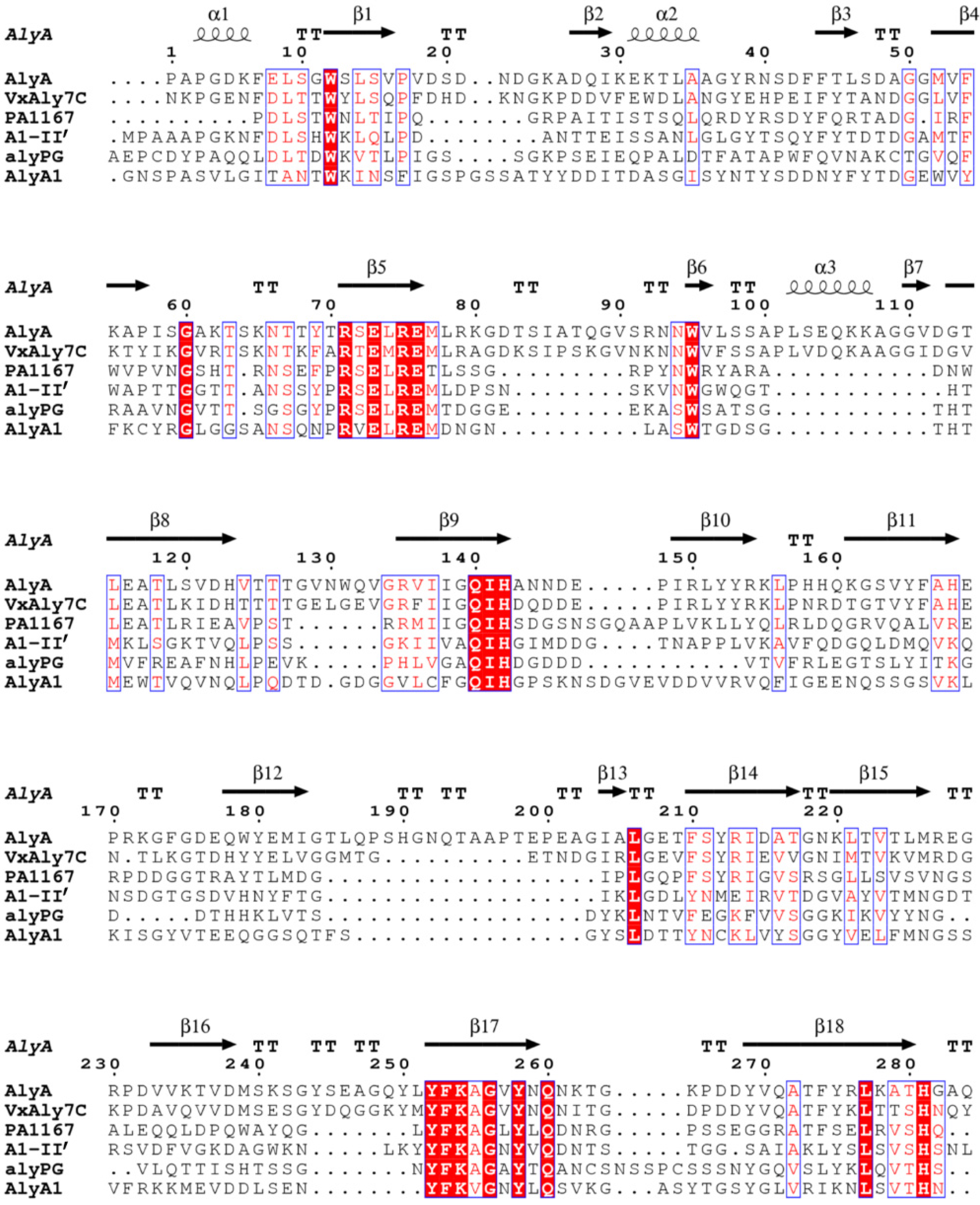
2.1. Cloning and Sequence Analysis of the Alginate Lyase Gene
2.2. Heterologous Expression and Purification of Recombinant VxAly7C-FL, VxAly7C-TM1, and VxAly7C-TM2
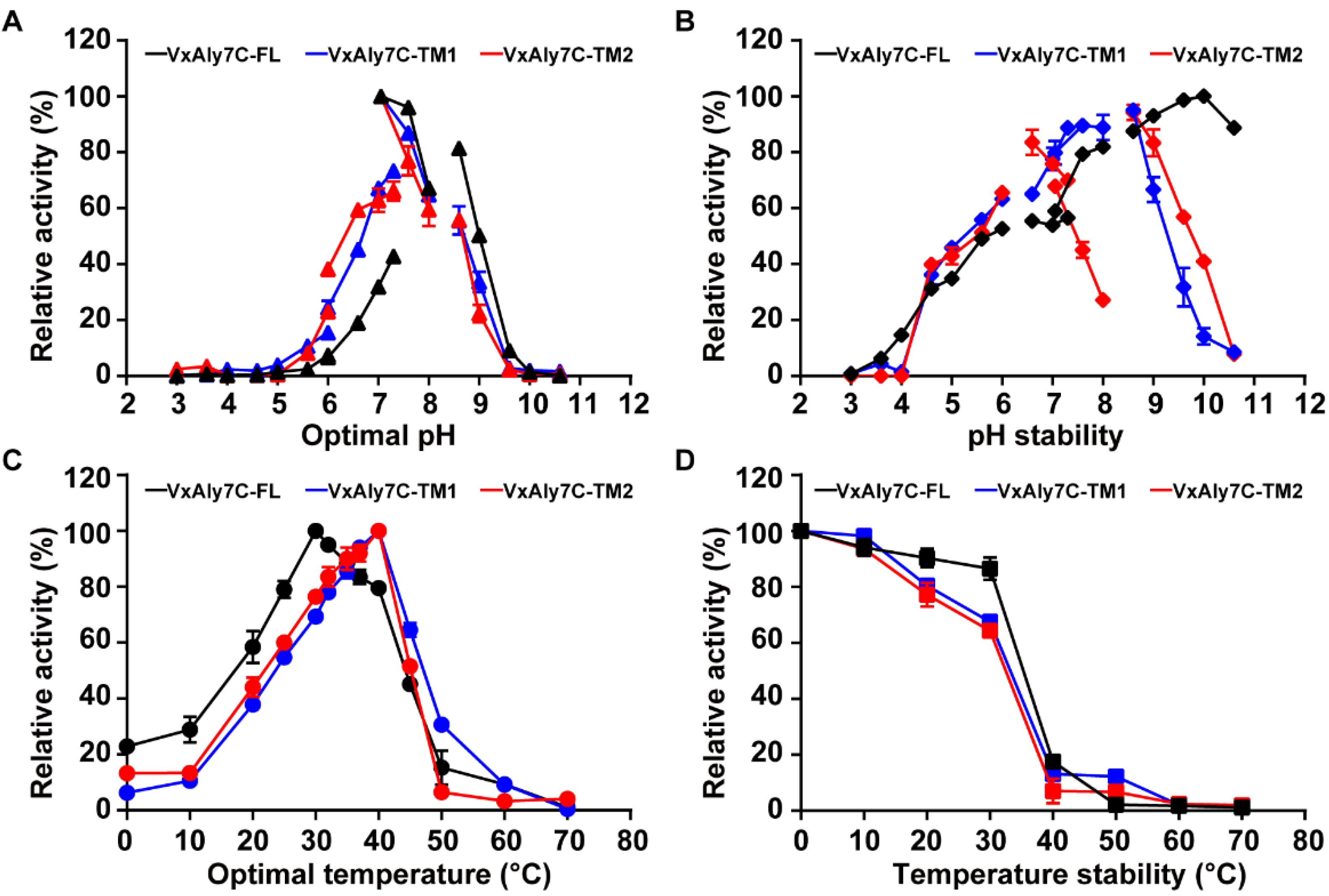
2.3. Biochemical Characteristics of Recombinant VxAly7C-FL, VxAly7C-TM1, and VxAly7C-TM2
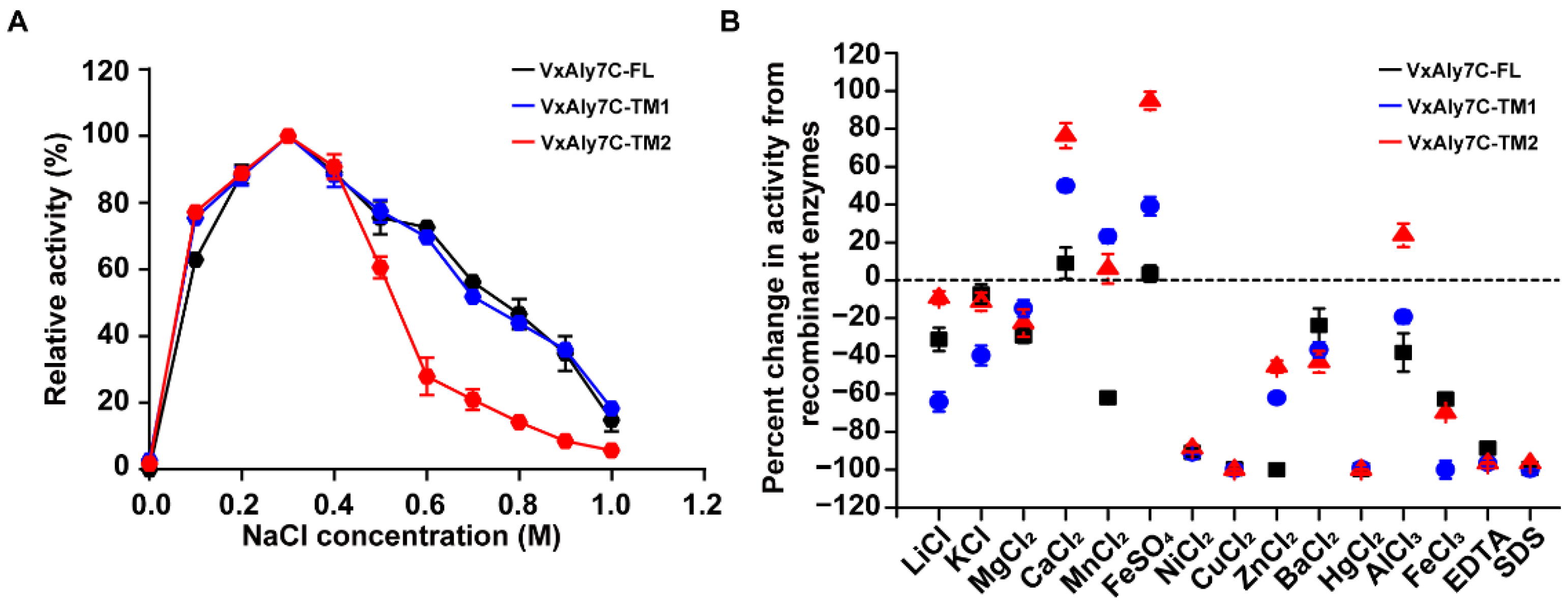
2.4. Effects of NaCl, Metal Ions, Chelators, and Detergents on Alginate Lyase Activity
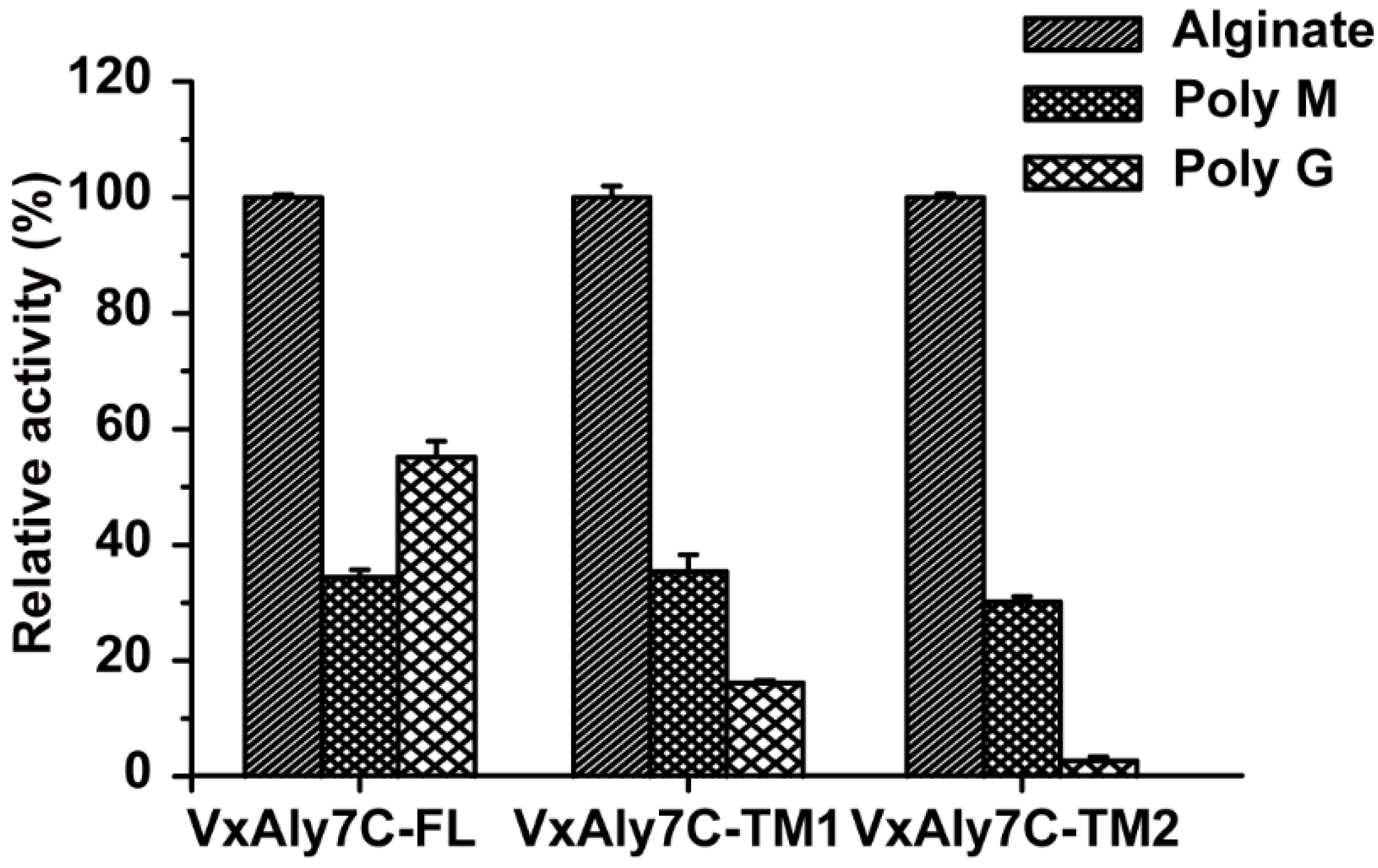
2.5. Substrate Specificity of Recombinant VxAly7C and Its Truncated Mutants
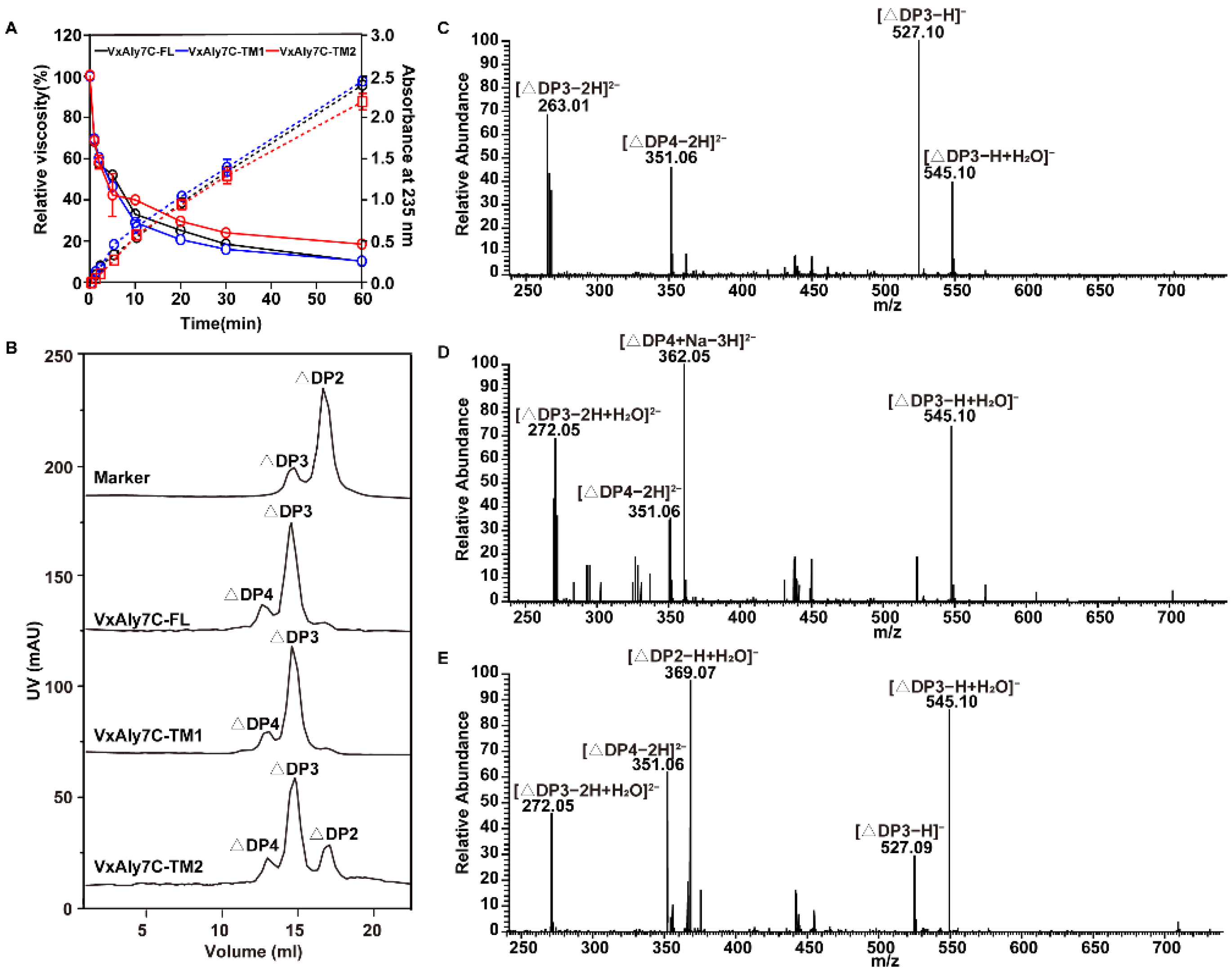
2.6. Analysis of the Mode of Action and End Product Distribution
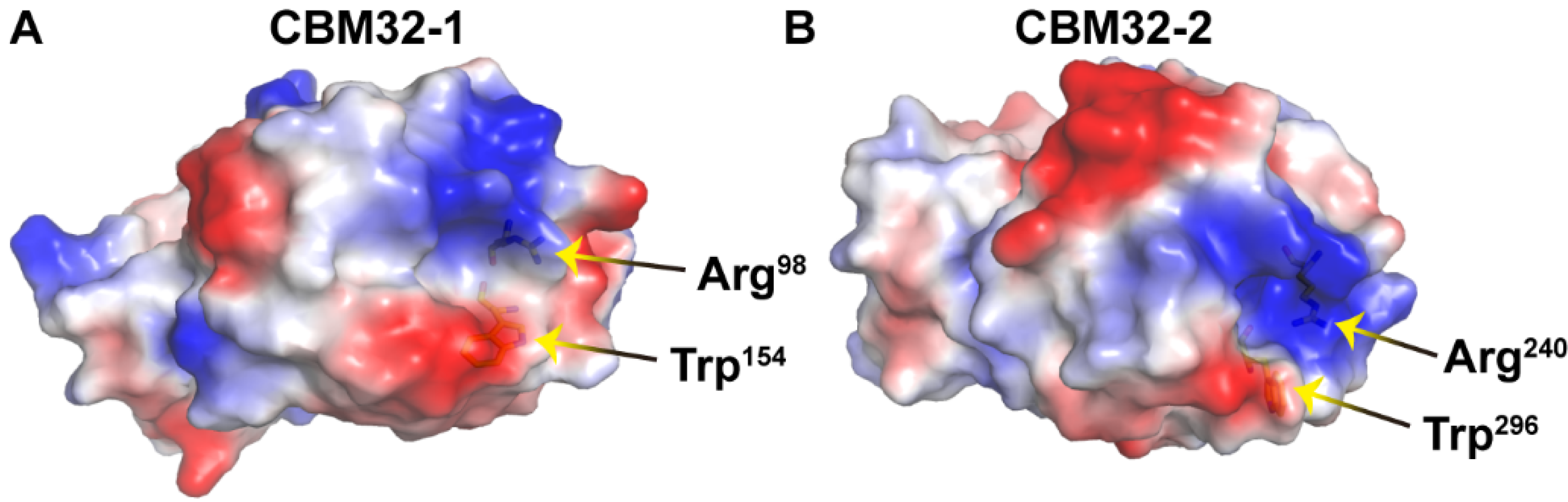
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmids, and Chemicals
4.2. Sequence Analysis
4.3. Construction of Recombinant VxAly7C and Its Truncated Mutants
4.4. Heterologous Expression and Purification of Recombinant Alginate Lyase
4.5. Enzyme Activity and Kinetic Parameter Assay
4.6. Biochemical Characteristics of Recombinant VxAly7C-FL and Its Truncated Mutants
4.7. Substrate Specificity, Mode of Action, and End Product Distribution Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, J.; Cui, D.; Ma, S.; Chen, W.; Chen, D.; Shen, H. Characterization of a novel PL 17 family alginate lyase with exolytic and endolytic cleavage activity from marine bacterium Microbulbifer sp. SH-1. Int. J. Biol. Macromol. 2021, 169, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Gacesa, P. Alginates. Carbohydr. Polym. 1988, 8, 161–182. [Google Scholar] [CrossRef]
- Aarstad, O.A.; Stanisci, A.; Saetrom, G.I.; Tondervik, A.; Sletta, H.; Aachmann, F.L.; Skjak-Braek, G. Biosynthesis and function of long guluronic acid-blocks in alginate produced by Azotobacter vinelandii. Biomacromolecules 2019, 20, 1613–1622. [Google Scholar] [CrossRef]
- Liu, J.; Yang, S.; Li, X.; Yan, Q.; Reaney, M.J.T.; Jiang, Z. Alginate oligosaccharides: Production, biological activities, and potential applications. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1859–1881. [Google Scholar] [CrossRef]
- Zhang, F.; Fu, Z.; Tang, L.; Zhang, Z.; Han, F.; Yu, W. Biochemical characterization of a novel exo-type PL7 alginate lyase VsAly7D from marine Vibrio sp. QY108. Int. J. Mol. Sci. 2021, 22, 8402. [Google Scholar] [CrossRef]
- Zhu, Y.; Wu, L.; Chen, Y.; Ni, H.; Xiao, A.; Cai, H. Characterization of an extracellular biofunctional alginate lyase from marine Microbulbifer sp. ALW1 and antioxidant activity of enzymatic hydrolysates. Microbiol. Res. 2016, 182, 49–58. [Google Scholar] [CrossRef]
- Wang, X.; Sun, G.; Feng, T.; Zhang, J.; Huang, X.; Wang, T.; Xie, Z.; Chu, X.; Yang, J.; Wang, H.; et al. Sodium oligomannate therapeutically remodels gut microbiota and suppresses gut bacterial amino acids-shaped neuroinflammation to inhibit Alzheimer’s disease progression. Cell Res. 2019, 29, 787–803. [Google Scholar] [CrossRef]
- Cheng, Y.; Wang, D.; Gu, J.; Li, J.; Liu, H.; Li, F.; Han, W. Biochemical characteristics and variable alginate-degrading modes of a novel bifunctional endolytic alginate lyase. Appl. Environ. Microbiol. 2017, 83, e01608-17. [Google Scholar] [CrossRef]
- Hu, F.; Cao, S.; Li, Q.; Zhu, B.; Yao, Z. Construction and biochemical characterization of a novel hybrid alginate lyase with high activity by module recombination to prepare alginate oligosaccharides. Int. J. Biol. Macromol. 2021, 166, 1272–1279. [Google Scholar] [CrossRef]
- La, A.L.T.Z.; Hu, Y. Advances in the preparation of alginate oligosaccharides and its biological functions. Sheng Wu Gong Cheng Xue Bao 2022, 38, 104–118. [Google Scholar] [CrossRef]
- Zhu, B.; Yin, H. Alginate lyase: Review of major sources and classification, properties, structure-function analysis and applications. Bioengineered 2015, 6, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Q.; Zhang, K.; Zhu, Q.; Li, Z.; Liu, Y.; Fitzek, E.; Yohe, T.; Zhao, L.; Li, W.; Liu, T.; et al. Structural and biochemical characterization of a multidomain alginate lyase reveals a novel role of CBM32 in CAZymes. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 1862–1869. [Google Scholar] [CrossRef]
- Tang, L.; Guo, E.; Zhang, L.; Wang, Y.; Gao, S.; Bao, M.; Han, F.; Yu, W. The function of CBM32 in alginate lyase VxAly7B on the activity on both soluble sodium alginate and alginate gel. Front. Microbiol. 2021, 12, 798819. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Wei, T.D.; Chen, X.L.; Li, C.Y.; Wang, P.; Xie, B.B.; Qin, Q.L.; Zhang, X.Y.; Pang, X.H.; Zhou, B.C.; et al. Molecular insight into the role of the N-terminal extension in the maturation, substrate recognition, and catalysis of a bacterial alginate lyase from polysaccharide lyase family 18. J. Biol. Chem. 2014, 289, 29558–29569. [Google Scholar] [CrossRef] [PubMed]
- Gilkes, N.R.; Warren, R.A.; Miller, R.C., Jr.; Kilburn, D.G. Precise excision of the cellulose binding domains from two Cellulomonas fimi cellulases by a homologous protease and the effect on catalysis. J. Biol. Chem. 1988, 263, 10401–10407. [Google Scholar] [CrossRef]
- Bernardes, A.; Pellegrini, V.O.A.; Curtolo, F.; Camilo, C.M.; Mello, B.L.; Johns, M.A.; Scott, J.L.; Guimaraes, F.E.C.; Polikarpov, I. Carbohydrate binding modules enhance cellulose enzymatic hydrolysis by increasing access of cellulases to the substrate. Carbohydr. Polym. 2019, 211, 57–68. [Google Scholar] [CrossRef]
- Zhang, Z.; Tang, L.; Bao, M.; Liu, Z.; Yu, W.; Han, F. Functional characterization of carbohydrate-binding modules in a new alginate lyase, TsAly7B, from Thalassomonas sp. LD5. Mar. Drugs 2019, 18, 25. [Google Scholar] [CrossRef]
- Peng, C.; Wang, Q.; Lu, D.; Han, W.; Li, F. A novel bifunctional endolytic alginate lyase with variable alginate-degrading modes and versatile monosaccharide-producing properties. Front. Microbiol. 2018, 9, 167. [Google Scholar] [CrossRef]
- Ji, S.; Dix, S.R.; Aziz, A.A.; Sedelnikova, S.E.; Baker, P.J.; Rafferty, J.B.; Bullough, P.A.; Tzokov, S.B.; Agirre, J.; Li, F.L.; et al. The molecular basis of endolytic activity of a multidomain alginate lyase from Defluviitalea phaphyphila, a representative of a new lyase family, PL39. J. Biol. Chem. 2019, 294, 18077–18091. [Google Scholar] [CrossRef]
- Chen, P.; Liu, R.; Huang, M.; Zhu, J.; Wei, D.; Castellino, F.J.; Dang, G.; Xie, F.; Li, G.; Cui, Z.; et al. A unique combination of glycoside hydrolases in Streptococcus suis specifically and sequentially acts on host-derived alphaGal-epitope glycans. J. Biol. Chem. 2020, 295, 10638–10652. [Google Scholar] [CrossRef] [PubMed]
- Angelov, A.; Loderer, C.; Pompei, S.; Liebl, W. Novel family of carbohydrate-binding modules revealed by the genome sequence of Spirochaeta thermophila DSM 6192. Appl. Environ. Microbiol. 2011, 77, 5483–5489. [Google Scholar] [CrossRef]
- Shinya, S.; Nishimura, S.; Kitaoku, Y.; Numata, T.; Kimoto, H.; Kusaoke, H.; Ohnuma, T.; Fukamizo, T. Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase. BioChem. J. 2016, 473, 1085–1095. [Google Scholar] [CrossRef]
- Mizutani, K.; Sakka, M.; Kimura, T.; Sakka, K. Essential role of a family-32 carbohydrate-binding module in substrate recognition by Clostridium thermocellum mannanase CtMan5A. FEBS Lett. 2014, 588, 1726–1730. [Google Scholar] [CrossRef]
- Grondin, J.M.; Chitayat, S.; Ficko-Blean, E.; Houliston, S.; Arrowsmith, C.H.; Boraston, A.B.; Smith, S.P. An unusual mode of galactose recognition by a family 32 carbohydrate-binding module. J. Mol. Biol. 2014, 426, 869–880. [Google Scholar] [CrossRef]
- Shimada, Y.; Watanabe, Y.; Wakinaka, T.; Funeno, Y.; Kubota, M.; Chaiwangsri, T.; Kurihara, S.; Yamamoto, K.; Katayama, T.; Ashida, H. Alpha-N-Acetylglucosaminidase from Bifidobacterium bifidum specifically hydrolyzes alpha-linked N-acetylglucosamine at nonreducing terminus of O-glycan on gastric mucin. Appl. Microbiol. Biotechnol. 2015, 99, 3941–3948. [Google Scholar] [CrossRef]
- Das, S.N.; Wagenknecht, M.; Nareddy, P.K.; Bhuvanachandra, B.; Niddana, R.; Balamurugan, R.; Swamy, M.J.; Moerschbacher, B.M.; Podile, A.R. Amino groups of chitosan are crucial for binding to a family 32 carbohydrate binding module of a chitosanase from Paenibacillus elgii. J. Biol. Chem. 2016, 291, 18977–18990. [Google Scholar] [CrossRef] [PubMed]
- Meng, D.D.; Ying, Y.; Chen, X.H.; Lu, M.; Ning, K.; Wang, L.S.; Li, F.L. Distinct roles for carbohydrate-binding modules of glycoside hydrolase 10 (GH10) and GH11 xylanases from Caldicellulosiruptor sp. strain F32 in thermostability and catalytic efficiency. Appl. Environ. Microbiol. 2015, 81, 2006–2014. [Google Scholar] [CrossRef]
- Yang, M.; Li, N.; Yang, S.; Yu, Y.; Han, Z.; Li, L.; Mou, H. Study on expression and action mode of recombinant alginate lyases based on conserved domains reconstruction. Appl. Microbiol. Biotechnol. 2019, 103, 807–817. [Google Scholar] [CrossRef]
- Matsushima, R.; Danno, H.; Uchida, M.; Ishihara, K.; Suzuki, T.; Kaneniwa, M.; Ohtsubo, Y.; Nagata, Y.; Tsuda, M. Analysis of extracellular alginate lyase and its gene from a marine bacterial strain, Pseudoalteromonas atlantica AR06. Appl. Microbiol. Biotechnol. 2010, 86, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yang, X.; Bao, M.; Wu, Y.; Yu, W.; Han, F. Family 13 carbohydrate-binding module of alginate lyase from Agarivorans sp. L11 enhances its catalytic efficiency and thermostability, and alters its substrate preference and product distribution. Fems Microbiol. Lett. 2015, 362, 10. [Google Scholar] [CrossRef]
- Sim, P.F.; Furusawa, G.; Teh, A.H. Functional and structural studies of a multidomain alginate lyase from Persicobacter sp. CCB-QB2. Sci. Rep. 2017, 7, 13656. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Chen, P.; Zeng, Y.; Men, Y.; Mu, S.; Zhu, Y.; Chen, Y.; Sun, Y. The characterization and modification of a novel bifunctional and robust alginate lyase derived from Marinimicrobium sp. H1. Mar. Drugs 2019, 17, 545. [Google Scholar] [CrossRef] [PubMed]
- Kadowaki, M.A.S.; Higasi, P.; de Godoy, M.O.; Prade, R.A.; Polikarpov, I. Biochemical and structural insights into a thermostable cellobiohydrolase from Myceliophthora thermophila. FEBS J. 2018, 285, 559–579. [Google Scholar] [CrossRef]
- de Kreij, A.; van den Burg, B.; Venema, G.; Vriend, G.; Eijsink, V.G.; Nielsen, J.E. The effects of modifying the surface charge on the catalytic activity of a thermolysin-like protease. J. Biol. Chem. 2002, 277, 15432–15438. [Google Scholar] [CrossRef]
- Zeng, Y.; Xu, J.; Fu, X.; Tan, M.; Liu, F.; Zheng, H.; Song, H. Effects of different carbohydrate-binding modules on the enzymatic properties of pullulanase. Int. J. Biol. Macromol. 2019, 137, 973–981. [Google Scholar] [CrossRef]
- Han, W.; Gu, J.; Cheng, Y.; Liu, H.; Li, Y.; Li, F. Novel alginate lyase (Aly5) from a polysaccharide-degrading marine bacterium, Flammeovirga sp. strain MY04: Effects of module truncation on biochemical characteristics, alginate degradation patterns, and oligosaccharide-yielding properties. Appl. Environ. Microbiol. 2016, 82, 364–374. [Google Scholar] [CrossRef]
- Jakob, F.; Martinez, R.; Mandawe, J.; Hellmuth, H.; Siegert, P.; Maurer, K.H.; Schwaneberg, U. Surface charge engineering of a Bacillus gibsonii subtilisin protease. Appl. Microbiol. Biotechnol. 2013, 97, 6793–6802. [Google Scholar] [CrossRef]
- Martinez, R.; Schwaneberg, U.; Roccatano, D. Temperature effects on structure and dynamics of the psychrophilic protease subtilisin S41 and its thermostable mutants in solution. Protein Eng. Des. Sel. 2011, 24, 533–544. [Google Scholar] [CrossRef][Green Version]
- Hu, F.; Zhu, B.; Li, Q.; Yin, H.; Sun, Y.; Yao, Z.; Ming, D. Elucidation of a unique pattern and the role of carbohydrate binding module of an alginate lyase. Mar. Drugs 2019, 18, 32. [Google Scholar] [CrossRef]
- Tang, L.; Wang, Y.; Gao, S.; Wu, H.; Wang, D.; Yu, W.; Han, F. Biochemical characteristics and molecular mechanism of an exo-type alginate lyase VxAly7D and its use for the preparation of unsaturated monosaccharides. Biotechnol. Biofuels 2020, 13, 99. [Google Scholar] [CrossRef] [PubMed]

| Protein | Specific Activity (U/mg) | Molecular Weight (kDa) | Specific Activity (U/nmol) |
|---|---|---|---|
| VxAly7C-FL | 557.82 | 72.47 | 40.41 |
| VxAly7C-TM1 | 946.34 | 56.72 | 53.73 |
| VxAly7C-TM2 | 1351.47 | 40.41 | 54.62 |
| VxAly7C-FL | VxAly7C-TM1 | VxAly7C-TM2 | |
|---|---|---|---|
| Km (mM) | 8.91 ± 0.11 | 12.09 ± 0.07 | 19.23 ± 0.16 |
| kcat (s−1) | 1734.86 ± 10.49 | 5523.81 ± 12.31 | 15,960 ± 5.14 |
| kcat/Km (s−1·mM−1) | 194.93 ± 2.25 | 456.89 ± 2.45 | 829.95 ± 9.77 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, L.; Bao, M.; Wang, Y.; Fu, Z.; Han, F.; Yu, W. Effects of Module Truncation of a New Alginate Lyase VxAly7C from Marine Vibrio xiamenensis QY104 on Biochemical Characteristics and Product Distribution. Int. J. Mol. Sci. 2022, 23, 4795. https://doi.org/10.3390/ijms23094795
Tang L, Bao M, Wang Y, Fu Z, Han F, Yu W. Effects of Module Truncation of a New Alginate Lyase VxAly7C from Marine Vibrio xiamenensis QY104 on Biochemical Characteristics and Product Distribution. International Journal of Molecular Sciences. 2022; 23(9):4795. https://doi.org/10.3390/ijms23094795
Chicago/Turabian StyleTang, Luyao, Mengmeng Bao, Ying Wang, Zheng Fu, Feng Han, and Wengong Yu. 2022. "Effects of Module Truncation of a New Alginate Lyase VxAly7C from Marine Vibrio xiamenensis QY104 on Biochemical Characteristics and Product Distribution" International Journal of Molecular Sciences 23, no. 9: 4795. https://doi.org/10.3390/ijms23094795
APA StyleTang, L., Bao, M., Wang, Y., Fu, Z., Han, F., & Yu, W. (2022). Effects of Module Truncation of a New Alginate Lyase VxAly7C from Marine Vibrio xiamenensis QY104 on Biochemical Characteristics and Product Distribution. International Journal of Molecular Sciences, 23(9), 4795. https://doi.org/10.3390/ijms23094795






