CRISPR/Cas9-Mediated Mutagenesis of the Granule-Bound Starch Synthase Gene in the Potato Variety Yukon Gold to Obtain Amylose-Free Starch in Tubers
Abstract
:1. Introduction
2. Results
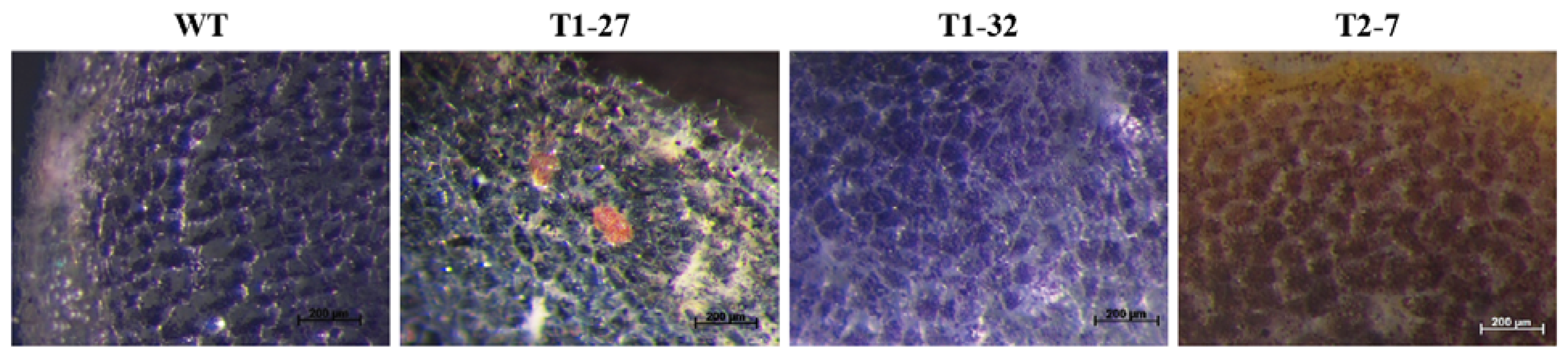
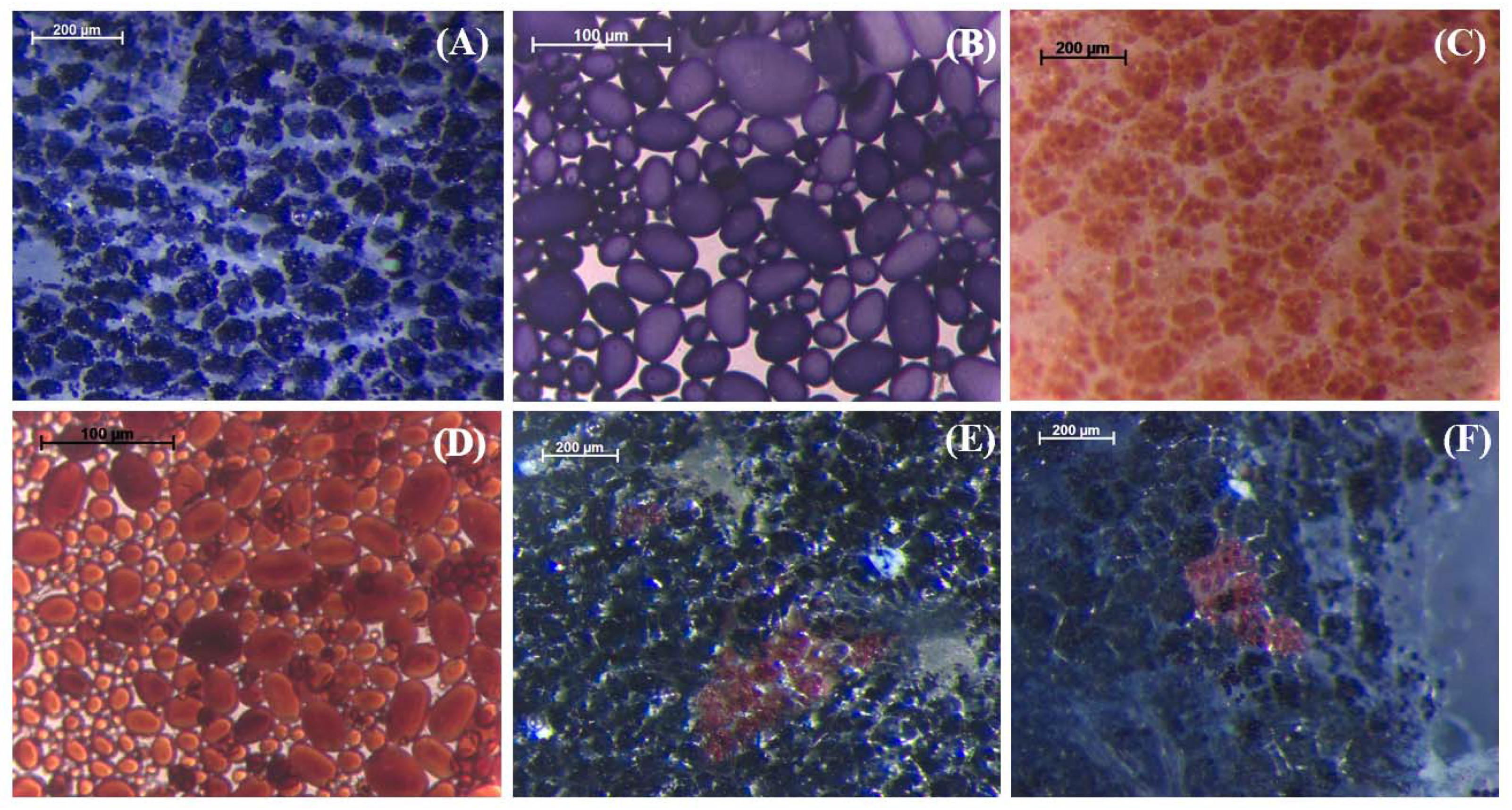
2.1. Qualitative Histochemical Analysis Showed Various Phenotypes in the Knockout Events
2.2. Molecular Characterization of Events Showed a Variety and Extent of Mutations
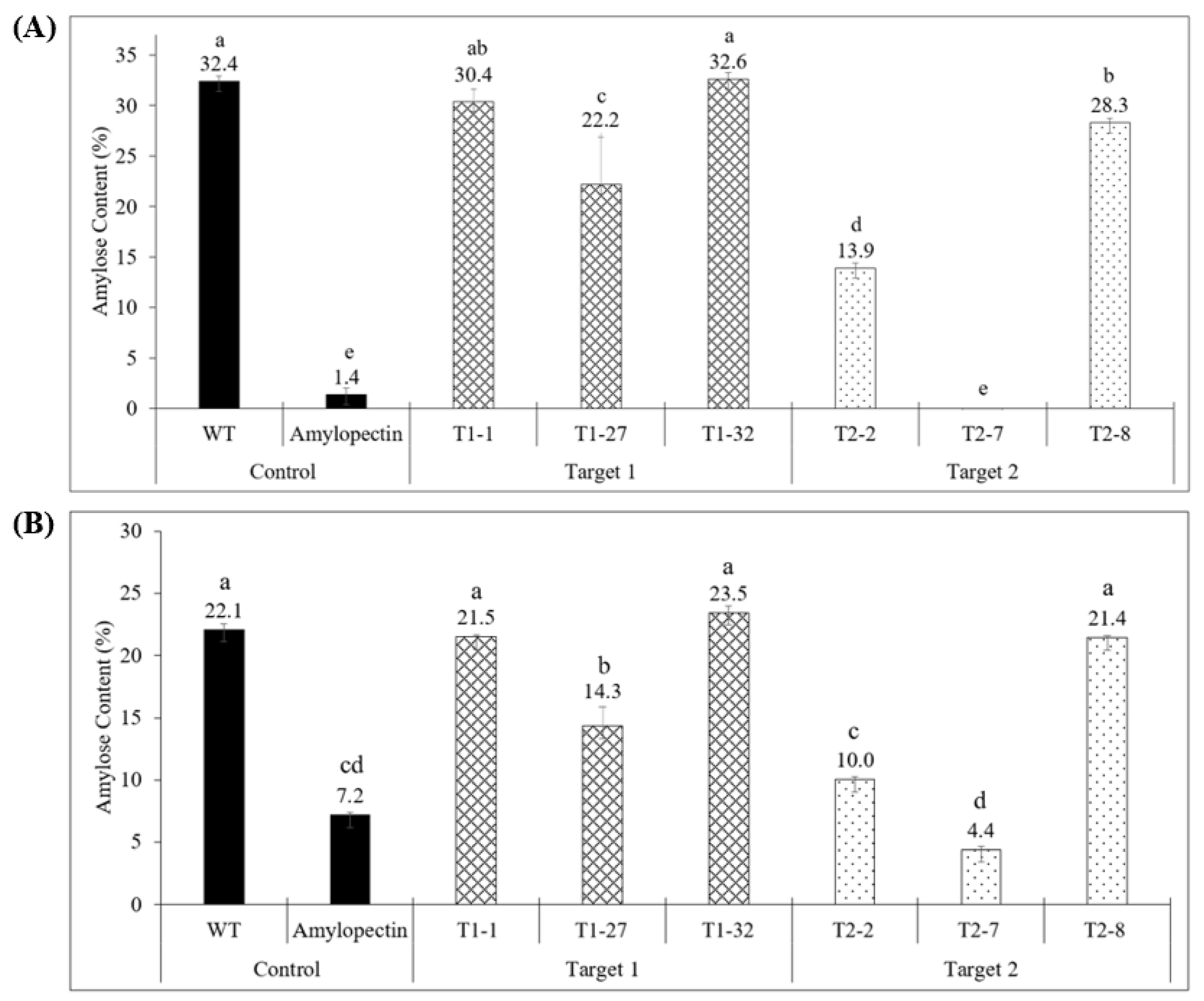
2.3. Quantitative Analysis of Starch Composition in Knockout Events Confirmed the Results of Histochemical Analysis
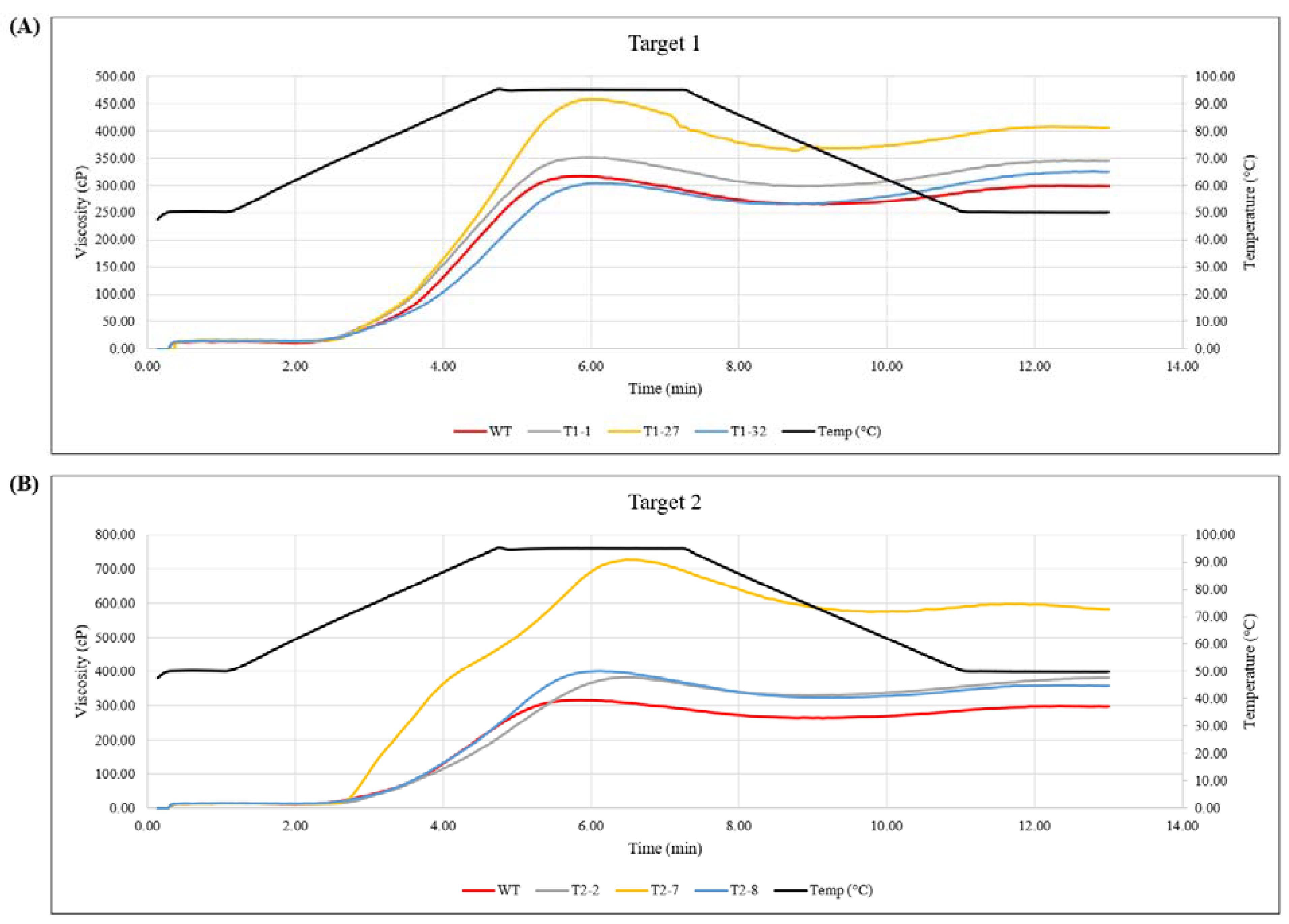
2.4. Viscosity Measurements in the Tuber Samples Confirmed Lack of Amylose in Event T2-7
3. Discussion
4. Materials and Methods
4.1. Plant Material and Binary Vector
4.2. Qualitative Analysis of Knockout Events
4.3. Molecular Characterization of Mutations
4.4. Specific Gravity Measurements and Quantitative Analysis of Starch Composition in the Putative Knockout Events
4.5. Starch Viscosity Measurements
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CRISPR/Cas9 | Clustered regularly interspaced short palindromic repeats and CRISPR-associated protein 9 |
| GBSS/gbssI | Granule-bound starch synthase |
| DSB | Double stranded break |
| NHEJ | Non-homologous end joining |
| INDELS | Insertions/Deletions |
| GFP/gfp | Green fluorescent protein |
| nptII | Neomycin phosphotransferase |
| WT | Wild-type |
| RVA | Rapid Visco Analyzer |
| EBN | Endosperm balance number |
| PV | Peak viscosity |
| cP | Centipoise |
References
- Hardigan, M.A.; Laimbeer, F.P.E.; Newton, L.; Crisovan, E.; Hamilton, J.P.; Vaillancourt, B.; Wiegert-Rininger, K.; Wood, J.C.; Douches, D.S.; Farré, E.M.; et al. Genome diversity of tuber-bearing Solanum uncovers complex evolutionary history and targets of domestication in the cultivated potato. Proc. Natl. Acad. Sci. USA 2017, 114, E9999–E10008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stokstad, E. The new potato. Science 2019, 363, 574–577. [Google Scholar] [CrossRef] [PubMed]
- Kraak, A. Industrial applications of potato starch products. J. Food Sci. Technol. 1992, 1, 107–112. [Google Scholar] [CrossRef]
- Fredriksson, H.; Silverio, J.; Andersson, R.; Eliasson, A.C.; Åman, P. The influence of amylose and amylopectin characteristics on gelatinization and retrogradation properties of different starches. Carbohydr. Polym. 1998, 35, 119–134. [Google Scholar] [CrossRef]
- Denyer, K.A.Y.; Johnson, P.; Zeeman, S.; Smith, A.M. The control of amylose synthesis. J. Plant Physiol. 2001, 158, 479–487. [Google Scholar] [CrossRef]
- Jobling, S. Improving starch for food and industrial applications. Curr. Opin. Plant Biol. 2004, 7, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, M.E.; Leeman, A.M.; Björck, I.M.E.; Eliasson, A.C. Some physical and nutritional characteristics of genetically modified potatoes varying in amylose/amylopectin ratios. Food Chem. 2007, 100, 136–146. [Google Scholar] [CrossRef]
- Buchanan, B.B.; Gruissem, W.; Jones, R.L. Biochemistry and Molecular Biology of Plants, 2nd ed.; John Wiley & Sons: West Sussex, UK, 2015; pp. 580–598. [Google Scholar]
- Liu, S. An Overview of Biological Basics. In Bioprocess Engineering, 2nd ed.; Liu, S., Ed.; Elsevier: New York, NY, USA, 2017; pp. 21–80. [Google Scholar]
- Lemos, P.V.F.; Barbosa, L.S.; Ramos, I.G.; Coelho, R.E.; Druzian, J.I. Characterization of amylose and amylopectin fractions separated from potato, banana, corn, and cassava starches. Int. J. Biol. Macromol. 2019, 132, 32–42. [Google Scholar] [CrossRef]
- Teng, L.Y.; Chin, N.L.; Yusof, Y.A. Rheological and textural studies of fresh and freeze-thawed native sago starch–sugar gels. II. Comparisons with other starch sources and reheating effects. Food Hydrocoll. 2013, 31, 156–165. [Google Scholar] [CrossRef]
- Jiao, M.; Gao, Y.; Tian, Y. Study the relationship between the microstructure and characteristics of quinoa starch by compared with common cereal starches. Am. J. Biochem. Biotechnol. 2020, 16, 561–567. [Google Scholar] [CrossRef]
- Yangcheng, H.; Jiang, H.; Blanco, M.; Jane, J.-L. Characterization of normal and waxy corn starch for bioethanol production. J. Agric. Food Chem. 2013, 61, 379–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puchta, H. The repair of double-strand breaks in plants: Mechanisms and consequences for genome evolution. J. Exp. Bot. 2005, 56, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, F.; Doudna, J.A. CRISPR–Cas9 structures and mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, Y. Plant Genome Editing with CRISPR Systems Methods and Protocols; Humana Press: New York, NY, USA, 2019. [Google Scholar]
- Andersson, M.; Turesson, H.; Nicolia, A.; Fält, A.-S.; Samuelsson, M.; Hofvander, P. Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep. 2017, 36, 117–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersson, M.A.-O.; Turesson, H.; Olsson, N.; Fält, A.S.; Ohlsson, P.; Gonzalez, M.N.; Samuelsson, M.; Hofvander, P.A.-O. Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery. Physiol. Plant. 2018, 164, 378–384. [Google Scholar] [CrossRef] [Green Version]
- Johansen, I.E.; Liu, Y.; Jørgensen, B.; Bennett, E.P.; Andreasson, E.; Nielsen, K.L.; Blennow, A.; Petersen, B.L. High efficacy full allelic CRISPR/Cas9 gene editing in tetraploid potato. Sci. Rep. 2019, 9, 17715. [Google Scholar] [CrossRef]
- Shepard, J.F.; Bidney, D.; Shahin, E. Potato protoplasts in crop improvement. Science 1980, 208, 17–24. [Google Scholar] [CrossRef]
- Kusano, H.; Ohnuma, M.; Mutsuro-Aoki, H.; Asahi, T.; Ichinosawa, D.; Onodera, H.; Asano, K.; Noda, T.; Horie, T.; Fukumoto, K.; et al. Establishment of a modified CRISPR/Cas9 system with increased mutagenesis frequency using the translational enhancer dMac3 and multiple guide RNAs in potato. Sci. Rep. 2018, 8, 13753. [Google Scholar] [CrossRef]
- Toinga-Villafuerte, S. CRISPR-Cas9 Targeted Mutagenesis of Green Fluorescent Protein Transgene and a Native, Granule Bound Starch Synthase, Gene in Potato. Ph.D. Thesis, Texas A&M University, College Station, TX, USA, 2021. [Google Scholar]
- Tsai, M.-L.; Li, C.-F.; Lii, C.-Y. Effects of granular structures on the pasting behaviors of starches. Cereal Chem. 1997, 74, 750–757. [Google Scholar] [CrossRef]
- Kumar, R.; Khatkar, B.S. Thermal, pasting and morphological properties of starch granules of wheat (Triticum aestivum L.) varieties. J. Food Sci. Technol. 2017, 54, 2403–2410. [Google Scholar] [CrossRef]
- Hui, Y.H.; Sherkat, F. Handbook of Food Science, Technology, and Engineering—4 Volume Set, 1st ed.; CRC Press: New York, NY, USA, 2005; pp. 1–17. [Google Scholar]
- Bradshaw, J.E. Introgression, base broadening and potato population improvements. In Potato Breeding: Theory and Practice; Bradshaw, J.E., Ed.; Springer International Publishing: Cham, Switzerland, 2021; pp. 341–403. [Google Scholar]
- Bonierbale, M.W.; Amoros, W.R.; Salas, E.; de Jong, W. Potato Breeding. In The Potato Crop: Its Agricultural, Nutritional and Social Contribution to Humankind; Campos, H., Ortiz, O., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 163–217. [Google Scholar]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [Green Version]
- Montecillo, J.A.V.; Chu, L.L.; Bae, H. CRISPR-Cas9 system for plant genome editing: Current approaches and emerging developments. Agronomy 2020, 10, 1033. [Google Scholar] [CrossRef]
- Janga, M.R.; Campbell, L.M.; Rathore, K.S. CRISPR/Cas9-mediated targeted mutagenesis in upland cotton (Gossypium hirsutum L.). Plant Mol. Biol. 2017, 94, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Voytas, D.F.; Gao, C. Precision genome engineering and agriculture: Opportunities and regulatory challenges. PLoS Biol. 2014, 12, e1001877. [Google Scholar] [CrossRef]
- Kunling, C.; Yanpeng, W.; Rui, Z.; Huawei, Z.; Caixia, G. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 2019, 70, 667–697. [Google Scholar]
- Ooms, G.; Bossen, M.E.; Burrell, M.M.; Karp, A. Genetic manipulation in potato with Agrobacterium rhizogenes. Potato Res. 1986, 29, 367–379. [Google Scholar] [CrossRef]
- Bruce, M.A.; Shoup Rupp, J.L. Agrobacterium-mediated transformation of Solanum tuberosum L., potato. In Transgenic Plants: Methods and Protocols; Kumar, S., Barone, P., Smith, M., Eds.; Springer: New York, NY, USA, 2019; pp. 203–223. [Google Scholar]
- Rathore, K.S.; Campbell, L.M.; Sherwood, S.; Nunes, E. Cotton (Gossypium hirsutum L.). In Agrobacterium Protocols, 3rd ed.; Wang, K., Ed.; Springer Science + Business Media: New York, NY, USA, 2015; Volume 2, pp. 267–279. [Google Scholar]
- Marcotrigiano, M. Origin of adventitious shoots regenerated from cultured tobacco leaf tissue. Am. J. Bot. 1986, 73, 1541–1547. [Google Scholar] [CrossRef]
- Zhang, H.; Cao, Y.; Zhang, H.; Xu, Y.; Zhou, C.; Liu, W.; Zhu, R.; Shang, C.; Li, J.; Shen, Z.; et al. Efficient generation of CRISPR/Cas9-mediated homozygous/biallelic Medicago truncatula mutants using a hairy root system. Front. Plant Sci. 2020, 11, 294. [Google Scholar] [CrossRef]
- Janga, M.R.; Pandeya, D.; Campbell, L.M.; Konganti, K.; Villafuerte, S.T.; Puckhaber, L.; Pepper, A.; Stipanovic, R.D.; Scheffler, J.A.; Rathore, K.S. Genes regulating gland development in the cotton plant. Plant Biotechnol. J. 2019, 17, 1142–1153. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Yuan, J.; Bizimungu, B.; Leblanc, D.; Lague, M. Effects of field selection parameters and specific gravity on culinary evaluation traits in a potato breeding programme. Potato Res. 2019, 62, 361–377. [Google Scholar] [CrossRef]
- Specific Gravity of Potato Tubers. Available online: https://www.agric.wa.gov.au/potatoes/specific-gravity-potato-tubers?page=0%2C0 (accessed on 30 January 2022).
- Juhász, R.; Salgó, A. Pasting behavior of amylose, amylopectin and their mixtures as determined by rva curves and first derivatives. Starch/Staerke 2008, 60, 70–78. [Google Scholar] [CrossRef]
- Zhao, S.S.; Dufour, D.; Sánchez, T.; Ceballos, H.; Zhang, F. Development of waxy cassava with different Biological and physico-chemical characteristics of starches for industrial applications. Biotechnol. Bioeng. 2011, 108, 10. [Google Scholar] [CrossRef] [PubMed]
- Toae, R.; Sriroth, K.; Rojanaridpiched, C.; Vichukit, V.; Chotineeranat, S.; Wansuksri, R.; Chatakanonda, P.; Piyachomkwan, K. Outstanding characteristics of Thai non-GM bred waxy cassava starches compared with normal cassava starch, waxy cereal starches and stabilized cassava starches. Plants 2019, 8, 447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez, T.; Dufour, D.; Moreno, I.X.; Ceballos, H. Comparison of pasting and gel stabilities of waxy and normal starches from potato, maize, and rice with those of a novel waxy cassava starch under thermal, chemical, and mechanical stress. J. Agric. Food Chem. 2010, 58, 5093–5099. [Google Scholar] [CrossRef] [PubMed]
- Allahgholipour, M.; Ali, A.J.; Alinia, F.; Nagamine, T.; Kojima, Y. Relationship between rice grain amylose and pasting properties for breeding better quality rice varieties. Plant Breed. 2006, 125, 6. [Google Scholar] [CrossRef]
- Park, S.; Kim, Y.-R. Clean label starch: Production, physicochemical characteristics, and industrial applications. Food Sci. Biotechnol. 2021, 30, 1–17. [Google Scholar] [CrossRef]
- Sandhya, D.; Jogam, P.; Allini, V.R.; Abbagani, S.; Alok, A. The present and potential future methods for delivering CRISPR/Cas9 components in plants. J. Genet. Eng. Biotechnol. 2020, 18, 25. [Google Scholar] [CrossRef]
- Bravo, J.P.K.; Liu, M.A.-O.; Hibshman, G.N.; Dangerfield, T.L.; Jung, K.; McCool, R.A.-O.; Johnson, K.A.-O.; Taylor, D.A.-O. Structural basis for mismatch surveillance by CRISPR-Cas9. Nature 2022, 603, 343–347. [Google Scholar] [CrossRef]
- Sharma, V.; Rausch, K.D.; Graeber, J.V.; Schmidt, S.J.; Buriak, P.; Tumbleson, M.E.; Singh, V. Effect of resistant starch on hydrolysis and fermentation of corn starch for ethanol. Appl. Biochem. Biotechnol. 2010, 160, 800–811. [Google Scholar] [CrossRef]
- Waltz, E. USDA approves next-generation GM potato. Nat. Biotechnol. 2015, 33, 12–13. [Google Scholar] [CrossRef] [PubMed]
- Ghislain, M.; Douches, D.S. The genes and genomes of the potato. In The Potato Crop: Its Agricultural, Nutritional and Social Contribution to Humankind; Campos, H., Ortiz, O., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 139–162. [Google Scholar]
- Johnston, G.R.; Rowberry, R.G. Yukon Gold: A new yellow-fleshed, medium-early, high quality table and French-fry cultivar. Am. Potato J. 1981, 58, 241–244. [Google Scholar] [CrossRef]
- Baltes, N.J.; Gil-Humanes, J.; Cermak, T.; Atkins, P.A.; Voytas, D.F. DNA Replicons for Plant Genome Engineering. Plant Cell 2014, 26, 151–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirsch, C.D.; Hamilton, J.P.; Childs, K.L.; Cepela, J.; Crisovan, E.; Vaillancourt, B.; Hirsch, C.N.; Habermann, M.; Neal, B.; Buell, C.R. Spud DB: A resource for mining sequences, genotypes, and phenotypes to accelerate potato breeding. Plant Genome 2014, 7, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Chari, R.; Yeo, N.C.; Chavez, A.; Church, G.M. sgRNA Scorer 2.0: A species-independent model to predict CRISPR/Cas9 activity. ACS Synth. Biol. 2017, 6, 902–904. [Google Scholar] [CrossRef] [Green Version]
- Wong, N.; Liu, W.; Wang, X. WU-CRISPR: Characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 2015, 16, 218. [Google Scholar] [CrossRef] [Green Version]
- Hao, L.; Yuduan, D.; Yanqing, Z.; Wenqi, J.; Kabin, X.; Ling-Ling, C. CRISPR-P 2.0: An improved CRISPR-Cas9 tool for genome editing in plants. Mol. Plant 2017, 10, 530–532. [Google Scholar]
- Cermak, T.; Starker, C.G.; Voytas, D.F. Efficient design and assembly of custom TALENs using the golden gate platform. In Chromosomal Mutagenesis. Methods in Molecular Biology (Methods and Protocols); Pruett-Miller, S.M., Ed.; Humana Press: New York, NY, USA, 2015; pp. 133–159. [Google Scholar]
- Chetty, V.J.; Narváez-Vásquez, J.; Orozco-Cárdenas, M.L. Potato (Solanum tuberosum L.). In Agrobacterium Protocols Volume 2; Wang, K., Ed.; Springer: New York, NY, USA, 2015; pp. 85–96. [Google Scholar]
- Fajardo, D.; Jayanty, S.S.; Jansky, S.H. Rapid high throughput amylose determination in freeze dried potato tuber samples. J. Vis. Exp. 2013, 80, 50407. [Google Scholar] [CrossRef] [Green Version]
- SAS Institute Inc. JMP® 16 Documentation Library; SAS Institute Inc.: Cary, NC, USA, 2021. [Google Scholar]


| Line | Specific Gravity | Dry Matter (%) |
|---|---|---|
| WT | 1.105 ab | 26.0 abc |
| T1-1 | 1.094 abc | 24.4 abc |
| T1-27 | 1.082 c | 26.3 abc |
| T1-32 | 1.063 d | 20.1 d |
| T2-2 | 1.109 a | 27.1 a |
| T2-7 | 1.094 bc | 23.5 c |
| T2-8 | 1.094 bc | 24.5 bc |
| Sample | Peak Viscosity (cP) | Trough Viscosity (cP) | Breakdown (cP) | Final Viscosity (cP) | Setback (cP) | Peak Time (min) | Pasting Temperature (°C) |
|---|---|---|---|---|---|---|---|
| WT | 317.0 d | 264.0 d | 53.0 c | 297.3 d | 33.3 ab | 5.8 d | 67.0 c |
| T1-1 | 352.5 cd | 297.5 cd | 55.0 c | 343.5 bcd | 46.0 a | 5.9 cd | 66.5 c |
| T1-27 | 457.3 b | 359.0 b | 98.3 b | 405.0 b | 46.0 a | 6.0 cd | 68.1 bc |
| T1-32 | 307.7 d | 265.0 d | 42.7 c | 324.7 cd | 59.7 a | 6.2 abc | 67.0 c |
| T2-2 | 386.0 c | 329.3 bc | 56.7 c | 380.7 bc | 51.3 a | 6.4 ab | 70.2 a |
| T2-7 | 726.0 a | 573.0 a | 153.0 a | 581.0 a | 8.0 b | 6.5 a | 69.1 ab |
| T2-8 | 402.7 c | 323.7 bc | 79.0 bc | 358.3 bc | 34.7 ab | 6.1 bcd | 67.2 c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toinga-Villafuerte, S.; Vales, M.I.; Awika, J.M.; Rathore, K.S. CRISPR/Cas9-Mediated Mutagenesis of the Granule-Bound Starch Synthase Gene in the Potato Variety Yukon Gold to Obtain Amylose-Free Starch in Tubers. Int. J. Mol. Sci. 2022, 23, 4640. https://doi.org/10.3390/ijms23094640
Toinga-Villafuerte S, Vales MI, Awika JM, Rathore KS. CRISPR/Cas9-Mediated Mutagenesis of the Granule-Bound Starch Synthase Gene in the Potato Variety Yukon Gold to Obtain Amylose-Free Starch in Tubers. International Journal of Molecular Sciences. 2022; 23(9):4640. https://doi.org/10.3390/ijms23094640
Chicago/Turabian StyleToinga-Villafuerte, Stephany, Maria Isabel Vales, Joseph M. Awika, and Keerti S. Rathore. 2022. "CRISPR/Cas9-Mediated Mutagenesis of the Granule-Bound Starch Synthase Gene in the Potato Variety Yukon Gold to Obtain Amylose-Free Starch in Tubers" International Journal of Molecular Sciences 23, no. 9: 4640. https://doi.org/10.3390/ijms23094640
APA StyleToinga-Villafuerte, S., Vales, M. I., Awika, J. M., & Rathore, K. S. (2022). CRISPR/Cas9-Mediated Mutagenesis of the Granule-Bound Starch Synthase Gene in the Potato Variety Yukon Gold to Obtain Amylose-Free Starch in Tubers. International Journal of Molecular Sciences, 23(9), 4640. https://doi.org/10.3390/ijms23094640