Abstract
Trifolium repens (T. repens) can accumulate significant amounts of heavy metal ions, and has strong adaptability to wide environmental conditions, and relatively large biomass, which is considered a potential plant for phytoremediation. However, the molecular mechanisms of T. repens involved in Cd tolerance have not yet been studied in detail. This study was conducted to examine the integrative responses of T. repens exposed to a high-level CdCl2 by investigating the physiological and transcriptomic analyses. The results suggested that T. repens seedlings had a high degree of tolerance to Cd treatment. The roots accumulated higher Cd concentration than leaves and were mainly distributed in the cell wall. The content of MDA, soluble protein, the relative electrolyte leakage, and three antioxidant enzymes (POD, SOD, and APX) was increased with the Cd treatment time increasing, but the CAT enzymes contents were decreased in roots. Furthermore, the transcriptome analysis demonstrated that the differentially expressed genes (DEGs) mainly enriched in the glutathione (GSH) metabolism pathway and the phenylpropanoid biosynthesis in the roots. Overexpressed genes in the lignin biosynthesis in the roots might improve Cd accumulation in cell walls. Moreover, the DEGs were also enriched in photosynthesis in the leaves, transferase activity, oxidoreductase activity, and ABA signal transduction, which might also play roles in reducing Cd toxicity in the plants. All the above, clearly suggest that T. repens employ several different mechanisms to protect itself against Cd stress, while the cell wall biosynthesis and GSH metabolism could be considered the most important specific mechanisms for Cd retention in the roots of T. repens.
1. Introduction
Heavy metal contamination has been a prominent environmental issue worldwide, mainly caused by industrial activities (electroplating, smelting, mining, etc.) and agricultural activities (phosphate fertilizer, agrochemicals, manure application, etc.) [1,2]. Cadmium (Cd) is one of the most universal and toxic heavy metals, due to it being readily absorbed, transported, and accumulated in plants as a non-essential element [3,4,5]. Approximately 30,000 tons of Cd were estimated to be discharged into the atmosphere worldwide every year [4]. Excessive Cd is toxic to plants in physiology and biochemistry aspects, such as inhibiting plants’ seed germination, seedling growth, organs development and damaging the membrane system, the antioxidant enzyme system, photosynthesis efficiency, and the other key biosynthetic pathways [3,6]. Furthermore, Cd cannot be biodegraded, and it can enter the food chain easily of its high mobility, which would cause harm to the health of humans and animals [7,8]. Therefore, it is important to explore further the regulatory mechanisms response of plants to Cd toxicity, which is essential to minimize its risk in the food chain and further facilitate decontamination of Cd-contaminated soils by phytoremediation [9,10].
The strategy of plants under heavy metal stress is determined by many aspects, such as heavy metal concentration, species, rhizosphere environment, and related metabolite level [11]. There are different sensitivities to heavy metal exposure were existed among different plant species. Till now, the response mechanism of various herbaceous species to Cd stress has been widely studied, such as Galega orientalis [12], Solanum nigrum (S. nigrum) [10], Brassica rapa [13], Festuca elata [14], and Hibiscus cannabinus [15]. However, the response mechanism of Cd has not been revealed in all the above species. In general, there are two strategies to resist Cd exposure [16,17]. One strategy is the avoidance mechanism, which tries to reduce Cd migration and avoid heavy metals entering roots [18]. Mycorrhiza and root exudates in the rhizosphere play a significant role to avoid heavy metal stress [19]. The other strategy relies on confinement and detoxification of heavy metals, which is the tolerance mechanism [16]. It reduces Cd toxicity mainly with chelation and antioxidant systems [20,21]. Glutathione (GSH) can combine Cd ions with the extending function of phytochelatins (PCs) synthesis, and the PCs-Cd complex is transported into vacuoles for detoxification, which is an efficient defense strategy in non-hyperaccumulator plants [22]. Reactive oxygen species (ROS) will produce under Cd stress, which can cause severe damage to plants. Malondialdehyde (MDA) is an important index of phytotoxicity [23], which indicated the toxicity level of plants [24]. To mitigate oxidative damage to plants, the enzymatic and non-enzymatic antioxidant systems are employed [25]. The most important enzymes include catalase (CAT), peroxidase (POD), superoxide dismutase (SOD) and glutathione reductase (GR) [25], and ascorbic acid (ASA), GSH is a non-enzyme and reduced GSH the main non-enzymatic antioxidant can be oxidized to glutathione disulfide (GSSG) to scavenge ROS [22,26]. Furthermore, the transporter family also has an important role under Cd stress, such as heavy metal ATPase (HMA), ATP-binding cassette (ABC), natural resistance-associated macrophage protein (Nramp), and cation diffusion facilitator (CDF) [27,28].
Trifolium repens (T. repens), an important perennial legume, has strong adaptability to different environmental conditions and relatively large biomass [29]. Previous studies reported that it has great potential for phytoremediation because it can accumulate significant amounts of heavy metal ions (Cd2+, Pb2+, Zn2+, and Cr2O72−) in both roots and shoots without visible damage [2,30,31]. For instance, it can accumulate 55.81 and 90.3 mg/kg Pb in 100 and 500 mg/kg Pb contained soil, respectively [31], and heavy metal ions (Cr2O72−, Cd2+ and Pb2+) 19.37–168.74 mg/kg in roots and 10.89–86.53 mg/kg in shoots [2]. The Cd concentrations in the below-ground part of T. repens are higher than that of Bidens pilosa (B. pilosa) and S. nigrum after seed plasma treatment, and the mean remediation efficiency of T. repens is also the highest among the three cadmium-tolerant plant species [29]. However, the preceding research mainly focused on the exogenous additive (e.g., Nitric oxide), microbial community [31], rhizosphere, and endosphere [2], the mechanism of T. repens response to Cd stress remains largely unknown. Since T. repens has great potential in phytoremediation, elucidating the molecular mechanism underlying T. repens Cd response is essential. In recent years, RNA sequencing is a reliable and economic technology, and transcriptomic has been widely used to reveal differential response mechanisms among plants under Cd stress, such as Zea mays (Z. mays) [32], Oryza sativa (O. sativa) [33], Lolium multiflorum [34], and Nicotiana species [35].
In this study, an integrated analysis of physiological and comparative transcriptomics was conducted to investigate the short-term response of T. repens to Cd toxicity. The objectives of this study were to (Ⅰ) evaluate the Cd uptake and translocation of T. repens under Cd stress; (Ⅱ) investigate the plant physiological response under Cd stress, including oxidative damage (MDA), antioxidant enzymes (APX, SOD, POD, and CAT), non-enzymatic products (soluble protein and chlorophyⅡ); (Ⅲ) identify pivotal Cd responsive differentially expressed genes (DEGs) and their involved essential pathways; (IV) unveil the molecular mechanisms of detoxification and tolerance of T. repens response to Cd stress. The present study’s findings will be in favor of novel perspectives of the mechanism involved in Cd tolerance and accumulation in T. repens.
2. Results
2.1. Cd Concentrations in Leaves and Roots
In the present study, T. repens plants were treated with 300 mg/L CdCl2·2.5H2O solution and the leaves began to turn yellow on the 5th day of treatment (Figure 1). Then, the physiological and transcriptome data were measured in the first 72 h to investigate the coordinated mechanisms of T. repens in response to high-level cadmium stress in the early stage.
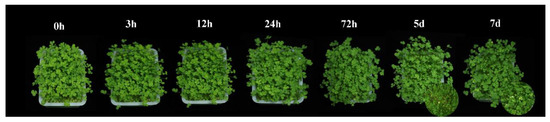
Figure 1.
The phenotypic changes of T. repens from 0 h to 7 d under the Cd stress.
Average leaf Cd concentrations at 0, 3, 12, 24, and 72 h after Cd treatment were 19.82, 129.95, 202.67, 260.56 and 564.72 mg/kg dry weight (DW), and the average root Cd concentration were 511.76, 1249.69, 1294.19, 1838.34 and 2773.95 mg/kg. The translocation factor is always less than 0.2 under Cd treatment in this study. The most dramatic change of Cd content occurred at 3 h after Cd treatment with an increase of 2.44 times in roots and 6.56 times in leaves. Moreover, between 24 h and 72 h, the increase in Cd content was still significant (1.51 times in roots and 2.17 times in leaves) (Figure 2a). Between 63.6% and 82.0% of Cd in leaves and roots was distributed in the cell wall fraction and 17.1–35.2% was distributed in the soluble fraction (cytoplasm). There was only a small amount of Cd (0.6–1.8%) distributed in organelles in T. repens (Figure 2b).

Figure 2.
(a) Cadmium (Cd) concentrations in roots and leaves, different letters (a–d) in the figure indicate significant differences at p < 0.05 level among different time points in the same tissue; (b) subcellular distribution of Cd in roots and leaves at 0, 3, 12, 24, and 72 h Cd treatment.
2.2. Changes in Physiology Characters under the Cd Stress
The MDA, SP, and EL were measured to assess the physiological response of white clover under Cd stress (Figure 3). With the increase in Cd treatment time, the MDA content was significantly increased in leaves and no significant difference in roots. The SP content was significantly increased in leaves and roots with the increase in Cd treatment time. The value of EL first goes down and then goes up during the 72 h treatment period. The EL value decreased significantly after 3 h and 24 h in the roots and leaves, respectively.
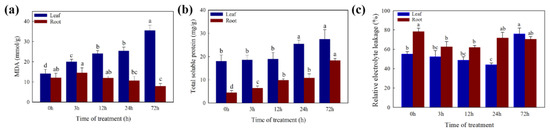
Figure 3.
Measurement of (a) MDA, (b) soluble protein, and (c) electrolyte leakage levels. Different letters (a–d) in the figures indicate significant differences at p < 0.05 level among different time points in the same tissue.
In addition, the content of SOD and APX (Figure 4) showed a significant increase after 72 h Cd treatment and no significant difference during the first 24 h in leaves. The CAT content (Figure 4) was increased significantly in the first 3 h and after 72 h Cd treatment in leaves. There was no significant difference in POD (Figure 4) during the 72 h Cd treatment in leaves. In roots, the content of SOD and CAT was increased significantly in the first 12 h, the POD was significantly increased in the first 3 h and the APX showed no significant difference.
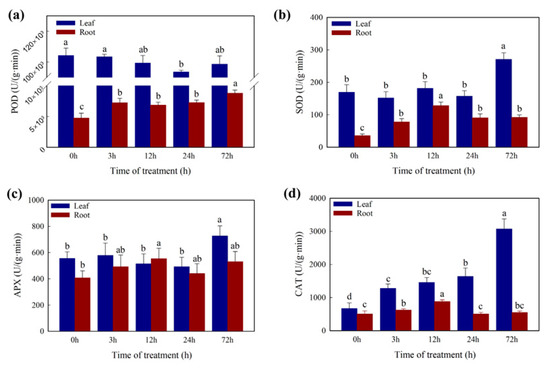
Figure 4.
Measurement of antioxidative enzyme activity (a) POD activity, (b) SOD activity, (c) APX activity, (d) CAT activity. Different letters (a–d) in the figures indicate significant differences at p < 0.05 level among different time points in the same tissue.
2.3. Transcriptome Sequencing and DEGs Identification
All the sequencing data were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database (accession no. RJNA771135). A total of 1,355,558,820 raw data was generated from the 30 samples. The raw reads ranged from 40,385,064 to 50,694,506. After the data filtering, the clean reads ranged from 38,718,352 to 48,295,400, and a total of 1,298,978,268 clean reads were generated. The data has a high level of quality, which is indicated by the Q20 and Q30 values, more than 97.89% and 93.90%, respectively (Table S2). The GC content ranged from 40.87% to 41.92%. Of the 30 to 48 million clean reads, 85.04% to 91.78% were mapped to the reference T. repens genome, and 75.84% to 79.85% were mapped uniquely to the genome [36] (Table S2). Around 75.06% to 81.04% were mapped to the exons, 3.73% to 4.77% were mapped to the introns, and 15.23% to 20.84% were mapped to the intergenic region (Table S2).
To further understand the dynamic expression changes of DEGs in T. repens at different time points after Cd treatment in leaves and roots, twenty-five comparison groups were analyzed (Figure 5). In leaves, the most remarkable gene expression change occurred in 72 h compared to 0 h. In the comparison, 2145 genes were altered (LT72 vs. LT0), with 1212 genes up-regulated and 933 genes down-regulated. In comparison with the other time points, the most remarkable changes were also found after 72 h Cd treatments. However, the most remarkable gene expression change (3665 genes altered) occurred during the first 3 h in roots, 2711 of which were up-regulated and 954 of which were down-regulated. Furthermore, a high gene alternation level existed between leaves and roots due to the different organs and complicated transcriptional responses to Cd. In the comparisons between leaves and roots, the DEGs at 3 h have been increased to 16,223 from 13,918 DEGs at 0 h, which indicated that Cd stress in the first 3 h can significantly impact the transcriptional profiles consistent with the above analysis.

Figure 5.
Summary of differentially expressed genes (DEGs) between organs and treatment duration.
In the analysis of the overlaps of DEGs in leaves, there was only one common gene among all the gene sets during the whole-time course (Figure S1). Almost all the Cd-induced genes and Cd-repressed genes in leaves at different gene sets were distinct. There were only 2 common genes between the up-regulated gene sets LT3 vs. LT0 and LT12 vs. LT3, 3 common genes between LT12 vs. LT3 and LT24 vs. LT12, and 4 common genes between LT24 vs. LT12 and LT72 vs. LT24. Among the down-regulated gene sets, there were 17 genes between LT3 vs. LT0 and LT12 vs. LT3, 1 common gene between LT12 vs. LT3 and LT24 vs. LT12, and 1 common gene between LT24 vs. LT12 and LT72 vs. LT24. In roots, there are no common genes among all the data sets, including the up-and down-regulated gene sets. Among the up-regulated gene sets, no common genes were found between RT3 vs. RT0 and RT12 vs. RT3, RT24 vs. RT12, and RT72 vs. RT24. There was only one common gene between up-regulated gene sets RT12 vs. RT3 and RT24 vs. RT12. Among the down-regulated gene sets, there was no common gene between RT12 vs. RT3 and RT24 vs. RT12, RT24 vs. RT12, and RT72 vs. RT24. Moreover, 22 common genes were found between the down-regulated gene sets RT3 vs. RT0 and RT12 vs. RT3. The overlapping genes between leaves and roots were also explored. Among all the data sets, 7385 common genes existed during the whole Cd treatment. There were 3309 common up-regulated genes, and 4071 common down-regulated genes were found among all the gene sets.
2.4. GO, KEGG Enrichment Analysis of DEGs, and WGCNA Analysis
The GO and KEGG enrichment analysis of identified DEGs under Cd stress in T. repens was conducted (Figure 6 and Figure S2). The results showed that the DEGs in leaves in the first 3 h Cd treatment were enriched in several general metabolic processes, such as alcohol metabolic process, polyol metabolic process, and the organic hydroxy compound metabolic process by GO analysis, and circadian rhythm-plant by KEGG analysis. After 12 h Cd treatment, the DEGs were mainly enriched in transferase activity, disulfide oxidoreductase activity, oxidoreductase activity, response to oxidative stress and response to the hormone by GO analysis, and photosynthesis-antenna proteins, phenylpropanoid biosynthesis, plant-pathogen interaction, MAPK signaling pathway-plant, Cysteine, and methionine metabolism by KEGG analysis (e.g., LT12 vs. LT3). In roots, the DEGs are mainly enriched in transferase activity, antioxidant activity, oxidoreductase activity, peroxidase activity, response to oxidative stress, membrane protein complex, defense response, response to the biotic stimulus by GO analysis, sulfur metabolism, glutathione metabolism, phenylpropanoid biosynthesis, Nitrogen metabolism, and MAPK signaling pathway-plant by KEGG analysis (e.g., RT72 vs. RT0).

Figure 6.
GO and KEGG enrichment analysis of all DEGs following CdCl2 exposure: LT12 vs. LT3 in leaves and RT72 vs. RT0 in roots.
A total of 9 modules were generated of the 38,147 genes from 30 samples (Figure 7) in the WGCNA analysis. Of which, 5 modules were remarkably related to the different tissues (leaves and roots) at multiple time points. The “blue”, “brown”, and “red” modules showed the expression specificity in roots, while the expression specificity of leaves was mainly shown in “turquoise” and “green” modules. In the present study, the top 5 genes with the highest Kwithin value of the identified DEGs under Cd stress in each module were considered as the hub genes in the corresponding pathway (Table S3). In roots, the hub genes are mainly related to the genes of phenylpropanoid biosynthesis in the “blue” module (chr6.jg878, chr11.jg7956, chr15.jg4577, chr8.jg2089, and chr7.jg6194). In the “brown” module, the hub genes were mainly related to the bZIP transcription factor (chr3.jg11413), the antioxidant enzyme (chr3.jg11201), and the transporters (chr7.jg6262, chr3.jg4277, and chr13.jg215). The hub genes in the “red” module were related to the bHLH transcription factor (chr4.jg5169) and the glutamine metabolism pathway (chr11.jg653, chr13.jg1149, and chr11.jg1125). In leaves, the hub genes were mainly related to the glutamine metabolism pathway (chr9.jg826, chr11.jg1220, and chr2.jg1989), the bHLH transcription factor (chr5.jg5695), and the antioxidant enzyme (chr6.jg5133) in the “turquoise” module. In the “green” module, the hub genes were mainly related to transporters (chr13.jg3472), the process of chlorophyll a/b biosynthesis (chr2.jg7790 and chr11.jg6504), the transcription factor (chr2.jg4404), and the ABA signal transduction pathway (chr15.jg4173). The co-expression networks of the hub genes of each module were shown in Figure 7d.

Figure 7.
Weighted gene co-expression network (WGCNA) analysis of DEGs of T. repens under Cd treatment. (a) Cluster dendrogram of 9 different expression modules; (b) module-sample relationship; (c) expression levels of genes among five selected modules most significantly correlated with roots and leaves under Cd stress; (d) the correlation networks of hub genes of the DEGs corresponding to the modules in WGCNA, the Kwithin value was showed in Table S3.
2.5. DEGs Involved in Lignin Biosynthesis and Heavy Metal Transporters in Cd Stress Response
The plant cell wall is the first barrier and has a significant fixation effect on heavy metal ions. Lignin is the main component of the cell walls and its biosynthesis can potentially increase the binding efficiency of the cell walls to the heavy metal ions [37]. In the present study, the DEGs were enriched in phenylpropanoid biosynthesis after 3 h and 72 h Cd treatment in roots and 12 h in leaves (Figure 6 and Figure S2), which was the main biosynthesis way of lignin in plants. A total of 11 DEGs were identified which involved in the lignin biosynthesis (Figure 8, Table S3), including 3 DEGs related to phenylalanine ammonia-lyase (PAL), 2 DEGs related to cinnamate 4-hydroxylase (C4H), 2 DEGs related to cinnamoyl-CoA reductase (CCR), 3 DEGs related to laccase (LAC) was induced in roots and leaves under Cd stress. The above DEGs were induced by Cd stress both in leaves and roots, and the highest expression level was shown after 3 h Cd treatment in roots. There was only one DEG related to cinnamyl alcohol dehydrogenase (CAD) that was induced both in leaves and roots, and the expression level was higher in leaves than in roots during 3 to 72 h under Cd treatment.
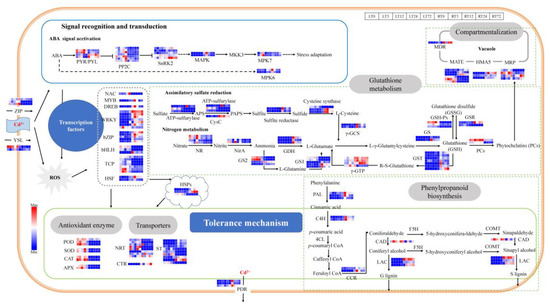
Figure 8.
Transcriptional changes of genes responsible for Cd response in leaves and roots of T. repens.
Furthermore, the Cd ions were absorbed into the plants mainly through the absorption pathway of divalent cations (e.g., Mn2+, Fe2+, Ca2+, Zn2+). Lots of transporters participate in the transfer processes of Cd in the plants. In the present study, 40 differentially expressed transport genes were identified which are possibly related to the Cd stress response (Figure 8, Table S3). The DEGs were induced by Cd stress both in leaves and roots. There were 3 DEGs related to Zinc/Iron-regulated transport protein (ZIP), which played an important role in the process of Cd entering into the cells. The Cd could also enter cells through Cd-chelates by yellow stripe 1-like (YSL) proteins (2 DEGs). The expression level of DEGs in leaves was higher than in roots. There were 8 DEGs related to the ATP-binding cassette (ABC) transport superfamily, including 3 DEGs related to pleiotropic drug resistance protein (PDR), 1 DEG related to multidrug resistance protein (MDR), and 5 DEGs related to multidrug resistance-associated protein (MRP). The DEGs expression level of MDR and PDR in roots was higher than in leaves. There were 2 DEGs expression levels related to MRP that was higher in leaves than in roots at all time points (chr8.jg2040 and chr3.jg2150), and the other 2 DEGs had higher expression level in roots (chr13.jg4938 and chr13.jg4939). There were 6 DEGs related to multidrug and toxic compound extrusion protein (MATE), 3 DEGs had higher expression levels in roots (chr1.jg10735, chr9.jg1754, and chr7.jg6262) and the other 3 DEGs had higher expression levels in leaves (chr4.jg4796, chr5.jg4276, and chr8.jg1983). There were 3 DEGs related to Heavy metal transporting ATPase (HMA) and 1 DGE related to Copper transporter (CTR), of which the expression levels were higher in roots. Moreover, there were 8 DEGs related to Nitrate transport protein (NRT) identified, and 5 DEGs had higher expression levels in leaves (chr13.jg3472, chr6.jg100, chr13.jg1054, chr5.jg1644, and chr11.jg4375) and the other 3 DEGs had higher expression levels in roots. There were 9 DEGs related to Sulfate transporter (ST), and 5 DEGs had higher expression levels in roots (chr2.jg57, chr2.jg8, chr7.jg512, chr4.jg12030, and chr12.jg1561) and the other 3 DEGs had higher expression levels in leaves. Most of the above genes were significantly expressed with the increase in treatment time.
2.6. DEGs Involved in Oxidation Resistance in Cd Stress Response
A large number of reactive oxygen species (ROS) could be generated under Cd stress, and the oxidation resistance systems in plants can be activated to prevent membrane lipid peroxidation and protect cells through scavenging ROS. The results of the present study identified 58 DEGs that were related to detoxification in white clover in response to Cd stress (Figure 8, Table S3). The antioxidant enzymatic genes, including CAT (2 DEGs), SOD (2 DEGs), POD (3 DEGs), APX (2 DEGs), glutathione reductase (GSR, 2 DEGs), glutathione peroxidase (GSH-Px, 3 DEGs) were induced both in leaves and roots. The GSR-related genes had higher expression levels in roots than in leaves, and the highest expression level was highest after 3 h Cd treatment in roots. Besides the GSR-related genes, the expression levels of the above genes were higher than in leaves.
As an important non-enzymatic antioxidant, Glutathione (GSH) has high efficiency in scavenging free radicals and resisting peroxidative damage in plants. The DEGs in the assimilatory sulfate reduction pathway and nitrogen metabolism pathway related to the GSH precursor biosynthesis have been identified in the study. In the assimilatory sulfate reduction pathway, the ATP-sulfurylase (3 DEGs), CysC (1 DEGs), sulfite reductase (2 DEGs), and cysteine synthase (3 DEGs), were induced in both leaves and roots. The expression level of the above genes was higher in roots and significantly up-regulated at 3 h Cd treatment in roots. In the nitrogen metabolism pathway, nitrate reductase (NR, 4 DEGs), Nitrite reductase (NirA, 2 DEGs), glutamate dehydrogenase (GDH, 3 DEGs), γ-glutamylcysteine synthetase (γ-GCS, 2DEGs), Glutamine synthetase (GS1, 5 DEGs; GS2, 4 DEGs) were induced both in leaves and roots. The expression levels of NR and NirA were higher in leaves and roots, and significantly up-regulated at 24 h Cd treatment in leaves. The GDH related genes have higher expression levels in roots and are significantly up-regulated at 3 h Cd treatment in roots. Four DEGs (chr2.jg2114, chr2.jg1989, chr6.jg3383, and chr11.jg1220) of GS had higher expression levels in leaves, and the other 5 DEGs (chr11.jg653, chr11.jg1125, chr4.jg11268, chr1.jg1885 and novel.837) express higher in roots. Furthermore, the glutathione S-transferase-related genes (GST, 7 DEGs) had high expression levels in roots than in leaves and were significantly up-regulated at 3 h and 72 h Cd treatment. The γ-glutamyl transpeptidase-related genes (γ-GTP, 4DEGs) were induced both in leaves and roots, which were expressed higher in roots. The PCs could also be catalyzed by phytochelatin synthase (PCs, 2DEGs). The induction expression of PCs related genes both in roots and leaves.
2.7. DEGs in Hormone Signal Transduction and Transcription Factors (TFs) in Cd Stress Response
In terms of signal recognition and transduction, the ABA signal transduction pathway (22 DGEs) was activated in white clover under Cd stress (Figure 8, Table S3). PYR/PYL (4 DEGs) in the pathway was induced both in leaves and roots, and 2 DEGs (chr8.jg428 and chr16.jg1328) were expressed higher in leaves and the other 2 DEGs (chr13.jg4773 and chr1.jg10967) expressed higher in roots. The other members, including PP2Cs (7 DEGs), SnRK2 (5 DEGs), MAPK (2 DEGs), MPK7 (2 DEGs), and MPK6 (DEGs) were also induced under Cd stress.
Moreover, the plant hormone signaling molecules could also impact the expression of transcription factors (TFs) and other defense proteins under the Cd stress. In the present study, a total of 27 DEGs were identified as TFs in T. repens under Cd stress (Figure 8, Table S3). The identified TF families include NAC (1 DEG), MYB (1 DEG), DREB (1 DEG), WRKY (7 DEGs), bZIP (5 DEGs), bHLH (3 DEGs), TCP (7 DEGs), and heat shock transcription factor (HSF, 2DEGs). The above genes were induced both in leaves and roots. The NAC TF family was significantly up-regulated at 72 h Cd treatment in leaves. The MYB and DREB TF families were up-regulated at 3 h Cd treatment in roots. Most of the genes of WRKY and bZIP TF families had higher expression levels in roots. Four DEGs (chr9.jg5416, chr7.jg7338, chr16.jg4775, and chr4.jg5478) of the WRKY TF family were significantly up-regulated at 3 h in roots, 2 DEGs (chr1.jg12558 and chr7.jg1916) at 72 h in roots, and 1 DEG (chr7.jg4527) at 3 h in leaves. Among the genes related to bZIP, 1 DEG (chr3.jg11413) was significantly up-regulated at 72 h and the other DEGs at 3 h Cd treatment in roots. The HSF TF family were significantly up-regulated at 3 h Cd treatment in roots, and most of the genes encoding HSP had higher expression level in roots.
2.8. Changes in Photosynthesis under Cd Stress
Furthermore, the total Chlorophyll content (Figure 9), Chlorophyll a, and Chlorophyll b were decreased significantly in the first 3 h and no significant difference in the subsequent processing time. The result of the KEGG enrichment analysis showed the DEGs were enriched in photosynthesis-antenna proteins at 12 h Cd treatment in leaves. A total of 25 DEGs were identified, including 11 DEGs in light reactions, and 14 DEGs in the Calvin-Benson cycle (Figure 9, Table S3). In the process of chlorophyll a/b biosynthesis and photosynthetic carbohydrate metabolism, the DEGs were down-regulated as the Cd treatment increased in leaves. The expression levels of the DEGs were extremely low in roots.

Figure 9.
The change of (a) chlorophyll contents, and different letters (a, b, c) in the figures indicate significant differences at p < 0.05 level among different time points in the same parameter; (b) the expression levels of the photosynthetic related genes under Cd stress.
2.9. Validation of Transcriptome Data by qRT-PCR Analyses
To confirm the RNA-seq results, 12 genes were randomly selected for qRT-PCR, including genes related to transporter protein, transcription factor, lignin synthesis, glutathione metabolism, hormone signaling, and chlorophyll biosynthesis (Table S1). Overall, the relative gene expression patterns of qRT-PCR were consistent with the RNA-seq (Figure 10) at all the treatment points in the roots and leaves of T. repens. The results showed that the RNA-seq result was reliable in the present study.

Figure 10.
Expression of the selected 12 genes inferred at 0, 3, 12, 24, and 72 h by RNA-seq and qRT-PCR.
3. Discussion
Based on the responses to the heavy metals, the plants can be divided into four types: metal-sensitive species, metal-tolerant nonhyperaccumulator species, metal-resistant excluder species, and metal hyper tolerant hyper-accumulator species [35]. T. repens could be classified into metal-tolerant species for it could grow well in metal-contaminated soils and has a significant accumulation of Cd, Cr, and Pb without visible damage [2,29,31,38]. The plant growth condition did not be affected by low concentration heavy metals, and the heavy metal accumulation ability of T. repens was significantly higher at a high background heavy metal concentration [2]. Moreover, the heavy metal accumulation ability does not appear to be limited, since the Cd concentration in plant tissues continues to rise with increasing exposure time [38]. Moreover, the highest Cd concentration 578 mg/kg has been found in China [39]. Previous studies [39] also found that the Cd concentration exceed the limit value (1 mg/kg) at about a quarter of the surveyed sites in China. Therefore, it is very worthwhile to study the tolerance of T. repens under high cadmium concentration in this study.
T. repens can accumulate 40 mg/kg in roots and no significant Cd in shoots after 3 days of exposure in the soil contaminated with 20 mg/kg. Moreover, for 56 days of exposure, the Cd concentration could be reached 106 and 3.8 mg/kg, in roots and shoots, respectively [38]. Moreover, the T. repens can accumulate Cd as high as 25.6 mg/kg (shoots) and 45.4 mg/kg (roots) after cold plasma seed treatment under the background Cd content is 3.02 mg/kg [29], while the Cd concentrations in the roots were even higher than two Cd hyperaccumulator, S. nigrum [40] and B. Pilosa [41]. In the current study, the results showed a high accumulation of Cd in the leaves and roots (Figure 1) under the high-level background Cd content and show no significant negative symptoms for 72 h exposure. The results showed the higher accumulation may be attributed to the quartz sand culture, which is similar to the results of Salvia sclarea (S. sclarea) [42], Malva parviflora [43], and Noccaea caerulescens [44] in hydroponic solution. Furthermore, the translocation factor is always less than 0.2 in our study, which is consistent with the previous study [38]. Besides, T. repens can also adapt to wide conditions with an extensive roots system and a large amount of biomass suggesting that T. repens is a candidate plant for Cd phytoextraction and phytostabilization [42].
As the first natural barrier of plant cells, the cell wall shows a significant fixation effect on heavy metal ions in keeping excess Cd out of the cell [37,45]. In our study, most of the Cd was bound to the plant cell wall and cytoplasm (Figure 1) and has a significant proportion in the cell wall. The results are consistent with that of Z. mays, Boehmeria nivea, Medicago sativa, and Nicotiana rustica, while the Cd was mainly accumulated in the soluble component in O. sativa, Hordeum vulgare, Phytolacca americana (P. americana), Amaranthus hypochondriacus, and Nicotiana tabacum [35,46]. The plant cell wall biosynthesis also is enhanced under the Cd stress to improve the Cd tolerant ability [47]. In our study, KEGG analysis showed that phenylpropanoid biosynthesis was over-presented in white clover under Cd exposure. The genes identified in the WGCNA are also hubs in phenylpropanoid biosynthesis. It is an important defense strategy for plants and can generate various metabolites, including guaiacyl and syringyl lignin, which are essential for cell wall biosynthesis [48,49]. In our study, 8 DEGs (PAL, C4H, CCR, and LAC) were up-regulated in roots related to lignin biosynthesis and achieved the highest expression level after 3 h Cd treatment. Increased lignin synthesis in roots to block Cd into the plant cells has also been identified in Vicia sativa [50], Brassica chinensis (B. chinensis) [47], Solanum melongena [51], Nicotiana species [52], and S. nigrum [10] under Cd stress. In addition, POD could regulate the biosynthesis of lignin and the related genes were upregulated in our studies [37,48]. The results indicated that the lignification biosynthesis would be strengthened once expose to Cd stress and improve the Cd fixed ability of the cell wall in T. repens.
Elevated ROS generation under Cd stress would disrupt the plant cell membranes with oxidative injuries by promoting the lipid peroxidation of membranes [53,54]. The amounts of MDA, EL, and SP were reported as the criteria for the identification of heavy metal tolerance [55]. However, the three indicators showed a strong increasing pattern in general, which showed that the damage to the membrane caused by Cd became more serious with the Cd treatment time increasing. Moreover, then the antioxidant defense system (POD, SOD, CAT, APX, etc.) was activated to mitigate H2O2 and lipid peroxidation damage [56,57]. For this study, the amounts of the antioxidant enzymes (POD, SOD, CAT, and APX) also showed an increasing pattern in general, besides the amount of CAT showed a decreasing pattern in the roots. The results suggest that the generated ROS might overwhelm the defense ability of the CAT enzyme in roots during the 72 h Cd treatment in T. repens. Furthermore, most of the related genes were up-regulated significantly in the leaves of T. repens, which were beneficial to remove excess ROS [35,53]. The HSP was also found to play a significant role in protecting the plant from adverse environments [45,58], and the expression level of HSP genes was also found up-regulated under Cd stress in T. repens.
GSH plays an important role in the detoxification of Cd in plants, which is related to both the chelation of Cd and scavenging ROS [59]. On the other hand, GSH is the precursor of PCs, and the PCs can bind most heavy metal ions [60]. It has been proven that the increased synthesis of GSH and PCs improves Cd tolerance in rice [61]. The expression level of PCs encoding genes was found up-regulated in T. repens (Figure 8) and then promoted the Cd accumulation in plants [37]. In the present study, KEGG analysis showed that the DEGs were enriched in sulfur metabolism, glutathione metabolism, and Nitrogen metabolism. The genes identified in the WGCNA are also hubs in glutathione metabolism. The DEGs related to the ATP sulfurylase, and cysteine synthase of the sulfur metabolism pathway were up-regulated both in the roots and leaves of T. repens. They are key enzymes for cysteine biosynthesis, which is the precursor for GSH biosynthesis [62]. Meanwhile, the genes related to NR, NirA, GS and GDH, and γ-GCS were also up-regulated under Cd stress in T. repens, which is consistent with the results of B. chinensis [63], Setaria italica [64], O. sativa [33]. In the glutathione metabolism, the DEGs related to GST were up-regulated in the roots of T. repens, which have unique functions in Cd detoxification [65]. The expression levels of GST genes were also up-regulated in O. sativa [66], Nicotiana species [35], and P. americana [67]. GST was able to catalyze GSH to be conjugated with ROS to protect plant cells from oxidative damage [68]. GST could catalyze the covalent binding of the cytotoxic substrates with GSH, and then formed S-glutathione conjugated which will be transferred into vacuolar to sequestrate the Cd [69].
The heavy metal ion transporters located on the plant membranes and the tonoplast take an important role in the transport and uptake of Cd [67]. The ABC transport family mainly (PDR, MDR, and MRP) is located in the tonoplast and can transport Cd conjugated complexes as Cd pumps [27,37]. In the current study, the ABC transporters related genes showed up-regulated after 3 h Cd treatment in T. repens. The up-regulated MRP and MDR genes can increase the transport of Cd amount into the vacuoles both in the leaves and roots, thereby reducing the Cd toxicity to the plants [70]. The up-regulated PDR genes can promote the Cd ion extrusion and improve the Cd resistance of T. repens, which is similar in Arabidopsis thaliana (A. thaliana) [71], Arachis hypogaea [72], and Lycopersicon esculentum [45]. We also identified 6 genes encoding MATE transporters up-regulated in T. repens, mainly transporting the exogenous toxic substances [73], which were consistent with the study on O. sativa [74] and Festuca arundinacea [14]. The YSL and ZIP genes were also up-regulated, which indicated the higher capacity of Cd2+ uptake and lower Cd symplastic loading into xylem in T. repens [52].
The response to high-dose Cd exposure could be ascribed to the plant signal perception and transduction [75,76] and reduction of photosynthetic carbon assimilation [77]. The calcium ion, ABA signal conduction pathway, and mitogen-activated protein kinase (MAPK) pathway were identified as the main signal perception pathway in response to stress tolerance of plants [28,75]. In this study, the member PYR/PYL and PP2Cs of the ABA signal transduction pathway were activated under Cd stress in T. repens. Moreover, then the plant hormone signaling molecules can be transmitted through their induced protein, which impacted the related gene regulation by modulating the expression level of TFs or defense proteins. NAC, MYB, DREB, WRKY, bZIP, bHLH, and TCP were identified as up/down-regulated under Cd stress in T. repens. Among them, the number of WRKY and TCP genes was the most, and their function in response to Cd stress has been verified in O. sativa [33] and A. thaliana [78]. The regulation of bHLH and bZIP can improve the Cd tolerance ability [79]. Only one gene was identified related to the other TFs, which showed they were also associated with the stress response in T. repens [28] NAC. Meanwhile, the thylakoid membranes of chloroplast would be dissolved under Cd stress, and the chlorophyll synthesis would be inhibited by destroying the chlorophyll photosynthesis enzyme activity [43,54,80]. In our study, the photosynthesis parameters (chlorophyll and chlorophyll a/b) decreased after 3 h of Cd treatment and resumes after 12 h. Except for the chlorophyll, the other two parameters continued to decrease after 72 h, which was also found in soybean plants [81] and S. sclarea [42]. The expression level of chlorophyll photosynthesis-related genes was down-regulated after 3 h Cd treatment and began to resume after 12 h. In the study of Brassica juncea [82] and P. americana [67], the expression level of photosynthesis-related genes decreased rapidly after 12 h of Cd treatment and began to restore after 48 h. The shorter time of this change in T. repens may be due to the high concentration of cadmium treated in this study. Taken together, our study contributes to the understanding of the molecular response mechanism in T. repens under high concentration Cd stress (Figure 8). At first, Cd caused oxidative stress to cells with excess ROS and induced the ABA hormone signaling pathway. The hormone signaling molecules can be transmitted through the induced protein, thus impacting the expression level of transcription factors (e.g., WRKY, bZIP, bHLH, HSF, etc.) and the other defense proteins [37]. Subsequently, the comprehensive defense system against Cd stress in T. repens was activated, including lignification biosynthesis, antioxidant enzyme system, chelation, and vacuolar compartmentalization. The Cd2+ was preliminarily blocked in the plant cell walls resulting in the alleviation of Cd toxicity. In the cells, the Cd2+ could be combined with various chelating substances (e.g., GSH and PCs) and further compartmentalized in vacuoles. Moreover, the GSH and other antioxidant enzymes would contribute to quenching ROS and protect cells from oxidative damage. In conclusion, T. repens showed strong tolerant capacity at a high Cd concentration and could be inferred as a Cd hyper-tolerant plant. Two major tolerance mechanisms under Cd exposure were identified: (1) enhanced the cell wall biosynthesis through the up-regulated genes of phenylpropanoid biosynthesis pathway to alleviate Cd toxicity; (2) enhanced GSH metabolism involved in Cd chelation and induction of antioxidant system against ROS accumulation. The results contribute to the understanding of the molecular physiological response mechanism of Cd stress in T. repens which will provide a profound theoretical basis for phytoremediation of Cd pollution. Moreover, a large number of candidate genes were provided in this study, which can be used to further investigate the molecular mechanism of plant tolerance to Cd.
4. Materials and Methods
4.1. Plant Material and Cd Treatments
The T. repens seeds (cv. Haifa) were sterilized in 5% sodium hypochlorite solution for 20 and rinsed several times with deionized water. Quartz sand was washed with tap water and deionized water thoroughly and then dried in the dry-heat oven at 65 °C; 0.2 g T. repens seeds per pot (20 cm × 15 cm × 12 cm) with 400 g quartz sand was sown. Seedling of T. repens was cultivated in a plant growth chamber (25 °C, 16 h light/8 h dark cycle) and irrigated with 1/2 strength Hoagland’s solution, the place of test posts was rotated every day.
After pre-test screening and analysis, T. repens was found to have a high tolerance to Cd. Because of this, the plants were treated with 0 mg/L and 300 mg/L CdCl2·2.5H2O solution 30 days after germination. Three replicates for each organ type (leaf and root) and time point (0 h, 3 h, 12 h, 24 h, and 72 h) were harvested after applying the Cd treatments. The samples were treated with liquid nitrogen immediately and stored at −80° for further analysis.
4.2. Cd Concentration and Subcellular Distribution in Leaves and Roots
The Cd concentration in leaves and roots was extracted with concentrated HNO3/HClO4 (4:1, v/v), and measured by an Inductively Coupled Plasma Optical Emission Spectrometer (ICP-OES, Optima 8300, CETAC Technologies®, Omaha, NE, USA) [83] according to the manufacturer’s recommendations. Cd translocation factor (TF) was defined as the ratio of Cd concentration in the leaves to that in the roots [5]. To determine the subcellular distribution of Cd in leaves and roots, the subcellular components were separated according to previous methods [33]. The Cd concentration of three isolated subcellular components, cell wall, organelle, and cytoplasm, was measured by ICP-OES. Statistical significance analysis was conducted with One-way Analysis of Variance (ANOVA) and the least significant difference (LSD) tests were conducted with SPSS 13.0 (SPSS Inc., Chicago, IL, USA).
4.3. Determination of Physiological Indexes
SOD activity was measured by the nitroblue tetrazole (NBT) method at 560 nm [84]. CAT activity was determined by the hydrolysis of the H2O2 method at 240 nm [85]. POD activity was investigated by guaiacol as a matrix according to [86] with some modifications at 470 nm [83]. APX activity was determined according to the method of previous studies [87]. The soluble protein (SP) was measured by the Coomassie brilliant blue method [88]. MDA content was measured with the thiobarbituric acid (TBA) method [89]. The electrolyte leakage (EL) was measured [42] with a conductivity meter (DDS-307, REX, Shanghai, China). The contents of photosynthetic pigments, total Chlorophyll content, Chlorophyll a, and Chlorophyll b, in fresh leaves were determined according to Arnon [90].
4.4. RNA Extraction, Transcriptome Sequencing, and Quality Control
Total RNA was extracted using the RNA prep Pure Plant Plus Kit (Polysaccharides & Polyphenolics-rich) (Tiangen, Beijing, China). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA).
Total RNA was used as input material for the RNA sample preparations. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in First Strand Synthesis Reaction Buffer (5×). First-strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase, then use RNaseH to degrade the RNA. Second strand cDNA synthesis was subsequently performed using DNA polymerase I and dNTP. The remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3′ ends of DNA fragments, Adaptor with a hairpin loop structure was ligated to prepare for hybridization. To select cDNA fragments of preferentially 370–420 bp in length, the library fragments were purified with the AMPure XP system (Beckman Coulter, Brea, CA, USA). Then PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers, and Index (X) Primer. At last, PCR products were purified (AMPure XP system, Beckman Coulter, Brea, CA, USA) and library quality was assessed on the Agilent Bioanalyzer 2100 system. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumia, San Diego, CA, USA) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Novaseq platform and 150 bp paired-end reads were generated (Novogene Co., Ltd. Beijing, China).
Raw data (raw reads) of fastq format were first processed through in-house Perl scripts. In this step, clean data (clean reads) were obtained by removing reads containing adapter, reads containing N base and low-quality reads from raw data. At the same time, Q20, Q30, and GC content of the clean data were calculated. All the downstream analyses were based on clean data with high quality. The clean reads were aligned to the T. repens reference genome [36] using HISAT software [91].
4.5. DEGs, GO, KEGG, and WGCNA Analysis
Differential expression analysis was performed using the DESeq2 R package (1.20.0) and FPKM (fragments per kilobase of transcripts sequence per million base pairs sequenced) method [92]. The resulting p-values were adjusted using Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with Padj < 0.05 and |log2FoldChange| > 2 were assigned as DEGs (differentially expressed genes) in the present study.
GO (Gene Ontology) enrichment and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis were implemented using the cluster profile R package to test the statistical (Padj < 0.05) enrichment of differential expression genes.
In addition, Weighted correlation network analysis (WGCNA) was conducted by R package WGCNA, which can be used for network construction, gene screening, gene cluster identification, topological feature calculation, data simulation, and visualization. Then, the networks were visualized by Cytoscape (v3.6.1).
4.6. Gene Expression Validation by qRT-PCR
To evaluate the reliability of the RNA-seq data, 12 candidate genes were selected for validation by quantitative real-time PCR (qRT-PCR) (Table S1). The RNA samples for RNA-seq experiments were also used for qRT-PCR. The primers were designed by the Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/, accessed on 25 February 2022) and listed in Table S1. Trβ-Actin gene was used as the internal control to quantify the gene expression level in white clover [93]. The first-strand cDNA was synthesized using All-in-One First-Strand Synthesis MasterMix (with dsDNase) (Yugong Biolabs Co., Ltd., Jiangsu, China). qRT-PCR was conducted by using Taq SYBR® Green qPCR Premix (Universal) (Yugong Biolabs Co., Ltd., Jiangsu, China) and performed at Bio-Rad CFX96 real-time PCR detection system. Each sample and gene had three technical replicates and the relative expression levels were evaluated using the 2−ΔΔCT method [94].
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23094612/s1.
Author Contributions
F.W. and G.N. designed the study, X.Z. provided the funding, F.W., J.F., X.Y. and L.Y. carried out the experiments, R.H., J.M., S.M., D.L. and J.Z. contributed reagents/materials/analysis tools; F.W. and G.N. drafted and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research work was funded by China Agriculture Research System of MOF and MARA, the Sichuan Province Breeding Research grant (2021YFYZ0013), and National Project on Sci-Tec Foundation Resources Survey (2017FY100602).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original sequencing data generated in the study have been deposited into the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database with the accession number RJNA771135.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Zhao, X.; Joo, J.C.; Lee, J.K.; Kim, J.Y. Mathematical estimation of heavy metal accumulations in Helianthus annuus L. with a sigmoid heavy metal uptake model. Chemosphere 2019, 220, 965–973. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Liu, C.; Li, B.; Dong, Y. Trifolium repens L. regulated phytoremediation of heavy metal contaminated soil by promoting soil enzyme activities and beneficial rhizosphere associated microorganisms. J. Hazard. Mater. 2021, 402, 123829. [Google Scholar] [CrossRef] [PubMed]
- El Rasafi, T.; Oukarroum, A.; Haddioui, A.; Song, H.; Kwon, E.E.; Bolan, N.; Tack, F.M.G.; Sebastian, A.; Prasad, M.N.V.; Rinklebe, J. Cadmium stress in plants: A critical review of the effects, mechanisms, and tolerance strategies. Crit. Rev. Environ. Sci. Technol. 2020, 52, 1–52. [Google Scholar] [CrossRef]
- Gallego, S.M.; Pena, L.B.; Barcia, R.A.; Azpilicueta, C.E.; Iannone, M.F.; Rosales, E.P.; Zawoznik, M.S.; Groppa, M.D.; Benavides, M.P. Unravelling cadmium toxicity and tolerance in plants: Insight into regulatory mechanisms. Environ. Exp. Bot. 2012, 83, 33–46. [Google Scholar] [CrossRef]
- Zuo, S.; Hu, S.; Rao, J.; Dong, Q.; Wang, Z. Zinc promotes cadmium leaf excretion and translocation in tall fescue (Festuca arundinacea). Chemosphere 2021, 276, 130186. [Google Scholar] [CrossRef] [PubMed]
- Sterckeman, T.; Thomine, S. Mechanisms of Cadmium Accumulation in Plants. Crit. Rev. Plant Sci. 2020, 39, 322–359. [Google Scholar] [CrossRef]
- Wang, P.; Chen, H.; Kopittke, P.M.; Zhao, F.-J. Cadmium contamination in agricultural soils of China and the impact on food safety. Environ. Pollut. 2019, 249, 1038–1048. [Google Scholar] [CrossRef]
- Haller, H.; Jonsson, A. Growing food in polluted soils: A review of risks and opportunities associated with combined phytoremediation and food production (CPFP). Chemosphere 2020, 254, 126826. [Google Scholar] [CrossRef]
- Khan, K.Y.; Ali, B.; Stoffella, P.J.; Cui, X.; Yang, X.; Guo, Y. Study amino acid contents, plant growth variables and cell ultrastructural changes induced by cadmium stress between two contrasting cadmium accumulating cultivars of Brassica rapa ssp. chinensis L. (pak choi). Ecotoxicol. Environ. Saf. 2020, 200, 110748. [Google Scholar] [CrossRef]
- Wang, J.; Chen, X.; Chu, S.; You, Y.; Chi, Y.; Wang, R.; Yang, X.; Hayat, K.; Zhang, D.; Zhou, P. Comparative cytology combined with transcriptomic and metabolomic analyses of Solanum nigrum L. in response to Cd toxicity. J. Hazard. Mater. 2021, 423 Pt B, 127168. [Google Scholar] [CrossRef]
- Małkowski, E.; Sitko, K.; Szopiński, M.; Gieroń, Ż.; Pogrzeba, M.; Kalaji, H.M.; Zieleźnik-Rusinowska, P. Hormesis in Plants: The Role of Oxidative Stress, Auxins and Photosynthesis in Corn Treated with Cd or Pb. Int. J. Mol. Sci. 2020, 21, 2099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Symanowicz, B.; Kalembasa, S.; Jaremko, D.; Niedbała, M. Effect of nitrogen application and year on concentration of Cu, Zn, Ni, Cr, Pb and Cd in herbage of Galega orientalis Lam. Plant Soil Environ. 2015, 61, 11–16. [Google Scholar] [CrossRef] [Green Version]
- Yu, R.; Li, D.; Du, X.; Xia, S.; Liu, C.; Shi, G. Comparative transcriptome analysis reveals key cadmium transport-related genes in roots of two pak choi (Brassica rapa L. ssp. chinensis) cultivars. BMC Genom. 2017, 18, 587. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Ai, H.; Cao, L.; Sui, R.; Ye, H.; Du, D.; Sun, J.; Yao, J.; Chen, K.; Chen, L. Transcriptome analysis providing novel insights for Cd-resistant tall fescue responses to Cd stress. Ecotoxicol. Environ. Saf. 2018, 160, 349–356. [Google Scholar] [CrossRef]
- Chen, P.; Chen, T.; Li, Z.; Jia, R.; Luo, D.; Tang, M.; Lu, H.; Hu, Y.; Yue, J.; Huang, Z. Transcriptome analysis revealed key genes and pathways related to cadmium-stress tolerance in Kenaf (Hibiscus cannabinus L.). Ind. Crops Prod. 2020, 158, 112970. [Google Scholar] [CrossRef]
- Lin, Y.F.; Aarts, M.G. The molecular mechanism of zinc and cadmium stress response in plants. Cell. Mol. Life Sci. 2012, 69, 3187–3206. [Google Scholar] [CrossRef] [PubMed]
- Anjum, N.A.; Hasanuzzaman, M.; Hossain, M.A.; Thangavel, P.; Roychoudhury, A.; Gill, S.S.; Rodrigo, M.A.; Adam, V.; Fujita, M.; Kizek, R.; et al. Jacks of metal/metalloid chelation trade in plants-an overview. Front. Plant Sci. 2015, 6, 192. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.S.; Dietz, K.J.; Mimura, T. Vacuolar compartmentalization as indispensable component of heavy metal detoxification in plants. Plant Cell Environ. 2016, 39, 1112–1126. [Google Scholar] [CrossRef] [PubMed]
- Bilal, S.; Shahzad, R.; Imran, M.; Jan, R.; Kim, K.M.; Lee, I.-J. Synergistic association of endophytic fungi enhances Glycine max L. resilience to combined abiotic stresses: Heavy metals, high temperature and drought stress. Ind. Crops Prod. 2020, 143, 111931. [Google Scholar] [CrossRef]
- Javed, M.T.; Akram, M.S.; Tanwir, K.; Javed Chaudhary, H.; Ali, Q.; Stoltz, E.; Lindberg, S. Cadmium spiked soil modulates root organic acids exudation and ionic contents of two differentially Cd tolerant maize (Zea mays L.) cultivars. Ecotoxicol. Environ. Saf. 2017, 141, 216–225. [Google Scholar] [CrossRef]
- Zha, Y.Q.; Zhang, K.K.; Pan, F.; Liu, X.; Han, S.M.; Guan, P. Cloning of PCS gene (TpPCS1) from Tagetes patula L. and expression analysis under cadmium stress. Plant Biol. 2021, 23, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Hou, D.; O’Connor, D.; Pan, S.; Zhu, J.; Bolan, N.S.; Mulder, J. Exogenous phosphorus treatment facilitates chelation-mediated cadmium detoxification in perennial ryegrass (Lolium perenne L.). J. Hazard. Mater. 2020, 389, 121849. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, S.; Jiang, W.; Liu, D. Cadmium accumulation, activities of antioxidant enzymes, and malondialdehyde (MDA) content in Pistia stratiotes L. Environ. Sci. Pollut. Res. 2013, 20, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Han, T.; Liang, Y.; Wu, Z.; Zhang, L.; Liu, Z.; Li, Q.; Chen, X.; Guo, W.; Jiang, L.; Pan, F.; et al. Effects of tetracycline on growth, oxidative stress response, and metabolite pattern of ryegrass. J. Hazard. Mater. 2019, 380, 120885. [Google Scholar] [CrossRef] [PubMed]
- Soares, C.; Carvalho, M.E.A.; Azevedo, R.A.; Fidalgo, F. Plants facing oxidative challenges—A little help from the antioxidant networks. Environ. Exp. Bot. 2019, 161, 4–25. [Google Scholar] [CrossRef]
- Rizwan, M.; Ali, S.; Adrees, M.; Ibrahim, M.; Tsang, D.C.W.; Zia-ur-Rehman, M.; Zahir, Z.A.; Rinklebe, J.; Tack, F.M.G.; Ok, Y.S. A critical review on effects, tolerance mechanisms and management of cadmium in vegetables. Chemosphere 2017, 182, 90–105. [Google Scholar] [CrossRef]
- Wang, H.; Liu, Y.; Peng, Z.; Li, J.; Huang, W.; Liu, Y.; Wang, X.; Xie, S.; Sun, L.; Han, E.; et al. Ectopic Expression of Poplar ABC Transporter PtoABCG36 Confers Cd Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 3293. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Mao, X.; Xu, Y.; Li, Y.; Zhao, N.; Yao, J.; Dong, Y.; Tigabu, M.; Zhao, X.; Li, S. Comparative transcriptomic analysis reveals the coordinated mechanisms of Populus × canadensis ‘Neva’ leaves in response to cadmium stress. Ecotoxicol. Environ. Saf. 2021, 216, 112179. [Google Scholar] [CrossRef]
- Zhao, L.; Deng, M.; Teng, Y.; Ren, W.; Wang, X.; Ma, W.; Luo, Y.; Christie, P. Enhanced biomass and cadmium accumulation by three cadmium-tolerant plant species following cold plasma seed treatment. J. Environ. Manag. 2021, 296, 113212. [Google Scholar] [CrossRef]
- Liu, S.L.; Yang, R.J.; Pan, Y.Z.; Wang, M.H.; Zhao, Y.; Wu, M.X.; Hu, J.; Zhang, L.L.; Ma, M.D. Exogenous NO depletes Cd-induced toxicity by eliminating oxidative damage, re-establishing ATPase activity, and maintaining stress-related hormone equilibrium in white clover plants. Environ. Sci. Pollut. Res. Int. 2015, 22, 16843–16856. [Google Scholar] [CrossRef]
- Liu, C.; Lin, H.; Dong, Y.; Li, B.; Liu, Y. Investigation on microbial community in remediation of lead-contaminated soil by Trifolium repens L. Ecotoxicol. Environ. Saf. 2018, 165, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Yue, R.; Lu, C.; Qi, J.; Han, X.; Yan, S.; Guo, S.; Liu, L.; Fu, X.; Chen, N.; Yin, H.; et al. Transcriptome Analysis of Cadmium-Treated Roots in Maize (Zea mays L.). Front. Plant Sci. 2016, 7, 1298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Yu, H.; Zhang, X.; Ye, D.; Huang, H.; Wang, Y.; Zheng, Z.; Li, T. A transcriptomic view of cadmium retention in roots of cadmium-safe rice line (Oryza sativa L.). J. Hazard. Mater. 2021, 418, 126379. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Zhang, Y.; He, Y.; Cao, Q.; Zhang, T.; Lou, L.; Cai, Q. Full-Length Transcriptome Assembly of Italian Ryegrass Root Integrated with RNA-Seq to Identify Genes in Response to Plant Cadmium Stress. Int. J. Mol. Sci. 2020, 21, 1067. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Chao, J.; Li, X.; Zhang, C.; Khan, R.; Du, S.; Xu, N.; Song, L.; Liu, H.; Shi, Y. Comparative transcriptome combined with biochemical and physiological analyses provide new insights toward cadmium accumulation with two contrasting Nicotiana species. Physiol. Plant. 2021, 173, 369–383. [Google Scholar] [CrossRef]
- Griffiths, A.G.; Moraga, R.; Tausen, M.; Gupta, V.; Bilton, T.P.; Campbell, M.A.; Ashby, R.L.; Nagy, I.; Khan, A.; Larking, A.; et al. Breaking Free: The Genomics of Allopolyploidy-facilitated Niche Expansion in White Clover. Plant Cell 2019, 31, 1466–1487. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Xia, B.; Meng, Y.; Yang, Z.; Pan, L.; Zhou, M.; Zhang, X. Transcriptome Analysis to Shed Light on the Molecular Mechanisms of Early Responses to Cadmium in Roots and Leaves of King Grass (Pennisetum americanum × P. purpureum). Int. J. Mol. Sci. 2019, 20, 2532. [Google Scholar] [CrossRef] [Green Version]
- Lanier, C.; Bernard, F.; Dumez, S.; Leclercq, J.; Lemiere, S.; Vandenbulcke, F.; Nesslany, F.; Platel, A.; Devred, I.; Cuny, D.; et al. Combined effect of Cd and Pb spiked field soils on bioaccumulation, DNA damage, and peroxidase activities in Trifolium repens. Environ. Sci. Pollut. Res. Int. 2016, 23, 1755–1767. [Google Scholar] [CrossRef]
- Duan, Q.; Lee, J.; Liu, Y.; Chen, H.; Hu, H. Distribution of Heavy Metal Pollution in Surface Soil Samples in China: A Graphical Review. Bull. Environ. Contam. Toxicol. 2016, 97, 303–309. [Google Scholar] [CrossRef]
- Wei, S.; Li, Y.; Zhou, Q.; Srivastava, M.; Chiu, S.; Zhan, J.; Wu, Z.; Sun, T. Effect of fertilizer amendments on phytoremediation of Cd-contaminated soil by a newly discovered hyperaccumulator Solanum nigrum L. J. Hazard. Mater. 2010, 176, 269–273. [Google Scholar] [CrossRef]
- Sun, Y.; Zhou, Q.; Wang, L.; Liu, W. Cadmium tolerance and accumulation characteristics of Bidens pilosa L. as a potential Cd-hyperaccumulator. J. Hazard. Mater. 2009, 161, 808–814. [Google Scholar] [CrossRef] [PubMed]
- Dobrikova, A.G.; Apostolova, E.L.; Hanć, A.; Yotsova, E.; Borisova, P.; Sperdouli, I.; Adamakis, I.S.; Moustakas, M. Cadmium toxicity in Salvia sclarea L.: An integrative response of element uptake, oxidative stress markers, leaf structure and photosynthesis. Ecotoxicol. Environ. Saf. 2021, 209, 111851. [Google Scholar] [CrossRef] [PubMed]
- Zoufan, P.; Jalali, R.; Hassibi, P.; Neisi, E.; Rastegarzadeh, S. Evaluation of antioxidant bioindicators and growth responses in Malva parviflora L. exposed to cadmium. Physiol. Mol. Biol. Plants 2018, 24, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Moustakas, M.; Bayçu, G.; Gevrek, N.; Moustaka, J.; Csatári, I.; Rognes, S.E. Spatiotemporal heterogeneity of photosystem II function during acclimation to zinc exposure and mineral nutrition changes in the hyperaccumulator Noccaea caerulescens. Environ. Sci. Pollut. Res. 2019, 26, 6613–6624. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Xie, Y.; He, Z.; Zhang, J.; Tang, Y.; Zhou, X. Network response of two cherry tomato (Lycopersicon esculentum) cultivars to Cadmium stress as revealed by transcriptome analysis. Ecotoxicol. Environ. Saf. 2021, 222, 112473. [Google Scholar] [CrossRef]
- Lu, H.; Li, Z.; Wu, J.; Shen, Y.; Li, Y.; Zou, B.; Tang, Y.; Zhuang, P. Influences of calcium silicate on chemical forms and subcellular distribution of cadmium in Amaranthus hypochondriacus L. Sci. Rep. 2017, 7, 40583. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Q.; Guo, J.-J.; He, C.-T.; Shen, C.; Huang, Y.-Y.; Chen, J.-X.; Guo, J.-h.; Yuan, J.-G.; Yang, Z.-Y. Comparative Transcriptome Analysis between Low- and High-Cadmium-Accumulating Genotypes of Pakchoi (Brassica chinensis L.) in Response to Cadmium Stress. Environ. Sci. Technol. 2016, 50, 6485–6494. [Google Scholar] [CrossRef]
- Chen, P.; Li, Z.; Luo, D.; Jia, R.; Lu, H.; Tang, M.; Hu, Y.; Yue, J.; Huang, Z. Comparative transcriptomic analysis reveals key genes and pathways in two different cadmium tolerance kenaf (Hibiscus cannabinus L.) cultivars. Chemosphere 2021, 263, 128211. [Google Scholar] [CrossRef]
- Gutsch, A.; Keunen, E.; Guerriero, G.; Renaut, J.; Cuypers, A.; Hausman, J.F.; Sergeant, K. Long-term cadmium exposure influences the abundance of proteins that impact the cell wall structure in Medicago sativa stems. Plant Biol. 2018, 20, 1023–1035. [Google Scholar] [CrossRef] [Green Version]
- Rui, H.; Chen, C.; Zhang, X.; Shen, Z.; Zhang, F. Cd-induced oxidative stress and lignification in the roots of two Vicia sativa L. varieties with different Cd tolerances. J. Hazard. Mater. 2016, 301, 304–313. [Google Scholar] [CrossRef]
- Li, B.; Chen, X.; Wu, Y.; Gu, A.; Zhang, J.; Luo, S.; Gao, X.; Zhao, J.; Pan, X.; Shen, S. Gene characterization and molecular pathway analysis of reverse thermosensitive genic male sterility in eggplant (Solanum melongena L.). Hortic. Res. 2019, 6, 118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, W.X.; Zhang, D.M.; Cao, Y.Q.; Dang, B.J.; Jia, W.; Xu, Z.C.; Han, D. Differential cadmium translocation and accumulation between Nicotiana tabacum L. and Nicotiana rustica L. by transcriptome combined with chemical form analyses. Ecotoxicol. Environ. Saf. 2021, 208, 111412. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Filardo, F.; Hu, X.; Zhao, X.; Fu, D. Cadmium stress alters the redox reaction and hormone balance in oilseed rape (Brassica napus L.) leaves. Environ. Sci. Pollut. Res. Int. 2016, 23, 3758–3769. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Guo, Z.; Li, R.; Ali, A.; Guo, D.; Lahori, A.H.; Wang, P.; Liu, X.; Wang, X.; Zhang, Z. Screening of Chinese mustard (Brassica juncea L.) cultivars for the phytoremediation of Cd and Zn based on the plant physiological mechanisms. Environ. Pollut. 2020, 261, 114213. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Mendoza, D.; Quiroz-Moreno, A.; Medrano, R.E.; Grimaldo-Juarez, O.; Zapata-Perez, O. Cell viability and leakage of electrolytes in Avicennia germinans exposed to heavy metals. Z. Nat. C J. Biosci. 2009, 64, 391–394. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Wu, Z.; Wang, T.; Mantri, N.; Huang, H.; Li, H.; Tao, Z.; Guo, Q. Physiological and transcriptomic analyses of cadmium stress response in Dendrobium officinale seedling. Plant Physiol. Biochem. 2020, 148, 152–165. [Google Scholar] [CrossRef]
- Xu, Z.; Dong, M.; Peng, X.; Ku, W.; Zhao, Y.; Yang, G. New insight into the molecular basis of cadmium stress responses of wild paper mulberry plant by transcriptome analysis. Ecotoxicol. Environ. Saf. 2019, 171, 301–312. [Google Scholar] [CrossRef]
- Song, G.; Yuan, S.; Wen, X.; Xie, Z.; Lou, L.; Hu, B.; Cai, Q.; Xu, B. Transcriptome analysis of Cd-treated switchgrass root revealed novel transcripts and the importance of HSF/HSP network in switchgrass Cd tolerance. Plant Cell Rep. 2018, 37, 1485–1497. [Google Scholar] [CrossRef]
- Hu, Z.; Fu, Q.; Zheng, J.; Zhang, A.; Wang, H. Transcriptomic and metabolomic analyses reveal that melatonin promotes melon root development under copper stress by inhibiting jasmonic acid biosynthesis. Hortic. Res. 2020, 7, 79. [Google Scholar] [CrossRef]
- Ding, S.; Ma, C.; Shi, W.; Liu, W.; Lu, Y.; Liu, Q.; Luo, Z.-B. Exogenous glutathione enhances cadmium accumulation and alleviates its toxicity in Populus × canescens. Tree Physiol. 2017, 37, 1697–1712. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Huang, X.Y.; Salt, D.E.; Zhao, F.J. Mutation in OsCADT1 enhances cadmium tolerance and enriches selenium in rice grain. New Phytol. 2020, 226, 838–850. [Google Scholar] [CrossRef] [PubMed]
- Baig, M.A.; Ahmad, J.; Ali, A.A.; Amna; Qureshi, M.I. Chapter 13—Role of Sulfur Metabolism in Cadmium Tolerance. In Cadmium Tolerance in Plants; Hasanuzzaman, M., Vara Prasad, M.N., Nahar, K., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 335–365. [Google Scholar]
- Liang, T.; Ding, H.; Wang, G.; Kang, J.; Pang, H.; Lv, J. Sulfur decreases cadmium translocation and enhances cadmium tolerance by promoting sulfur assimilation and glutathione metabolism in Brassica chinensis L. Ecotoxicol. Environ. Saf. 2016, 124, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wu, M.; Hao, L.; Yi, H. Sulfur dioxide derivatives alleviate cadmium toxicity by enhancing antioxidant defence and reducing Cd2+ uptake and translocation in foxtail millet seedlings. Ecotoxicol. Environ. Saf. 2018, 157, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.K.; Chen, B.; Li, X.X.; Wang, J.P.; Zhang, Y.; Wang, X.F.; Yan, Y.Y.; Ke, H.F.; Yang, J.; Wu, J.H.; et al. A newly identified cluster of glutathione S-transferase genes provides Verticillium wilt resistance in cotton. Plant J. 2019, 98, 213–227. [Google Scholar] [CrossRef]
- Jing, X.-Q.; Zhou, M.-R.; Nie, X.-M.; Zhang, L.; Shi, P.-T.; Shalmani, A.; Miao, H.; Li, W.-Q.; Liu, W.-T.; Chen, K.-M. OsGSTU6 Contributes to Cadmium Stress Tolerance in Rice by Involving in Intracellular ROS Homeostasis. J. Plant Growth Regul. 2021, 40, 945–961. [Google Scholar] [CrossRef]
- Zhao, L.; Zhu, Y.H.; Wang, M.; Ma, L.G.; Han, Y.G.; Zhang, M.J.; Li, X.C.; Feng, W.S.; Zheng, X.K. Comparative transcriptome analysis of the hyperaccumulator plant Phytolacca americana in response to cadmium stress. 3 Biotech 2021, 11, 327. [Google Scholar] [CrossRef]
- Hossain, M.A.; Piyatida, P.; Silva, J.; Fujita, M. Molecular Mechanism of Heavy Metal Toxicity and Tolerance in Plants: Central Role of Glutathione in Detoxification of Reactive Oxygen Species and Methylglyoxal and in Heavy Metal Chelation. J. Bot. 2012, 2012, 872875. [Google Scholar] [CrossRef]
- Kumar, S.; Trivedi, P.K. Glutathione S-Transferases: Role in Combating Abiotic Stresses Including Arsenic Detoxification in Plants. Front. Plant Sci. 2018, 9, 751. [Google Scholar] [CrossRef] [Green Version]
- Wojas, S.; Hennig, J.; Plaza, S.; Geisler, M.; Siemianowski, O.; Skłodowska, A.; Ruszczyńska, A.; Bulska, E.; Antosiewicz, D.M. Ectopic expression of Arabidopsis ABC transporter MRP7 modifies cadmium root-to-shoot transport and accumulation. Environ. Pollut. 2009, 157, 2781–2789. [Google Scholar] [CrossRef]
- Kim, D.-Y.; Bovet, L.; Maeshima, M.; Martinoia, E.; Lee, Y. The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J. 2007, 50, 207–218. [Google Scholar] [CrossRef]
- Yu, R.; Jiang, Q.; Xv, C.; Li, L.; Bu, S.; Shi, G. Comparative proteomics analysis of peanut roots reveals differential mechanisms of cadmium detoxification and translocation between two cultivars differing in cadmium accumulation. BMC Plant Biol. 2019, 19, 137. [Google Scholar] [CrossRef] [PubMed]
- Dong, B.; Niu, L.; Meng, D.; Song, Z.; Wang, L.; Jian, Y.; Fan, X.; Dong, M.; Yang, Q.; Fu, Y. Genome-wide analysis of MATE transporters and response to metal stress in Cajanus cajan. J. Plant Interact. 2019, 14, 265–275. [Google Scholar] [CrossRef] [Green Version]
- Ogawa, I.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Time course analysis of gene regulation under cadmium stress in rice. Plant Soil 2009, 325, 97. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Qin, S.; Rengel, Z.; Gao, W.; Nie, Z.; Liu, H.; Li, C.; Zhao, P. Cadmium stress increases antioxidant enzyme activities and decreases endogenous hormone concentrations more in Cd-tolerant than Cd-sensitive wheat varieties. Ecotoxicol. Environ. Saf. 2019, 172, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Saijo, Y.; Loo, E.P.-i. Plant immunity in signal integration between biotic and abiotic stress responses. New Phytol. 2020, 225, 87–104. [Google Scholar] [CrossRef] [Green Version]
- Ali, B.; Gill, R.A.; Yang, S.; Gill, M.B.; Ali, S.; Rafiq, M.T.; Zhou, W. Hydrogen sulfide alleviates cadmium-induced morpho-physiological and ultrastructural changes in Brassica napus. Ecotoxicol. Environ. Saf. 2014, 110, 197–207. [Google Scholar] [CrossRef]
- Opdenakker, K.; Remans, T.; Keunen, E.; Vangronsveld, J.; Cuypers, A. Exposure of Arabidopsis thaliana to Cd or Cu excess leads to oxidative stress mediated alterations in MAPKinase transcript levels. Environ. Exp. Bot. 2012, 83, 53–61. [Google Scholar] [CrossRef]
- Yao, X.; Cai, Y.; Yu, D.; Liang, G. bHLH104 confers tolerance to cadmium stress in Arabidopsis thaliana. J. Integr. Plant Biol. 2018, 60, 691–702. [Google Scholar] [CrossRef]
- Kapoor, D.; Singh, M.P.; Kaur, S.; Bhardwaj, R.; Zheng, B.; Sharma, A. Modulation of the Functional Components of Growth, Photosynthesis, and Anti-Oxidant Stress Markers in Cadmium Exposed Brassica juncea L. Plants 2019, 8, 260. [Google Scholar] [CrossRef] [Green Version]
- Andresen, E.; Lyubenova, L.; Hubáček, T.; Bokhari, S.N.H.; Matoušková, Š.; Mijovilovich, A.; Rohovec, J.; Küpper, H. Chronic exposure of soybean plants to nanomolar cadmium reveals specific additional high-affinity targets of cadmium toxicity. J. Exp. Bot. 2019, 71, 1628–1644. [Google Scholar] [CrossRef]
- Zhang, D.; Du, Y.; He, D.; Zhou, D.; Wu, J.; Peng, J.; Liu, L.; Liu, Z.; Yan, M. Use of Comparative Transcriptomics Combined With Physiological Analyses to Identify Key Factors Underlying Cadmium Accumulation in Brassica juncea L. Front. Genet. 2021, 12, 655885. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Ma, Q.; Li, S.; Zhu, M.; Xia, Z.; Yu, W. Toxicological effects of single and joint sulfamethazine and cadmium stress in soil on pakchoi (Brassica chinensis L.). Chemosphere 2021, 263, 128296. [Google Scholar] [CrossRef] [PubMed]
- Kazemi Shahandashti, S.S.; Maali Amiri, R.; Zeinali, H.; Ramezanpour, S.S. Change in membrane fatty acid compositions and cold-induced responses in chickpea. Mol. Biol. Rep. 2013, 40, 893–903. [Google Scholar] [CrossRef] [PubMed]
- Beers, R.F., Jr.; Sizer, I.W. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J. Biol. Chem. 1952, 195, 133–140. [Google Scholar] [CrossRef]
- Zhou, W.; Leul, M. Uniconazole-induced tolerance of rape plants to heat stress in relation to changes in hormonal levels, enzyme activities and lipid peroxidation. Plant Growth Regul. 1999, 27, 99–104. [Google Scholar] [CrossRef]
- AbdElgawad, H.; Zinta, G.; Hamed, B.A.; Selim, S.; Beemster, G.; Hozzein, W.N.; Wadaan, M.A.M.; Asard, H.; Abuelsoud, W. Maize roots and shoots show distinct profiles of oxidative stress and antioxidant defense under heavy metal toxicity. Environ. Pollut. 2020, 258, 113705. [Google Scholar] [CrossRef]
- Cheng, Y.; Wei, H.; Sun, R.; Tian, Z.; Zheng, X. Rapid method for protein quantitation by Bradford assay after elimination of the interference of polysorbate 80. Anal. Biochem. 2016, 494, 37–39. [Google Scholar] [CrossRef]
- Heidarvand, L.; Maali-Amiri, R. Physio-biochemical and proteome analysis of chickpea in early phases of cold stress. J. Plant Physiol. 2013, 170, 459–469. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, T.; Hou, J.; Iqbal, M.Z.; Zhang, Y.; Cheng, B.; Feng, H.; Li, Z.; Liu, L.; Zhou, J.; Feng, G.; et al. Overexpression of the white clover TrSAMDC1 gene enhanced salt and drought resistance in Arabidopsis thaliana. Plant Physiol. Biochem. PPB 2021, 165, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).