Ectopic Expression of OsJAZs Alters Plant Defense and Development
Abstract
1. Introduction
2. Results
2.1. Phylogenetic Analysis of OsJAZs and AtJAZs
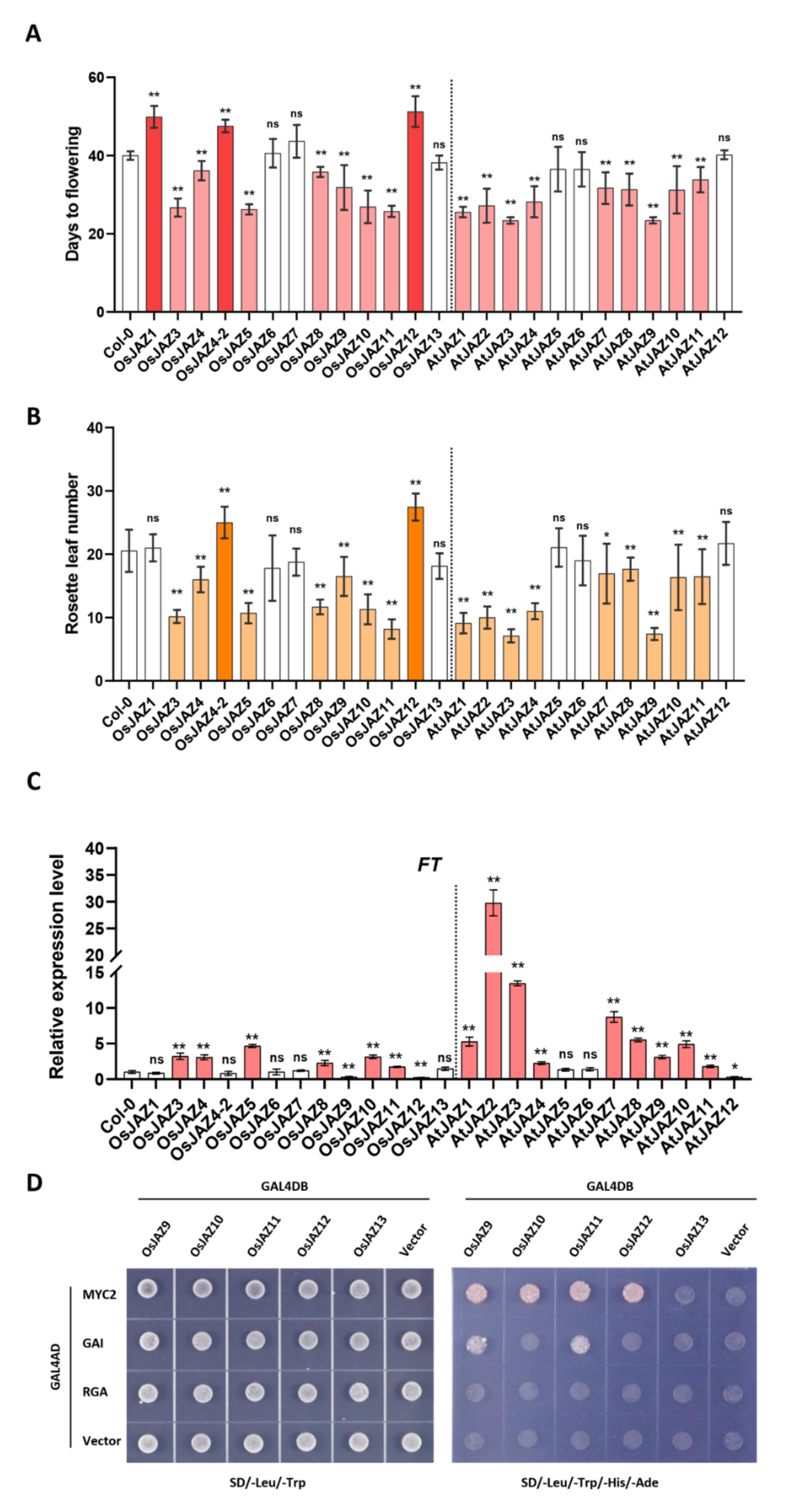
2.2. The Majority of OsJAZs Affect Flowering Time via Coordination with AtFT Expression
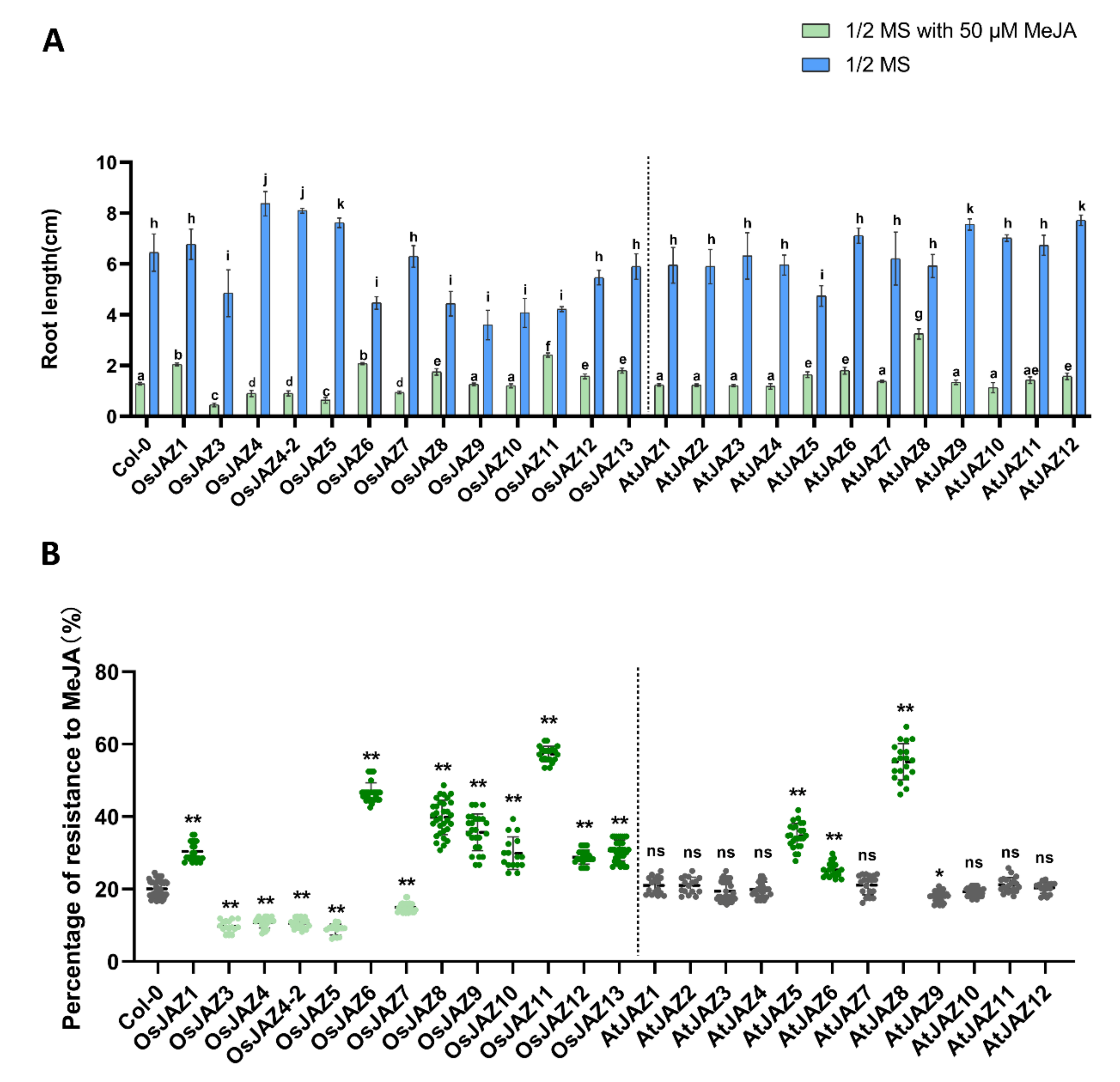
2.3. OsJAZs Regulate Root Growth in Arabidopsis
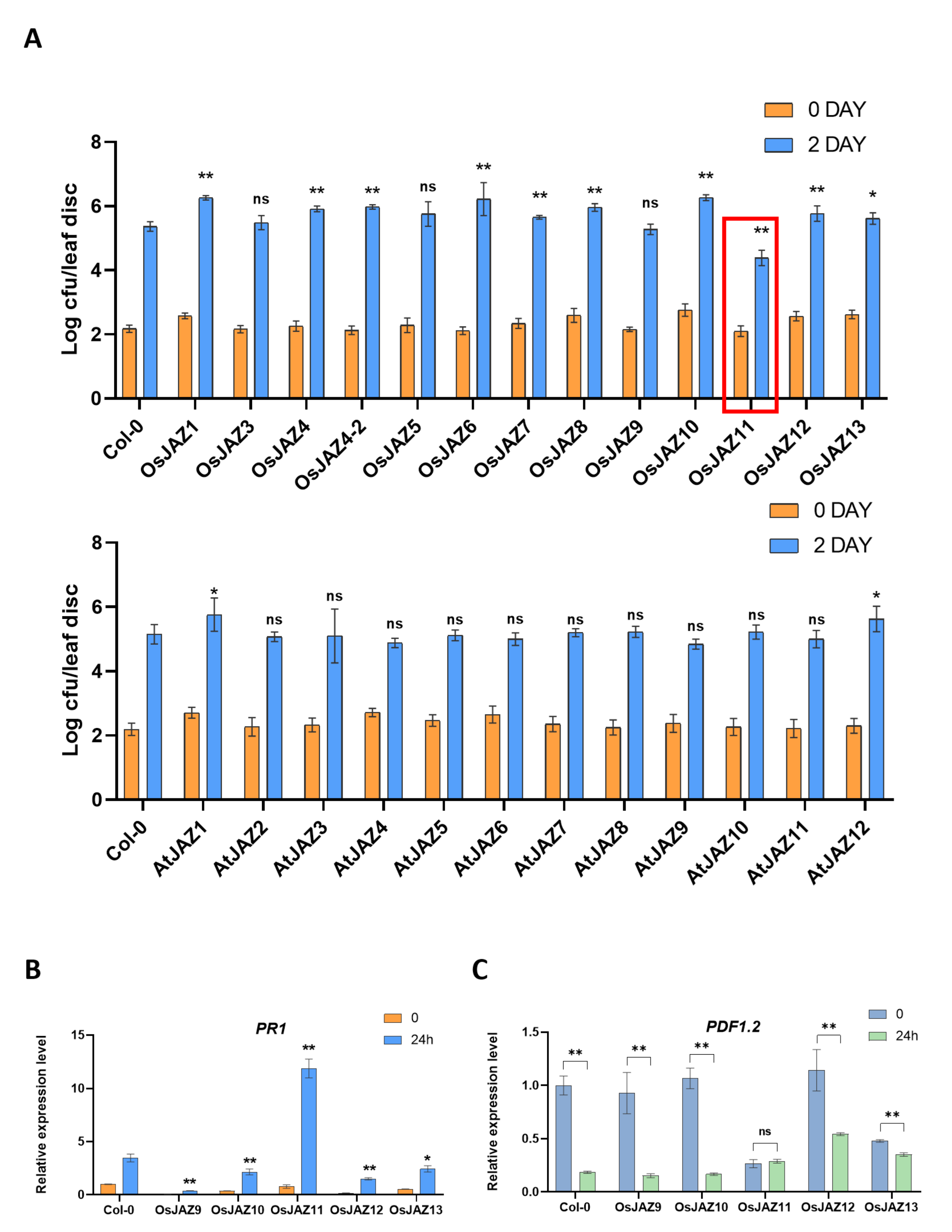
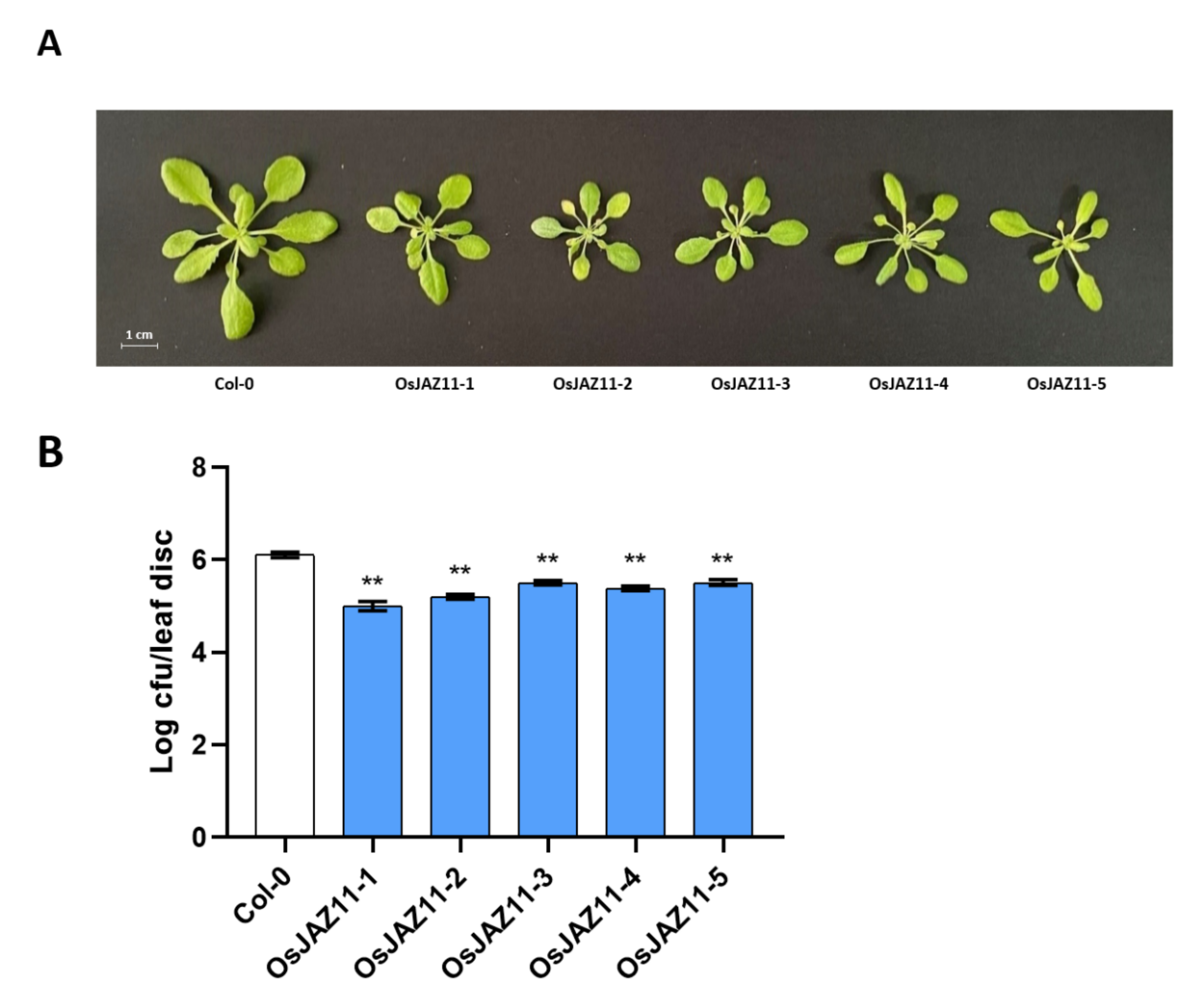
2.4. Various OsJAZs Are Involved in Plant Immunity, and Overexpression of Only OsJAZ11 Enhances Plant Resistance to Pst DC3000
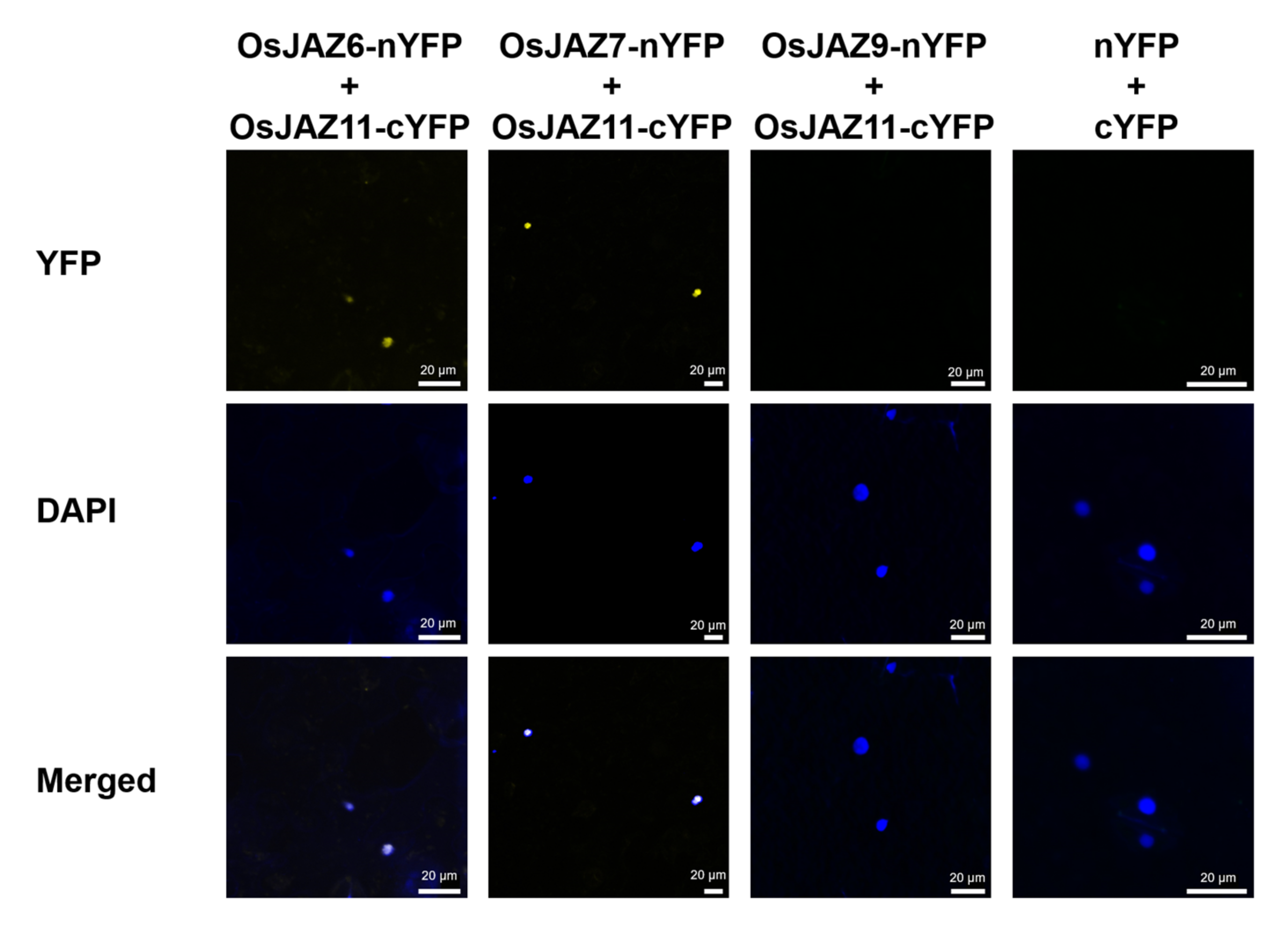
2.5. The Majority of OsJAZs Can Form Homodimers and Heterodimers
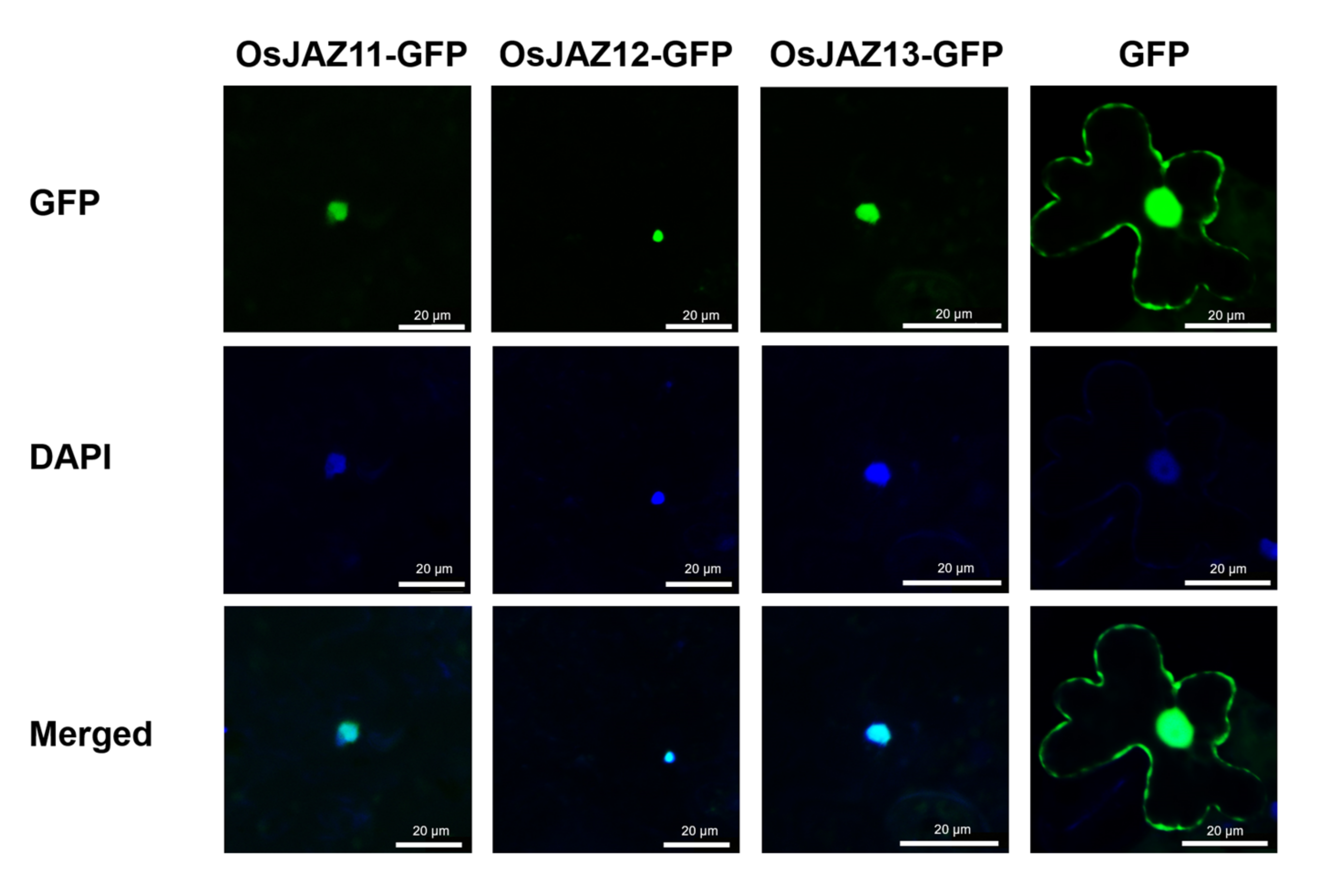
2.6. OsJAZ11 Functions in the Nucleus
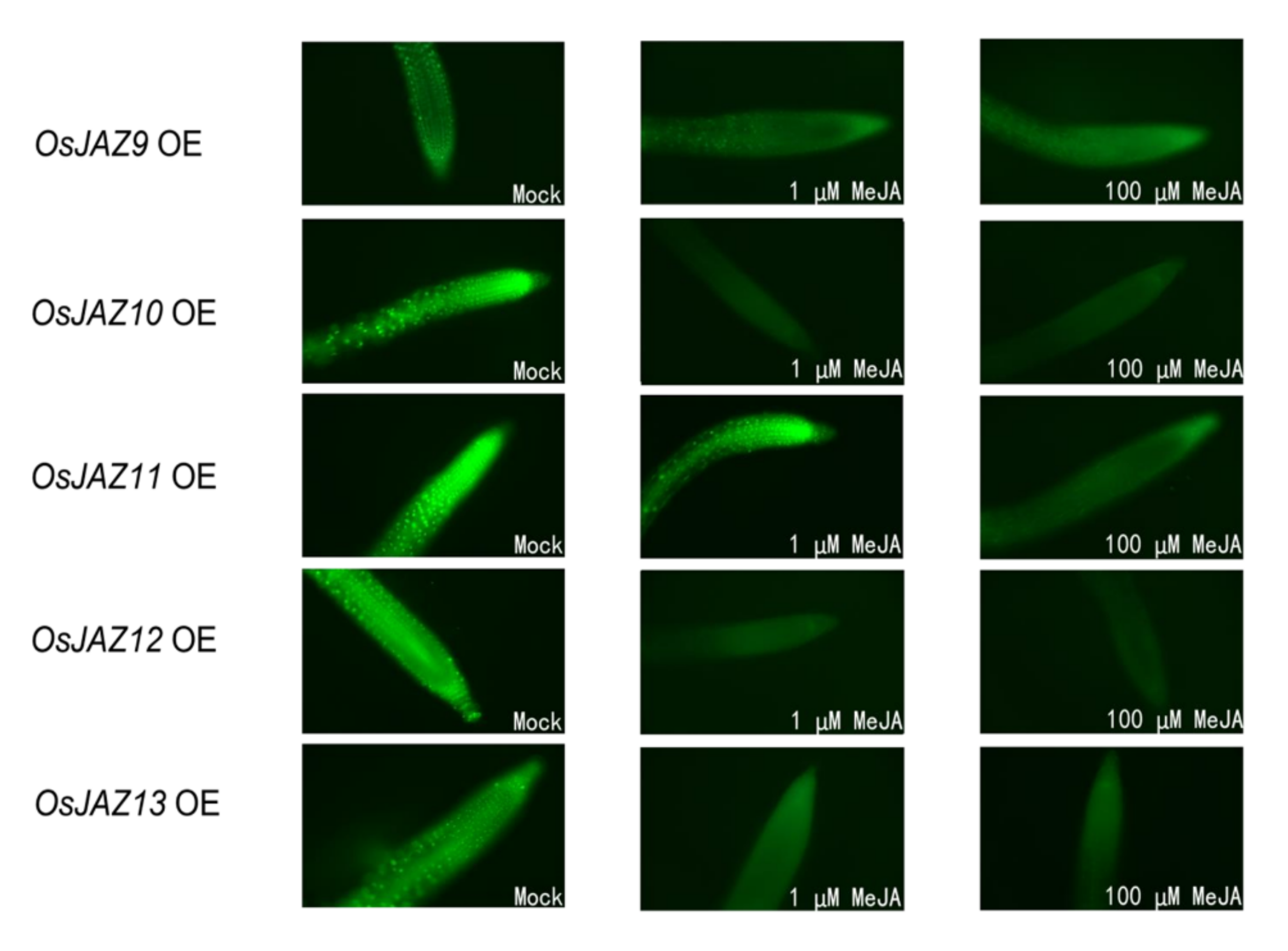
2.7. Different JAZ Proteins Show Different Sensitivity to JA
3. Discussion
4. Materials and Methods
4.1. Construction of Vectors
4.2. Plant Material and Growth Conditions
4.3. Construction of Transgenic Plants
4.4. Phylogenetic Analysis
4.5. Quantification of Flowering Time
4.6. Y2H Assays
4.7. JA Resistance Analysis
4.8. Pathogen Infection Assays
4.9. Gene Transcription Assays
4.10. BiFC Assays
4.11. Subcellular Protein Localization
4.12. In Vivo OsJAZ-GFP Degradation Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Qi, T.; Wasternack, C.; Xie, D. Jasmonate signaling and crosstalk with gibberellin and ethylene. Curr. Opin. Plant Biol. 2014, 21, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Zhang, D. Roles of jasmonate signalling in plant inflorescence and flower development. Curr. Opin. Plant Biol. 2015, 27, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, S.; Chini, A.; Hamberg, M.; Adie, B.; Porzel, A.; Kramell, R.; Miersch, O.; Wasternack, C.; Solano, R. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 2009, 5, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Wager, A.; Browse, J. Social Network: JAZ Protein Interactions Expand Our Knowledge of Jasmonate Signaling. Front. Plant Sci. 2012, 3, 41. [Google Scholar] [CrossRef] [PubMed]
- Vanholme, B.; Grunewald, W.; Bateman, A.; Kohchi, T.; Gheysen, G. The tify family previously known as ZIM. Trends Plant Sci. 2007, 12, 239–244. [Google Scholar] [CrossRef]
- Chung, H.S.; Howe, G.A. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 2009, 21, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Fonseca, S.; Chico, J.M.; Fernandez-Calvo, P.; Solano, R. The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 2009, 59, 77–87. [Google Scholar] [CrossRef]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Perez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E.; et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef]
- Chini, A.; Fonseca, S.; Fernandez, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; Garcia-Casado, G.; Lopez-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Melotto, M.; Mecey, C.; Niu, Y.; Chung, H.S.; Katsir, L.; Yao, J.; Zeng, W.; Thines, B.; Staswick, P.; Browse, J.; et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 2008, 55, 979–988. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Figueroa, P.; Browse, J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 2011, 62, 2143–2154. [Google Scholar] [CrossRef]
- Fernandez-Calvo, P.; Chini, A.; Fernandez-Barbero, G.; Chico, J.M.; Gimenez-Ibanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; Franco-Zorrilla, J.M.; et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 2011, 23, 701–715. [Google Scholar] [CrossRef]
- Katsir, L.; Chung, H.S.; Koo, A.J.; Howe, G.A. Jasmonate signaling: A conserved mechanism of hormone sensing. Curr. Opin. Plant Biol. 2008, 11, 428–435. [Google Scholar] [CrossRef]
- Ju, L.; Jing, Y.; Shi, P.; Liu, J.; Chen, J.; Yan, J.; Chu, J.; Chen, K.M.; Sun, J. JAZ proteins modulate seed germination through interaction with ABI5 in bread wheat and Arabidopsis. New Phytol. 2019, 223, 246–260. [Google Scholar] [CrossRef]
- Oh, Y.; Baldwin, I.T.; Galis, I. NaJAZh regulates a subset of defense responses against herbivores and spontaneous leaf necrosis in Nicotiana attenuata plants. Plant Physiol. 2012, 159, 769–788. [Google Scholar] [CrossRef]
- Yu, X.; Chen, G.; Tang, B.; Zhang, J.; Zhou, S.; Hu, Z. The Jasmonate ZIM-domain protein gene SlJAZ2 regulates plant morphology and accelerates flower initiation in Solanum lycopersicum plants. Plant Sci. 2018, 267, 65–73. [Google Scholar] [CrossRef]
- Zhu, D.; Bai, X.; Chen, C.; Chen, Q.; Cai, H.; Li, Y.; Ji, W.; Zhai, H.; Lv, D.; Luo, X.; et al. GsTIFY10, a novel positive regulator of plant tolerance to bicarbonate stress and a repressor of jasmonate signaling. Plant Mol. Biol. 2011, 77, 285–297. [Google Scholar] [CrossRef]
- Jing, Y.; Liu, J.; Liu, P.; Ming, D.; Sun, J. Overexpression of TaJAZ1 increases powdery mildew resistance through promoting reactive oxygen species accumulation in bread wheat. Sci. Rep. 2019, 9, 5691. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Yan, S.; Sun, C.; Li, S.; Li, J.; Xu, M.; Liu, X.; Zhang, S.; Zhao, Q.; Li, Y.; et al. A maize jasmonate Zim-domain protein, ZmJAZ14, associates with the JA, ABA, and GA signaling pathways in transgenic Arabidopsis. PLoS ONE 2015, 10, e0121824. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Gimenez-Ibanez, S.; Goossens, A.; Solano, R. Redundancy and specificity in jasmonate signalling. Curr. Opin. Plant Biol. 2016, 33, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Campos, M.L.; Yoshida, Y.; Major, I.T.; de Oliveira Ferreira, D.; Weraduwage, S.M.; Froehlich, J.E.; Johnson, B.F.; Kramer, D.M.; Jander, G.; Sharkey, T.D.; et al. Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat. Commun. 2016, 7, 12570. [Google Scholar] [CrossRef]
- Guo, Q.; Yoshida, Y.; Major, I.T.; Wang, K.; Sugimoto, K.; Kapali, G.; Havko, N.E.; Benning, C.; Howe, G.A. JAZ repressors of metabolic defense promote growth and reproductive fitness in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E10768–E10777. [Google Scholar] [CrossRef]
- Cai, Q.; Yuan, Z.; Chen, M.; Yin, C.; Luo, Z.; Zhao, X.; Liang, W.; Hu, J.; Zhang, D. Jasmonic acid regulates spikelet development in rice. Nat. Commun. 2014, 5, 3476. [Google Scholar] [CrossRef]
- Seo, J.S.; Joo, J.; Kim, M.J.; Kim, Y.K.; Nahm, B.H.; Song, S.I.; Cheong, J.J.; Lee, J.S.; Kim, J.K.; Choi, Y.D. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 2011, 65, 907–921. [Google Scholar] [CrossRef]
- Yamada, S.; Kano, A.; Tamaoki, D.; Miyamoto, A.; Shishido, H.; Miyoshi, S.; Taniguchi, S.; Akimitsu, K.; Gomi, K. Involvement of OsJAZ8 in jasmonate-induced resistance to bacterial blight in rice. Plant Cell Physiol. 2012, 53, 2060–2072. [Google Scholar] [CrossRef]
- Ye, H.; Du, H.; Tang, N.; Li, X.; Xiong, L. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol. Biol 2009, 71, 291–305. [Google Scholar] [CrossRef]
- Wu, H.; Ye, H.; Yao, R.; Zhang, T.; Xiong, L. OsJAZ9 acts as a transcriptional regulator in jasmonate signaling and modulates salt stress tolerance in rice. Plant Sci. 2015, 232, 1–12. [Google Scholar] [CrossRef]
- Feng, X.; Zhang, L.; Wei, X.; Zhou, Y.; Dai, Y.; Zhu, Z. OsJAZ13 Negatively Regulates Jasmonate Signaling and Activates Hypersensitive Cell Death Response in Rice. Int. J. Mol. Sci. 2020, 21, 4379. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.S.; Koo, A.J.; Gao, X.; Jayanty, S.; Thines, B.; Jones, A.D.; Howe, G.A. Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 2008, 146, 952–964. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.L.; Yao, J.; Mei, C.S.; Tong, X.H.; Zeng, L.J.; Li, Q.; Xiao, L.T.; Sun, T.P.; Li, J.; Deng, X.W.; et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 2012, 109, E1192–E1200. [Google Scholar] [CrossRef] [PubMed]
- Fukazawa, J.; Ohashi, Y.; Takahashi, R.; Nakai, K.; Takahashi, Y. DELLA degradation by gibberellin promotes flowering via GAF1-TPR-dependent repression of floral repressors in Arabidopsis. Plant Cell 2021, 33, 2258–2272. [Google Scholar] [CrossRef]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef] [PubMed]
- Turck, F.; Fornara, F.; Coupland, G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 2008, 59, 573–594. [Google Scholar] [CrossRef]
- Li, C.; Luo, L.; Fu, Q.; Niu, L.; Xu, Z.F. Isolation and functional characterization of JcFT, a FLOWERING LOCUS T (FT) homologous gene from the biofuel plant Jatropha curcas. BMC Plant Biol. 2014, 14, 125. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, Y.; Zhang, K.; Guo, D.; Cui, B.; Wang, X.; Huang, X. Promoting flowering, lateral shoot outgrowth, leaf development, and flower abscission in tobacco plants overexpressing cotton FLOWERING LOCUS T (FT)-like gene GhFT1. Front. Plant Sci. 2015, 6, 454. [Google Scholar] [CrossRef]
- Gao, X.; Walworth, A.E.; Mackie, C.; Song, G.Q. Overexpression of blueberry FLOWERING LOCUS T is associated with changes in the expression of phytohormone-related genes in blueberry plants. Hortic. Res. 2016, 3, 16053. [Google Scholar] [CrossRef]
- Pan, J.; Hu, Y.; Wang, H.; Guo, Q.; Chen, Y.; Howe, G.A.; Yu, D. Molecular Mechanism Underlying the Synergetic Effect of Jasmonate on Abscisic Acid Signaling during Seed Germination in Arabidopsis. Plant Cell 2020, 32, 3846–3865. [Google Scholar] [CrossRef]
- Staswick, P.E.; Su, W.; Howell, S.H. Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 1992, 89, 6837–6840. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Sonobe, K.; Ogawa, N.; Masuda, S.; Nagatani, A.; Kobayashi, Y.; Ohta, H. Inhibition of arabidopsis hypocotyl elongation by jasmonates is enhanced under red light in phytochrome B dependent manner. J. Plant Res. 2013, 126, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Ben-Romdhane, W.; Hassairi, A.; Aboul-Soud, M.A.M. Identification of TIFY/JAZ family genes in Solanum lycopersicum and their regulation in response to abiotic stresses. PLoS ONE 2017, 12, e0177381. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Araki, T.; Arteaga-Vazquez, M.A.; Berger, F.; Dolan, L.; Haseloff, J.; Ishizaki, K.; Kyozuka, J.; Lin, S.S.; Nagasaki, H.; et al. The Naming of Names: Guidelines for Gene Nomenclature in Marchantia. Plant Cell Physiol. 2016, 57, 257–261. [Google Scholar] [CrossRef] [PubMed]
- Ebel, C.; BenFeki, A.; Hanin, M.; Solano, R.; Chini, A. Characterization of wheat (Triticum aestivum) TIFY family and role of Triticum Durum TdTIFY11a in salt stress tolerance. PLoS ONE 2018, 13, e0200566. [Google Scholar] [CrossRef] [PubMed]
- Gimenez-Ibanez, S.; Boter, M.; Ortigosa, A.; Garcia-Casado, G.; Chini, A.; Lewsey, M.G.; Ecker, J.R.; Ntoukakis, V.; Solano, R. JAZ2 controls stomata dynamics during bacterial invasion. New Phytol. 2017, 213, 1378–1392. [Google Scholar] [CrossRef]
- Boter, M.; Golz, J.F.; Gimenez-Ibanez, S.; Fernandez-Barbero, G.; Franco-Zorrilla, J.M.; Solano, R. FILAMENTOUS FLOWER Is a Direct Target of JAZ3 and Modulates Responses to Jasmonate. Plant Cell 2015, 27, 3160–3174. [Google Scholar] [CrossRef]
- Oblessuc, P.R.; Obulareddy, N.; DeMott, L.; Matiolli, C.C.; Thompson, B.K.; Melotto, M. JAZ4 is involved in plant defense, growth, and development in Arabidopsis. Plant J. 2020, 101, 371–383. [Google Scholar] [CrossRef]
- Zhang, T.; Meng, L.; Kong, W.; Yin, Z.; Wang, Y.; Schneider, J.D.; Chen, S. Quantitative proteomics reveals a role of JAZ7 in plant defense response to Pseudomonas syringae DC3000. J. Proteom. 2018, 175, 114–126. [Google Scholar] [CrossRef]
- Jung, J.H.; Seo, Y.H.; Seo, P.J.; Reyes, J.L.; Yun, J.; Chua, N.H.; Park, C.M. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 2007, 19, 2736–2748. [Google Scholar] [CrossRef]
- Zhai, Q.; Zhang, X.; Wu, F.; Feng, H.; Deng, L.; Xu, L.; Zhang, M.; Wang, Q.; Li, C. Transcriptional Mechanism of Jasmonate Receptor COI1-Mediated Delay of Flowering Time in Arabidopsis. Plant Cell 2015, 27, 2814–2828. [Google Scholar] [CrossRef] [PubMed]
- Nakata, M.; Mitsuda, N.; Herde, M.; Koo, A.J.; Moreno, J.E.; Suzuki, K.; Howe, G.A.; Ohme-Takagi, M. A bHLH-type transcription factor, aba-inducible bhlh-type transcription factor/ja-associated myc2-like1, acts as a repressor to negatively regulate jasmonate signaling in arabidopsis. Plant Cell 2013, 25, 1641–1656. [Google Scholar] [CrossRef] [PubMed]
- Pouteau, S.; Ferret, V.; Gaudin, V.; Lefebvre, D.; Sabar, M.; Zhao, G.; Prunus, F. Extensive phenotypic variation in early flowering mutants of Arabidopsis. Plant Physiol. 2004, 135, 201–211. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wei, J.; Liu, D.; Liu, G.; Tang, J.; Chen, Y. Molecular Cloning, Characterization, and Expression of MiSOC1: A Homolog of the Flowering Gene suppressor of overexpression of constans1 from Mango (Mangifera indica L). Front. Plant Sci. 2016, 7, 1758. [Google Scholar] [CrossRef]
- Bostock, R.M. Signal crosstalk and induced resistance: Straddling the line between cost and benefit. Annu. Rev. Phytopathol. 2005, 43, 545–580. [Google Scholar] [CrossRef]
- Stout, M.J.; Thaler, J.S.; Thomma, B.P. Plant-mediated interactions between pathogenic microorganisms and herbivorous arthropods. Annu. Rev. Entomol. 2006, 51, 663–689. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef]
- Koornneef, A.; Leon-Reyes, A.; Ritsema, T.; Verhage, A.; Den Otter, F.C.; Van Loon, L.C.; Pieterse, C.M. Kinetics of salicylate-mediated suppression of jasmonate signaling reveal a role for redox modulation. Plant Physiol. 2008, 147, 1358–1368. [Google Scholar] [CrossRef]
- Brooks, D.M.; Bender, C.L.; Kunkel, B.N. The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in Arabidopsis thaliana. Mol. Plant Pathol. 2005, 6, 629–639. [Google Scholar] [CrossRef]
- Zheng, X.Y.; Spivey, N.W.; Zeng, W.; Liu, P.P.; Fu, Z.Q.; Klessig, D.F.; He, S.Y.; Dong, X. Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 2012, 11, 587–596. [Google Scholar] [CrossRef]
- Block, A.; Schmelz, E.; Jones, J.B.; Klee, H.J. Coronatine and salicylic acid: The battle between Arabidopsis and Pseudomonas for phytohormone control. Mol. Plant Pathol. 2005, 6, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Scala, A.; Mirabella, R.; Mugo, C.; Matsui, K.; Haring, M.A.; Schuurink, R.C. E-2-hexenal promotes susceptibility to Pseudomonas syringae by activating jasmonic acid pathways in Arabidopsis. Front. Plant Sci. 2013, 4, 74. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Deng, L.; Zhai, Q.; Zhao, J.; Chen, Q.; Li, C. Mediator Subunit MED25 Couples Alternative Splicing of JAZ Genes with Fine-Tuning of Jasmonate Signaling. Plant Cell 2020, 32, 429–448. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Liu, Q.; Xue, Q. Computational identification and phylogenetic analysis of the MAPK gene family in Oryza sativa. Plant Physiol. Biochem. 2007, 45, 6–14. [Google Scholar] [CrossRef]
- Chen, L.; Hu, W.; Tan, S.; Wang, M.; Ma, Z.; Zhou, S.; Deng, X.; Zhang, Y.; Huang, C.; Yang, G.; et al. Genome-wide identification and analysis of MAPK and MAPKK gene families in Brachypodium distachyon. PLoS ONE 2012, 7, e46744. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, L.; Xu, X.; Cai, C.; Guo, W. Genome-wide identification of mitogen-activated protein kinase gene family in Gossypium raimondii and the function of their corresponding orthologs in tetraploid cultivated cotton. BMC Plant Biol. 2014, 14, 345. [Google Scholar] [CrossRef]
- Jiang, M.; Wen, F.; Cao, J.; Li, P.; She, J.; Chu, Z. Genome-wide exploration of the molecular evolution and regulatory network of mitogen-activated protein kinase cascades upon multiple stresses in Brachypodium distachyon. BMC Genom. 2015, 16, 228. [Google Scholar] [CrossRef]
- Wang, J.; Pan, C.; Wang, Y.; Ye, L.; Wu, J.; Chen, L.; Zou, T.; Lu, G. Genome-wide identification of MAPK, MAPKK, and MAPKKK gene families and transcriptional profiling analysis during development and stress response in cucumber. BMC Genom. 2015, 16, 386. [Google Scholar] [CrossRef]
- Song, S.; Qi, T.; Huang, H.; Ren, Q.; Wu, D.; Chang, C.; Peng, W.; Liu, Y.; Peng, J.; Xie, D. The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell 2011, 23, 1000–1013. [Google Scholar] [CrossRef]
| A D | OsJAZ1 | OsJAZ3 | OsJAZ4 | OsJAZ4-2 | OsJAZ5 | OsJAZ6 | OsJAZ7 | OsJAZ8 | OsJAZ9 | OsJAZ10 | OsJAZ11 | OsJAZ12 | OsJAZ13 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B D | |||||||||||||
| OsJAZ1 | - | - | - | - | - | - | - | - | ++ | ++ | ++ | - | - |
| OsJAZ3 | - | - | - | - | - | - | - | - | - | ++ | - | - | - |
| OsJAZ4 | - | - | - | - | - | - | - | - | ++ | ++ | ++ | ++ | - |
| OsJAZ4-2 | - | ++ | ++ | ++ | ++ | ++ | ++ | - | ++ | ++ | ++ | ++ | - |
| OsJAZ5 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| OsJAZ6 | + | - | - | - | ++ | - | ++ | - | ++ | ++ | ++ | - | - |
| OsJAZ7 | - | - | - | + | ++ | - | ++ | + | ++ | ++ | ++ | ++ | - |
| OsJAZ8 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| OsJAZ9 | - | - | - | - | - | - | - | ++ | - | - | - | - | |
| OsJAZ10 | - | - | + | - | ++ | - | - | - | ++ | ++ | - | ++ | + |
| OsJAZ11 | ++ | - | - | ++ | ++ | ++ | ++ | - | - | ++ | ++ | - | + |
| OsJAZ12 | - | - | - | ++ | - | - | - | ++ | - | - | - | - | - |
| OsJAZ13 | - | ++ | ++ | ++ | - | - | ++ | ++ | - | + | ++ | - | - |
| Phylogenetic Group | Days to Flowering | MeJA Resistance | Bacterial Resistance | |
|---|---|---|---|---|
| OsJAZ8 | I | |||
| OsJAZ6 | I | |||
| OsJAZ7 | I | |||
| OsJAZ11 | I | |||
| OsJAZ12 | I | |||
| OsJAZ9 | I | |||
| OsJAZ10 | I | |||
| OsJAZ13 | I | |||
| OsJAZ1 | II | |||
| OsJAZ3 | II | |||
| OsJAZ4 | II | |||
| OsJAZ5 | II | |||
| AtJAZ10 | II | |||
| AtJAZ3 | II | |||
| AtJAZ4 | II | |||
| AtJAZ9 | II | |||
| AtJAZ11 | II | |||
| AtJAZ12 | II | |||
| AtJAZ1 | I | |||
| AtJAZ2 | I | |||
| AtJAZ5 | I | |||
| AtJAZ6 | I | |||
| AtJAZ7 | III | |||
| AtJAZ8 | III |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, B.; Shang, L.; Li, Y.; Zhang, Q.; Chu, Z.; He, S.; Yang, W.; Ding, X. Ectopic Expression of OsJAZs Alters Plant Defense and Development. Int. J. Mol. Sci. 2022, 23, 4581. https://doi.org/10.3390/ijms23094581
Sun B, Shang L, Li Y, Zhang Q, Chu Z, He S, Yang W, Ding X. Ectopic Expression of OsJAZs Alters Plant Defense and Development. International Journal of Molecular Sciences. 2022; 23(9):4581. https://doi.org/10.3390/ijms23094581
Chicago/Turabian StyleSun, Baolong, Luyue Shang, Yang Li, Qiang Zhang, Zhaohui Chu, Shengyang He, Wei Yang, and Xinhua Ding. 2022. "Ectopic Expression of OsJAZs Alters Plant Defense and Development" International Journal of Molecular Sciences 23, no. 9: 4581. https://doi.org/10.3390/ijms23094581
APA StyleSun, B., Shang, L., Li, Y., Zhang, Q., Chu, Z., He, S., Yang, W., & Ding, X. (2022). Ectopic Expression of OsJAZs Alters Plant Defense and Development. International Journal of Molecular Sciences, 23(9), 4581. https://doi.org/10.3390/ijms23094581







