Antisense Oligonucleotides Conjugated with Lipophilic Compounds: Synthesis and In Vitro Evaluation of Exon Skipping in Duchenne Muscular Dystrophy
Abstract
:1. Introduction
2. Results and Discussion
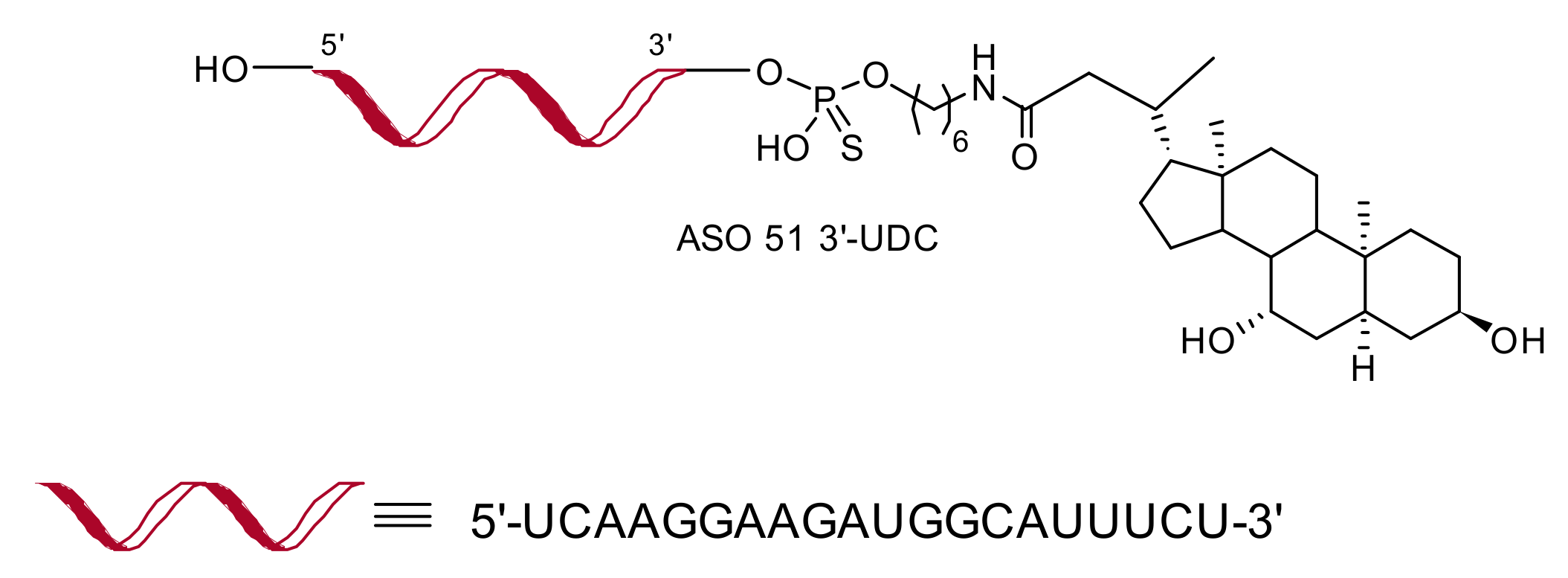
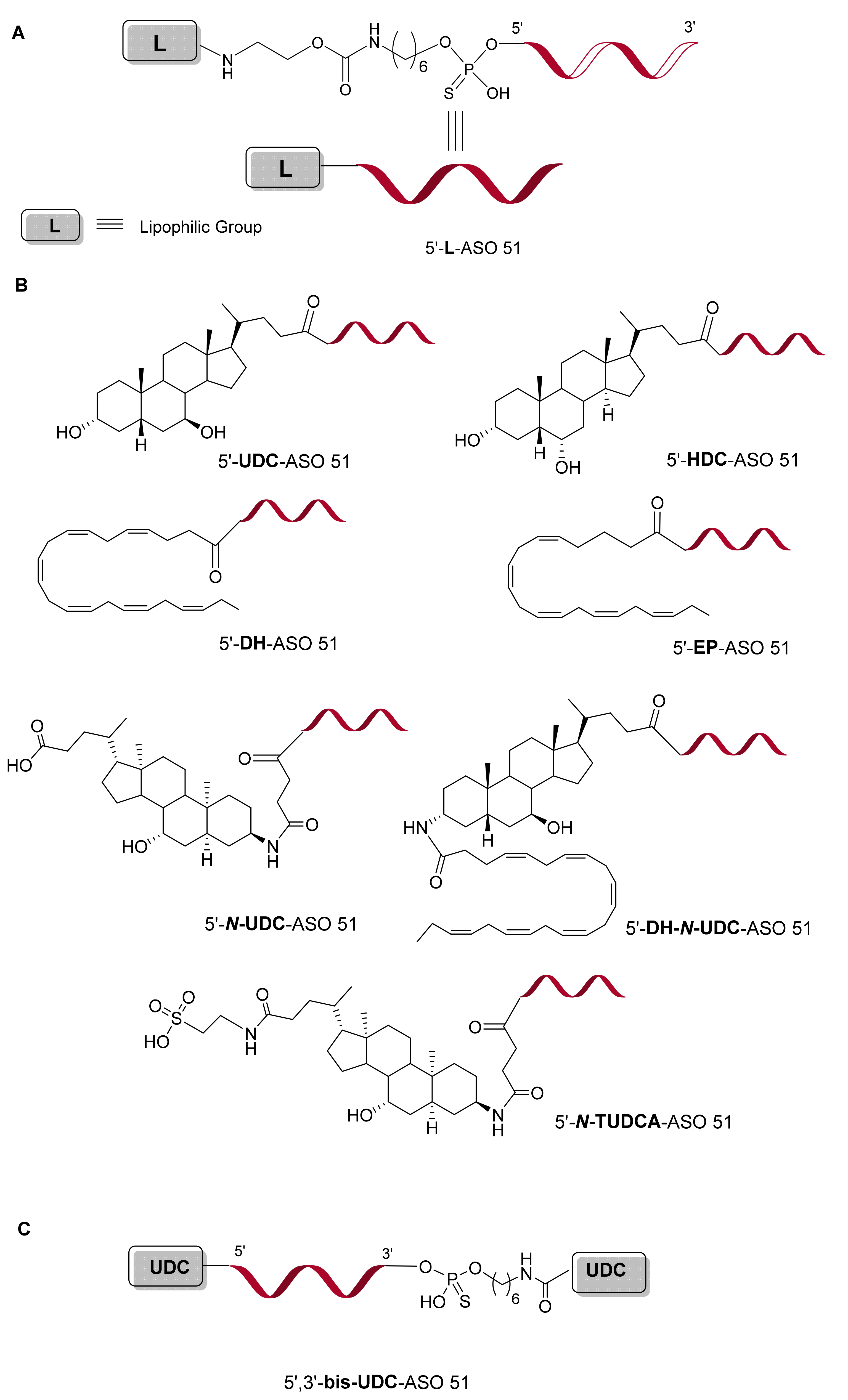
2.1. Design and Synthesis of ASO 51 Conjugated with Lipophilic Compounds
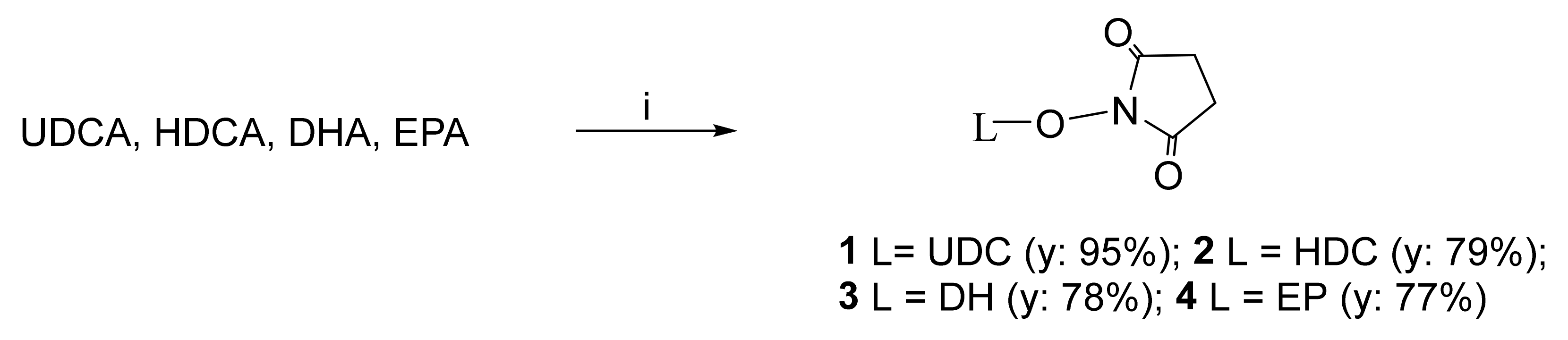
2.1.1. Conversion of UDCA, HDCA, DHA, and EPA into Their N-Hydroxysuccinimide Esters
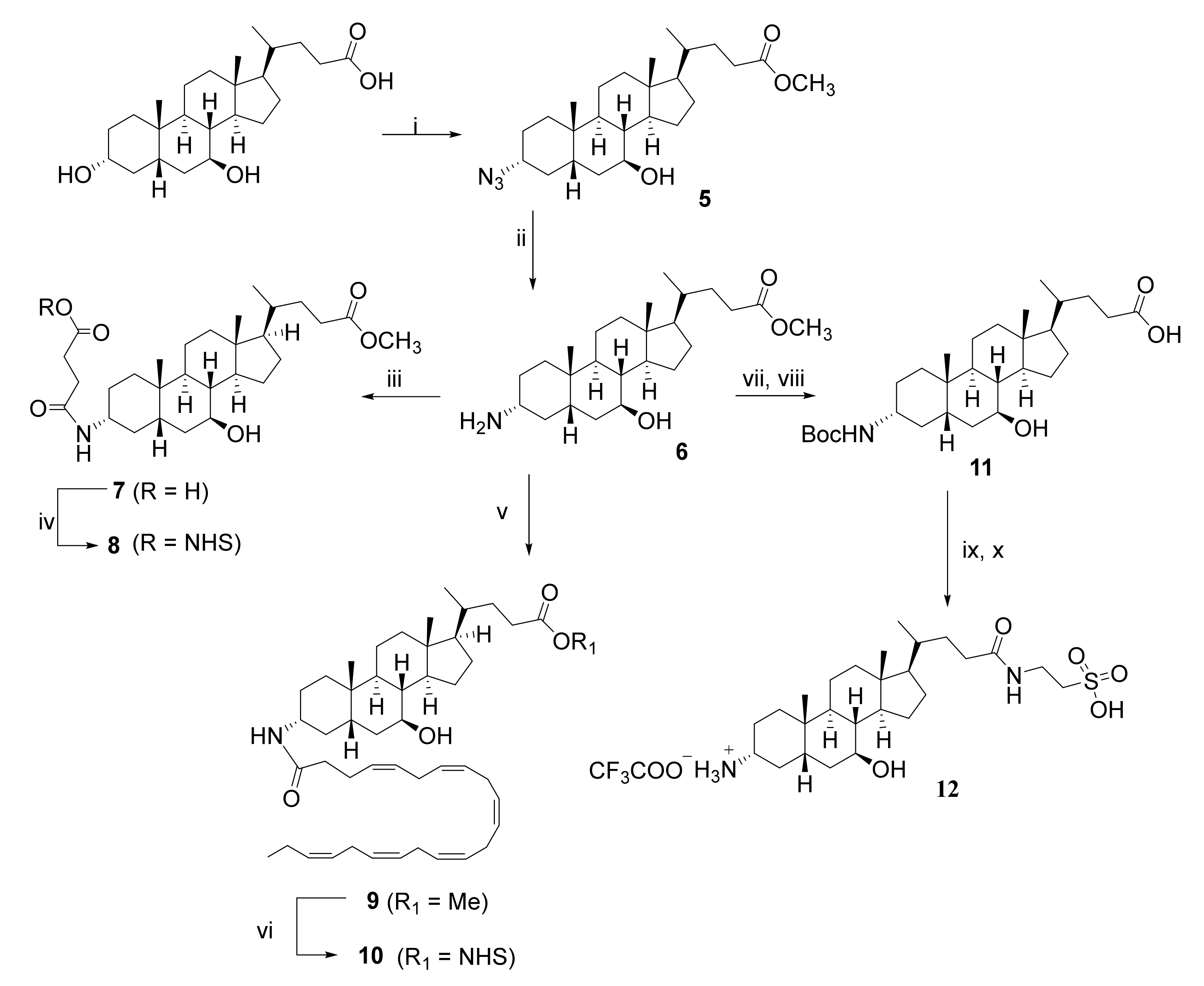
2.1.2. Preparation of NHS Esters 8 and 10, and of 3-α-Amino TUDCA 12 via the Common Intermediate 3-α-Amino UDCMe 6
2.1.3. Solid-Phase Synthesis of Lipophilic-Modified ASO 51
2.1.4. Synthesis of 5′-UDC-ASO Carrying Different Amounts of PS Linkages
2.2. Lipophilic-Modified ASO 51 Formed Stable Duplex with Their Complementary RNA
2.3. In Vitro Tests of ASO 51 Conjugated with Lipophilic Compounds
2.3.1. 5′-UDC Conjugation Further Increases Potency of ASO 51 Compared to 3′-UDC
2.3.2. Evaluation of the Variable PS Content in the 5′-UDC-ASO 51
2.3.3. Evaluation of Variable Lipophilic Compounds Conjugated to ASO 51
2.3.4. Gymnotic Delivery of Selected Conjugated ASO 51
2.4. Lipophilic-Modified ASO 51 Spontaneously Formed Nanoparticle Aggregates in Acqueous Media
3. Materials and Methods
3.1. Chemistry
3.1.1. General Procedure for the Synthesis of NHS-Esters
Characterization of NHS-Ester of HDCA 2
Characterization of NHS-Ester of DHA 3
Characterization of NHS-Ester of EPA 4
3.1.2. Synthesis of 3-α-Amino UDCMe 6
3.1.3. Synthesis of NHS Ester 8
3.1.4. Synthesis of Ester 9
3.1.5. Synthesis of NHS-Ester 10
3.1.6. Synthesis of 3-α-N-BOC UDCA 11
3.1.7. Synthesis of 3-α-Amino TUDCA 12
3.1.8. General Procedure for Solid Phase Synthesis of Lipophilic-Modified ASO Targeting Human DMD Exon 51
3.1.9. General Procedure for Oligonucleotide Purifications
3.1.10. UV Melting Studies
3.1.11. Photon Correlation Spectroscopy (PCS)
3.1.12. Transmission Electron Microscopy (TEM) Studies
3.2. Exon Skipping Studies
Immunofluorescence Analysis Dystrophin Expression in Myotubes Treated with ASO
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahn, A.H.; Kunkel, L.M. The structural and functional diversity of dystrophin. Nat. Genet. 1993, 3, 283–291. [Google Scholar] [CrossRef]
- Melacini, P.; Vianello, A.; Villanova, C.; Fanin, M.; Miorin, M.; Angelini, C.; Dalla Volta, S. Cardiac and respiratory involvement in advanced stage Duchenne muscular dystrophy. Neuromuscul. Disord. 1996, 6, 367–376. [Google Scholar] [CrossRef]
- Verhaart, I.E.C.; Aartsma-Rus, A. Therapeutic developments for Duchenne muscular dystrophy. Nat. Rev. Neurol. 2019, 15, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Shen, L.; Zhang, Z.; Xie, X. Therapeutic strategies for duchenne muscular dystrophy: An update. Genes 2020, 11, 837. [Google Scholar] [CrossRef] [PubMed]
- Hanson, B.; Wood, M.J.A.; Roberts, T.C. Molecular correction of Duchenne muscular dystrophy by splice modulation and gene editing. RNA Biol. 2021, 18, 1048–1062. [Google Scholar] [CrossRef]
- Goemans, N.M.; Tulinius, M.; van den Hauwe, M.; Kroksmark, A.K.; Buyse, G.; Wilson, R.J.; van Deutekom, J.C.; de Kimpe, S.J.; Lourbakos, A.; Campion, G. Long-term efficacy, safety, and pharmacokinetics of drisapersen in duchenne muscular dystrophy: Results from an open-label extension study. PLoS ONE 2016, 11, e0161955. [Google Scholar] [CrossRef] [PubMed]
- Goyenvalle, A.; Griffith, G.; Babbs, A.; El Andaloussi, S.; Ezzat, K.; Avril, A.; Dugovic, B.; Chaussenot, R.; Ferry, A.; Voit, T.; et al. Functional correction in mouse models of muscular dystrophy using exon-skipping tricyclo-DNA oligomers. Nat. Med. 2015, 21, 270–275. [Google Scholar] [CrossRef]
- Aartsma-Rus, A.; Kaman, W.E.; Bremmer-Bout, M.; Janson, A.A.M.; den Dunnen, J.T.; van Ommen, G.J.B.; van Deutekom, J.C.T. Comparative analysis of antisense oligonucleotide analogs for targeted DMD exon 46 skipping in muscle cells. Gene Ther. 2004, 11, 1391–1398. [Google Scholar] [CrossRef] [Green Version]
- Yin, H.; Lu, Q.; Wood, M. Effective exon skipping and restoration of dystrophin expression by peptide nucleic acid antisense oligonucleotides in mdx mice. Mol. Ther. 2008, 16, 38–45. [Google Scholar] [CrossRef]
- Ito, K.; Takakusa, H.; Kakuta, M.; Kanda, A.; Takagi, N.; Nagase, H.; Watanabe, N.; Asano, D.; Goda, R.; Masuda, T.; et al. Renadirsen, a novel 2′omerna/ena® chimera antisense oligonucleotide, induces robust exon 45 skipping for dystrophin in vivo. Curr. Issues Mol. Biol. 2021, 43, 90. [Google Scholar] [CrossRef]
- Kandasamy, P.; McClorey, G.; Shimizu, M.; Kothari, N.; Alam, R.; Iwamoto, N.; Kumarasamy, J.; Bommineni, G.R.; Bezigian, A.; Chivatakarn, O.; et al. Control of backbone chemistry and chirality boost oligonucleotide splice switching activity. Nucleic Acids Res. 2022. [Google Scholar] [CrossRef] [PubMed]
- Hoy, S.M. Nusinersen: First Global Approval. Drugs 2017, 77, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Hu, C.; Moufawad El Achkar, C.; Black, L.E.; Douville, J.; Larson, A.; Pendergast, M.K.; Goldkind, S.F.; Lee, E.A.; Kuniholm, A.; et al. Patient-Customized Oligonucleotide Therapy for a Rare Genetic Disease. N. Engl. J. Med. 2019, 381, 1644–1652. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Zhao, J.; Pearson, Z.J.; Boskovic, Z.V.; Wang, J. RNA-Targeting Splicing Modifiers: Drug Development and Screening Assays. Molecules 2021, 26, 2263. [Google Scholar] [CrossRef]
- Hammond, S.M.; Aartsma-Rus, A.; Alves, S.; Borgos, S.E.; Buijsen, R.A.M.; Collin, R.W.J.; Covello, G.; Denti, M.A.; Desviat, L.R.; Echevarría, L.; et al. Delivery of oligonucleotide-based therapeutics: Challenges and opportunities. EMBO Mol. Med. 2021, 13, e13243. [Google Scholar] [CrossRef]
- Godfrey, C.; Desviat, L.R.; Smedsrød, B.; Piétri-Rouxel, F.; Denti, M.A.; Disterer, P.; Lorain, S.; Nogales-Gadea, G.; Sardone, V.; Anwar, R.; et al. Delivery is key: Lessons learnt from developing splice-switching antisense therapies. EMBO Mol. Med. 2017, 9, 545–557. [Google Scholar] [CrossRef]
- Juliano, R.L. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016, 44, 6518–6548. [Google Scholar] [CrossRef]
- Relizani, K.; Goyenvalle, A. Use of Tricyclo-DNA Antisense Oligonucleotides for Exon Skipping. Methods Mol. Biol. 2018, 1828, 381–394. [Google Scholar] [CrossRef]
- Goyenvalle, A.; Leumann, C.; Garcia, L. Therapeutic Potential of Tricyclo-DNA antisense oligonucleotides. J. Neuromuscul. Dis. 2016, 3, 157–167. [Google Scholar] [CrossRef] [Green Version]
- Relizani, K.; Griffith, G.; Echevarría, L.; Zarrouki, F.; Facchinetti, P.; Vaillend, C.; Leumann, C.; Garcia, L.; Goyenvalle, A. Efficacy and Safety Profile of Tricyclo-DNA Antisense Oligonucleotides in Duchenne Muscular Dystrophy Mouse Model. Mol. Ther. Nucleic Acids 2017, 8, 144. [Google Scholar] [CrossRef] [Green Version]
- Roberts, T.C.; Langer, R.; Wood, M.J.A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020, 19, 673–694. [Google Scholar] [CrossRef] [PubMed]
- Sarepta Therapeutics Reports Positive Clinical Results from Phase 2 MOMENTUM Study of SRP-5051 in Patients with Duchenne Muscular Dystrophy Amenable to Skipping Exon 51|Sarepta Therapeutics, Inc. Available online: https://investorrelations.sarepta.com/news-releases/news-release-details/sarepta-therapeutics-reports-positive-clinical-results-phase-2 (accessed on 9 March 2022).
- Relizani, K.; Echevarría, L.; Zarrouki, F.; Gastaldi, C.; Dambrune, C.; Aupy, P.; Haeberli, A.; Komisarski, M.; Tensorer, T.; Larcher, T.; et al. Palmitic acid conjugation enhances potency of tricyclo-DNA splice switching oligonucleotides. Nucleic Acids Res. 2022, 50, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Østergaard, M.E.; Jackson, M.; Low, A.; Chappell, A.E.; Lee, R.G.; Peralta, R.Q.; Yu, J.; Kinberger, G.A.; Dan, A.; Carty, R.; et al. Conjugation of hydrophobic moieties enhances potency of antisense oligonucleotides in the muscle of rodents and non-human primates. Nucleic Acids Res. 2019, 47, 6045–6058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seth, P.P.; Tanowitz, M.; Frank Bennett, C. Selective tissue targeting of synthetic nucleic acid drugs. J. Clin. Investig. 2019, 129, 915–925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Allen, N.; Prakash, T.P.; Liang, X.H.; Crooke, S.T. Lipid Conjugates Enhance Endosomal Release of Antisense Oligonucleotides into Cells. Nucleic Acid Ther. 2019, 29, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, E.; Bovolenta, M.; Preti, L.; Capobianco, M.L.; Mamchaoui, K.; Bertoldo, M.; Perrone, D. Synthesis and exon-skipping properties of a 3′-ursodeoxycholic acid-conjugated oligonucleotide targeting dmd pre-mrna: Pre-synthetic versus post- synthetic approach. Molecules 2021, 26, 7662. [Google Scholar] [CrossRef] [PubMed]
- Pascual-Morena, C.; Cavero-Redondo, I.; Álvarez-Bueno, C.; Mesas, A.E.; Pozuelo-Carrascosa, D.; Martínez-Vizcaíno, V. Restorative treatments of dystrophin expression in Duchenne muscular dystrophy: A systematic review. Ann. Clin. Transl. Neurol. 2020, 7, 1738–1752. [Google Scholar] [CrossRef] [PubMed]
- Van Deutekom, J.C.; Janson, A.A.; Ginjaar, I.B.; Frankhuizen, W.S.; Aartsma-Rus, A.; Bremmer-Bout, M.; den Dunnen, J.T.; Koop, K.; van der Kooi, A.J.; Goemans, N.M.; et al. Local Dystrophin Restoration with Antisense Oligonucleotide PRO051. N. Engl. J. Med. 2007, 357, 2677–2686. [Google Scholar] [CrossRef] [Green Version]
- Pavlović, N.; Goločorbin-Kon, S.; Ðanić, M.; Stanimirov, B.; Al-Salami, H.; Stankov, K.; Mikov, M. Bile Acids and Their Derivatives as Potential Modifiers of Drug Release and Pharmacokinetic Profiles. Front. Pharmacol. 2018, 9, 1283. [Google Scholar] [CrossRef]
- Goossens, J.F.; Bailly, C. Ursodeoxycholic acid and cancer: From chemoprevention to chemotherapy. Pharmacol. Ther. 2019, 203, 107396. [Google Scholar] [CrossRef]
- Fiorucci, S.; Distrutti, E. The pharmacology of bile acids and their receptors. In Handbook of Experimental Pharmacology; Springer: Cham, Swizterland, 2019; Volume 256, pp. 3–18. [Google Scholar]
- Virtanen, E.; Kolehmainen, E. Use of bile acids in pharmacological and supramolecular applications. Eur. J. Org. Chem. 2004, 2004, 3385–3399. [Google Scholar] [CrossRef]
- Stillwell, W.; Wassall, S.R. Docosahexaenoic acid: Membrane properties of a unique fatty acid. Chem. Phys. Lipids 2003, 126, 1–27. [Google Scholar] [CrossRef]
- Nikan, M.; Osborn, M.F.; Coles, A.H.; Godinho, B.M.; Hall, L.M.; Haraszti, R.A.; Hassler, M.R.; Echeverria, D.; Aronin, N.; Khvorova, A. Docosahexaenoic Acid Conjugation Enhances Distribution and Safety of siRNA upon Local Administration in Mouse Brain. Mol. Ther. Nucleic Acids 2016, 5, e344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Schacky, C. The role of omega-3 fatty acids in cardiovascular disease. Curr. Atheroscler. Rep. 2003, 5, 139–145. [Google Scholar] [CrossRef]
- Kondrackiene, J.; Beuers, U.; Kupcinskas, L. Efficacy and safety of ursodeoxycholic acid versus cholestyramine in intrahepatic cholestasis of pregnancy. Gastroenterology 2005, 129, 894–901. [Google Scholar] [CrossRef]
- Yuen, L.H.; Franzini, R.M. Stability of Oligonucleotide-Small Molecule Conjugates to DNA-Deprotection Conditions. Bioconjug. Chem. 2017, 28, 1076–1083. [Google Scholar] [CrossRef]
- Li, Y.; Gabriele, E.; Samain, F.; Favalli, N.; Sladojevich, F.; Scheuermann, J.; Neri, D. Optimized Reaction Conditions for Amide Bond Formation in DNA-Encoded Combinatorial Libraries. ACS Comb. Sci. 2016, 18, 438–443. [Google Scholar] [CrossRef] [Green Version]
- Franzini, R.M.; Samain, F.; Abd Elrahman, M.; Mikutis, G.; Nauer, A.; Zimmermann, M.; Scheuermann, J.; Hall, J.; Neri, D. Systematic evaluation and optimization of modification reactions of oligonucleotides with amines and carboxylic acids for the synthesis of dna-encoded chemical libraries. Bioconjug. Chem. 2014, 25, 1453–1461. [Google Scholar] [CrossRef]
- Massarenti, C.; Bortolini, O.; Fantin, G.; Cristofaro, D.; Ragno, D.; Perrone, D.; Marchesi, E.; Toniolo, G.; Massi, A. Fluorous-tag assisted synthesis of bile acid-bisphosphonate conjugates: Via orthogonal click reactions: An access to potential anti-resorption bone drugs. Org. Biomol. Chem. 2017, 15, 4907–4920. [Google Scholar] [CrossRef]
- Kojima, N.; Takebayashi, T.; Mikami, A.; Ohtsuka, E.; Komatsu, Y. Efficient synthesis of oligonucleotide conjugates on solid-support using an (aminoethoxycarbonyl)aminohexyl group for 5′-terminal modification. Bioorganic Med. Chem. Lett. 2009, 19, 2144–2147. [Google Scholar] [CrossRef]
- Echevarría, L.; Aupy, P.; Relizani, K.; Bestetti, T.; Griffith, G.; Blandel, F.; Komisarski, M.; Haeberli, A.; Svinartchouk, F.; Garcia, L.; et al. Evaluating the Impact of Variable Phosphorothioate Content in Tricyclo-DNA Antisense Oligonucleotides in a Duchenne Muscular Dystrophy Mouse Model. Nucleic Acid Ther. 2019, 29, 148–160. [Google Scholar] [CrossRef] [PubMed]
- Ezzat, K.; Aoki, Y.; Koo, T.; McClorey, G.; Benner, L.; Coenen-Stass, A.; O’Donovan, L.; Lehto, T.; Garcia-Guerra, A.; Nordin, J.; et al. Self-Assembly into Nanoparticles Is Essential for Receptor Mediated Uptake of Therapeutic Antisense Oligonucleotides. Nano Lett. 2015, 15, 4364–4373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riccardi, C.; Musumeci, D.; Russo Krauss, I.; Piccolo, M.; Irace, C.; Paduano, L.; Montesarchio, D. Exploring the conformational behaviour and aggregation properties of lipid-conjugated AS1411 aptamers. Int. J. Biol. Macromol. 2018, 118, 1384–1399. [Google Scholar] [CrossRef] [PubMed]
- Pecora, R. Dynamic light scattering measurement of nanometer particles in liquids. J. Nanoparticle Res. 2000, 2, 123–131. [Google Scholar] [CrossRef]



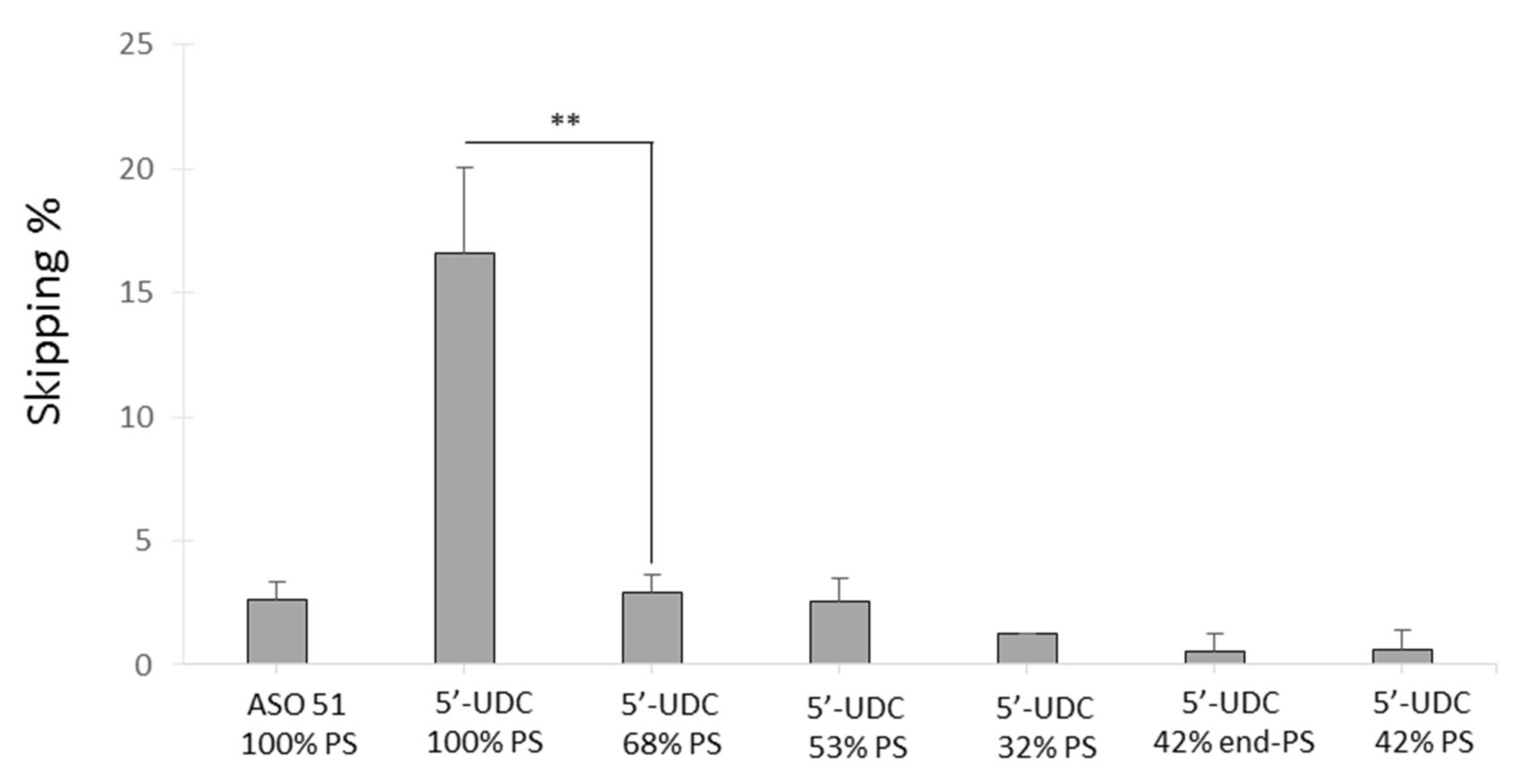
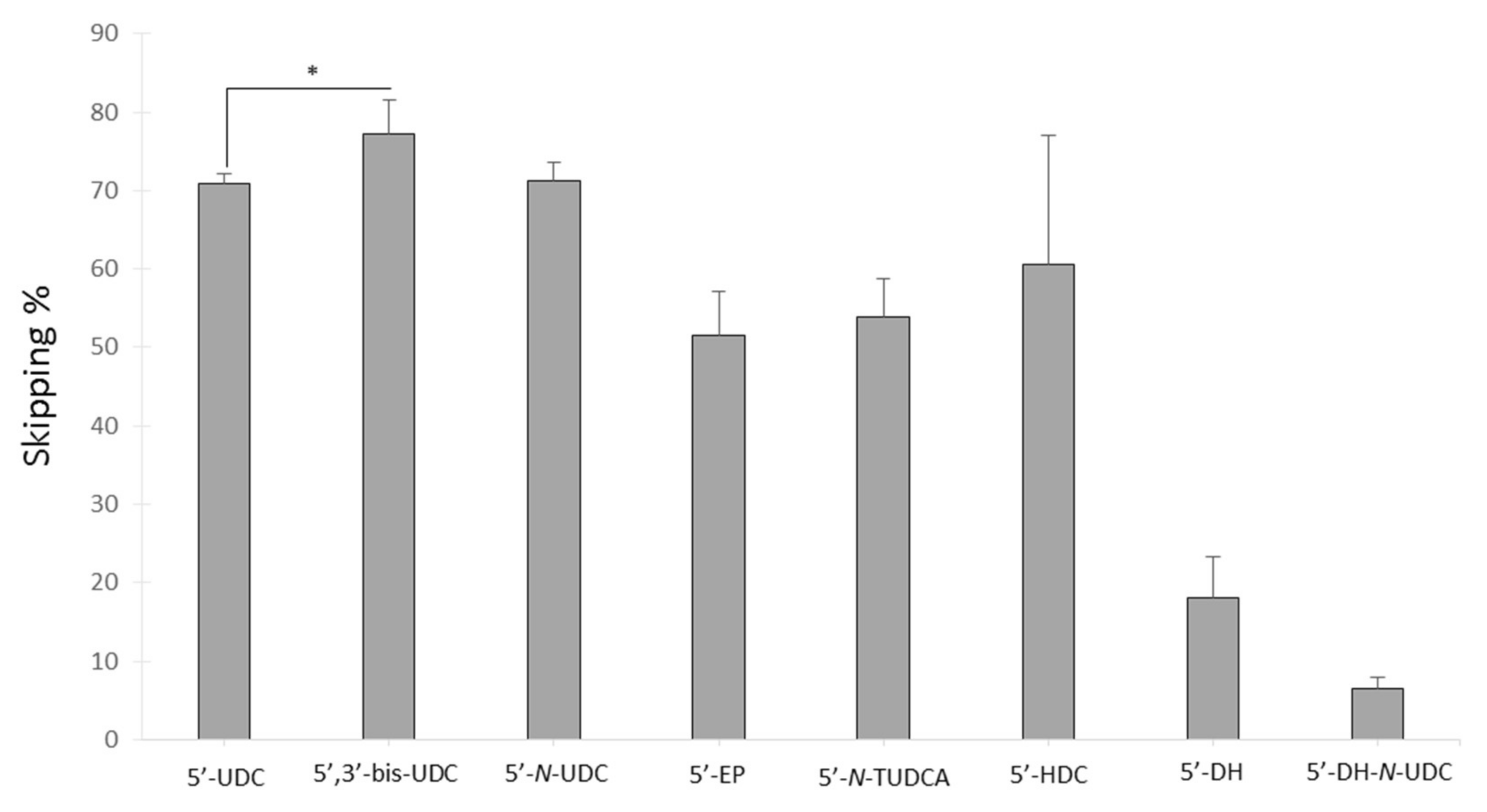


| Name | Sequence 5′-3′ |
|---|---|
| 5′-UDC-ASO 51 | U*C*A*A*G*G*A*A*G*A*U*G*G*C*A*U*U*U*C*U |
| 5′-UDC-ASO 51–68% PS | U*C*A*AG*GA*AG*AU*GG*CA*U*U*U*C*U |
| 5′-UDC-ASO 51–53% PS | U*CA*AG*GA*AG*AU*GG*CA*UU*U*C*U |
| 5′-UDC-ASO 51–42% end-PS | U*C*A*A*GGAAGAUGGCAU*U*U*C*U |
| 5′-UDC-ASO 51–42% PS | U*C*AAG*GAA*GAU*GGC*AUUU*C*U |
| 5′-UDC-ASO 51–32% PS | U*C*A*AGGAAGAUGGCAUU*U*C*U |
| Name | Tm * |
|---|---|
| ASO 51 | 71.02 |
| 5′-UDC-ASO 51 | 70.25 |
| 5′,3′-bis-UDC-ASO 51 | 72.02 |
| 5′-HDC-ASO 51 | 69.02 |
| 5′-DH-ASO 51 | 69.04 |
| 5′-EP-ASO 51 | 69.47 |
| 5′-N-UDC-ASO 51 | 69.02 |
| 5′-N-TUDCA-ASO 51 | 69.02 |
| 5′-DH-N-UDC-ASO 51 | 67.50 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marchesi, E.; Cortesi, R.; Preti, L.; Rimessi, P.; Sguizzato, M.; Bovolenta, M.; Perrone, D. Antisense Oligonucleotides Conjugated with Lipophilic Compounds: Synthesis and In Vitro Evaluation of Exon Skipping in Duchenne Muscular Dystrophy. Int. J. Mol. Sci. 2022, 23, 4270. https://doi.org/10.3390/ijms23084270
Marchesi E, Cortesi R, Preti L, Rimessi P, Sguizzato M, Bovolenta M, Perrone D. Antisense Oligonucleotides Conjugated with Lipophilic Compounds: Synthesis and In Vitro Evaluation of Exon Skipping in Duchenne Muscular Dystrophy. International Journal of Molecular Sciences. 2022; 23(8):4270. https://doi.org/10.3390/ijms23084270
Chicago/Turabian StyleMarchesi, Elena, Rita Cortesi, Lorenzo Preti, Paola Rimessi, Maddalena Sguizzato, Matteo Bovolenta, and Daniela Perrone. 2022. "Antisense Oligonucleotides Conjugated with Lipophilic Compounds: Synthesis and In Vitro Evaluation of Exon Skipping in Duchenne Muscular Dystrophy" International Journal of Molecular Sciences 23, no. 8: 4270. https://doi.org/10.3390/ijms23084270
APA StyleMarchesi, E., Cortesi, R., Preti, L., Rimessi, P., Sguizzato, M., Bovolenta, M., & Perrone, D. (2022). Antisense Oligonucleotides Conjugated with Lipophilic Compounds: Synthesis and In Vitro Evaluation of Exon Skipping in Duchenne Muscular Dystrophy. International Journal of Molecular Sciences, 23(8), 4270. https://doi.org/10.3390/ijms23084270











