Development and Evolution of DNA-Dependent Protein Kinase Inhibitors toward Cancer Therapy
Abstract
1. Introduction
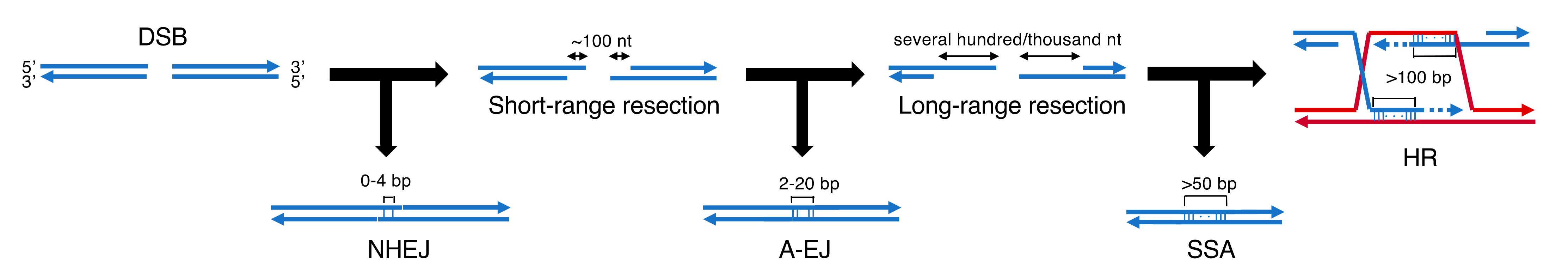
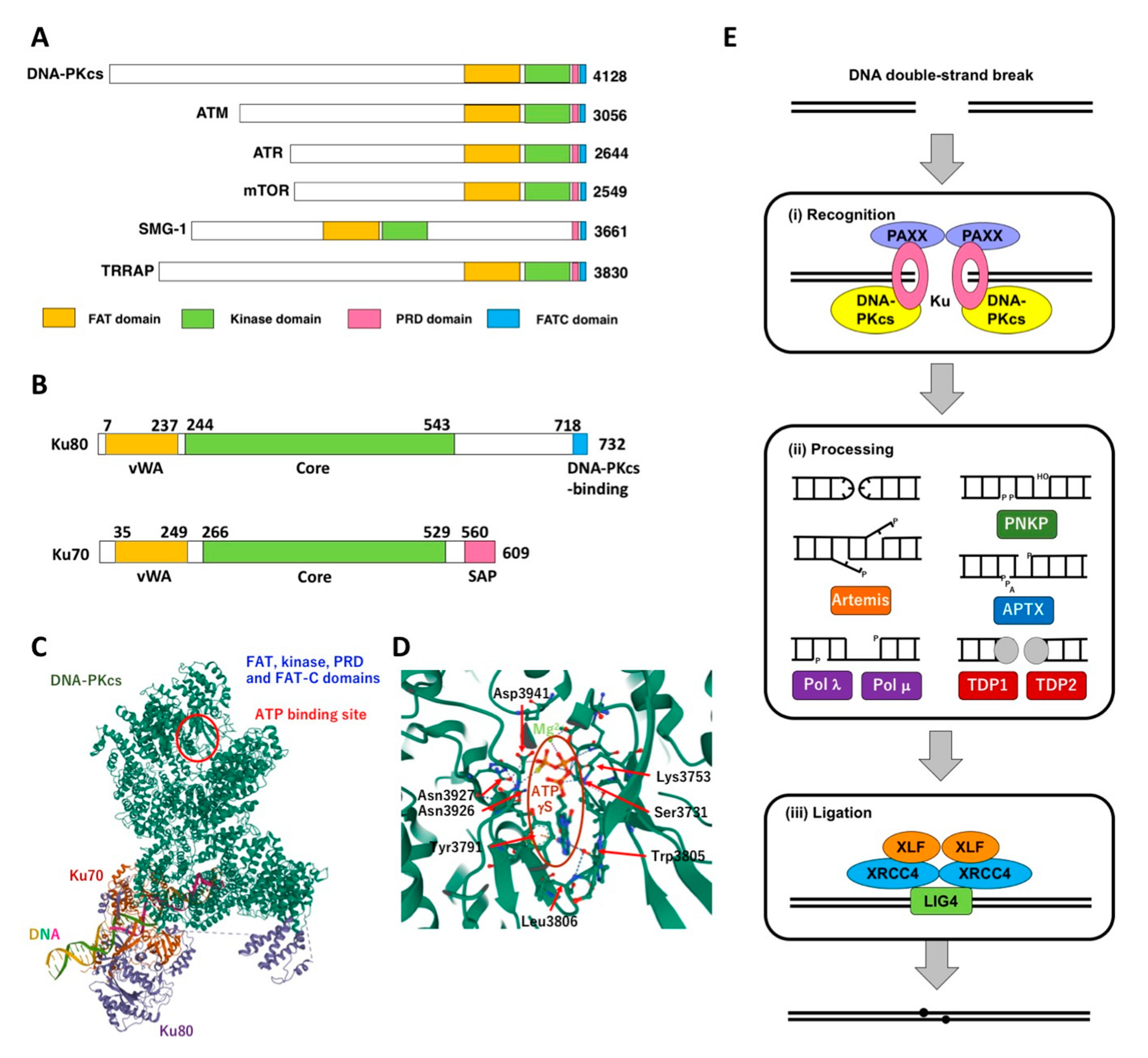
2. DNA-PK and Its Role NHEJ
3. Development and Evolution of DNA-PK Inhibitors–Pursuit for Potency and Selectivity
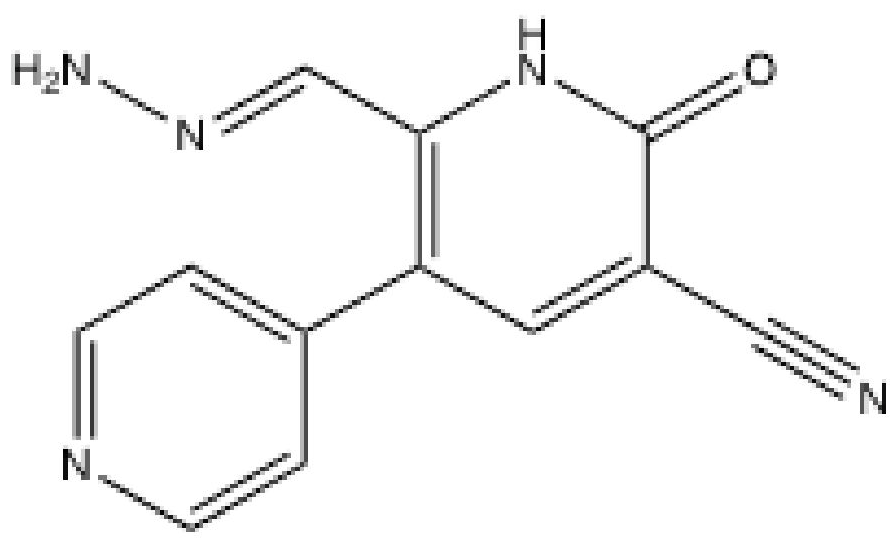
3.1. OK-1035
3.2. Wortmannin
3.3. LY294002-Derived Inhibitors
3.4. Arylmorpholine-Based Inhibitors
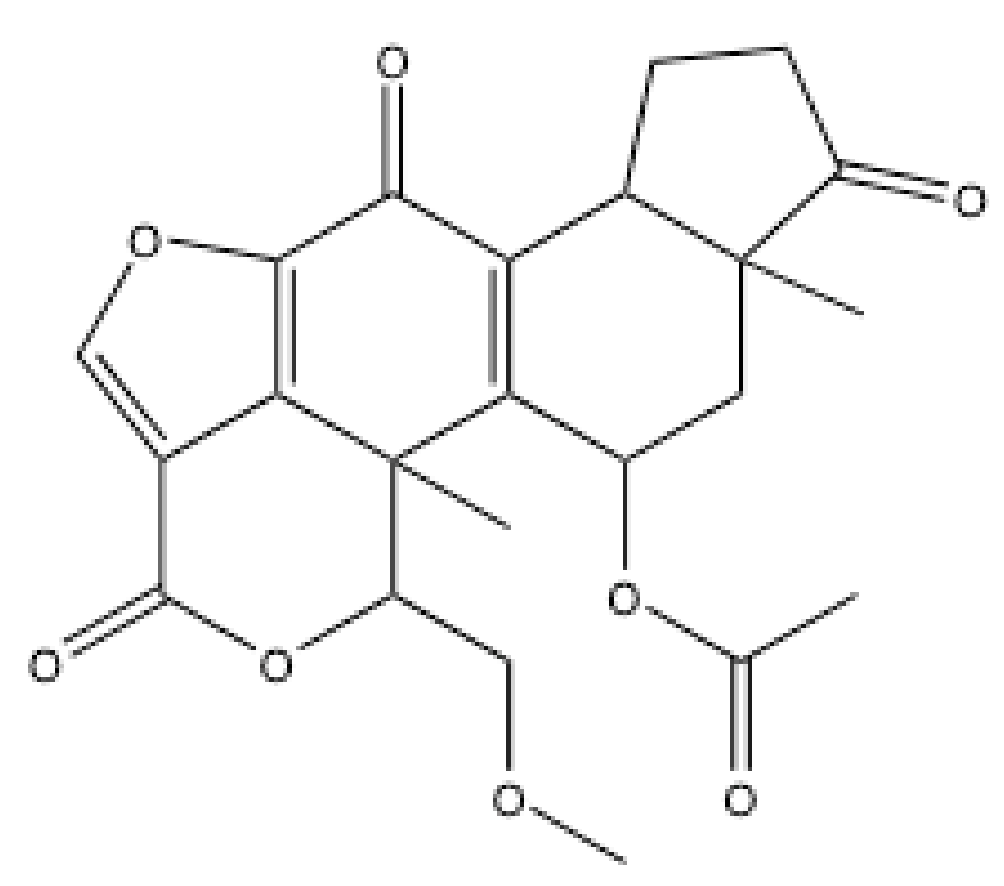
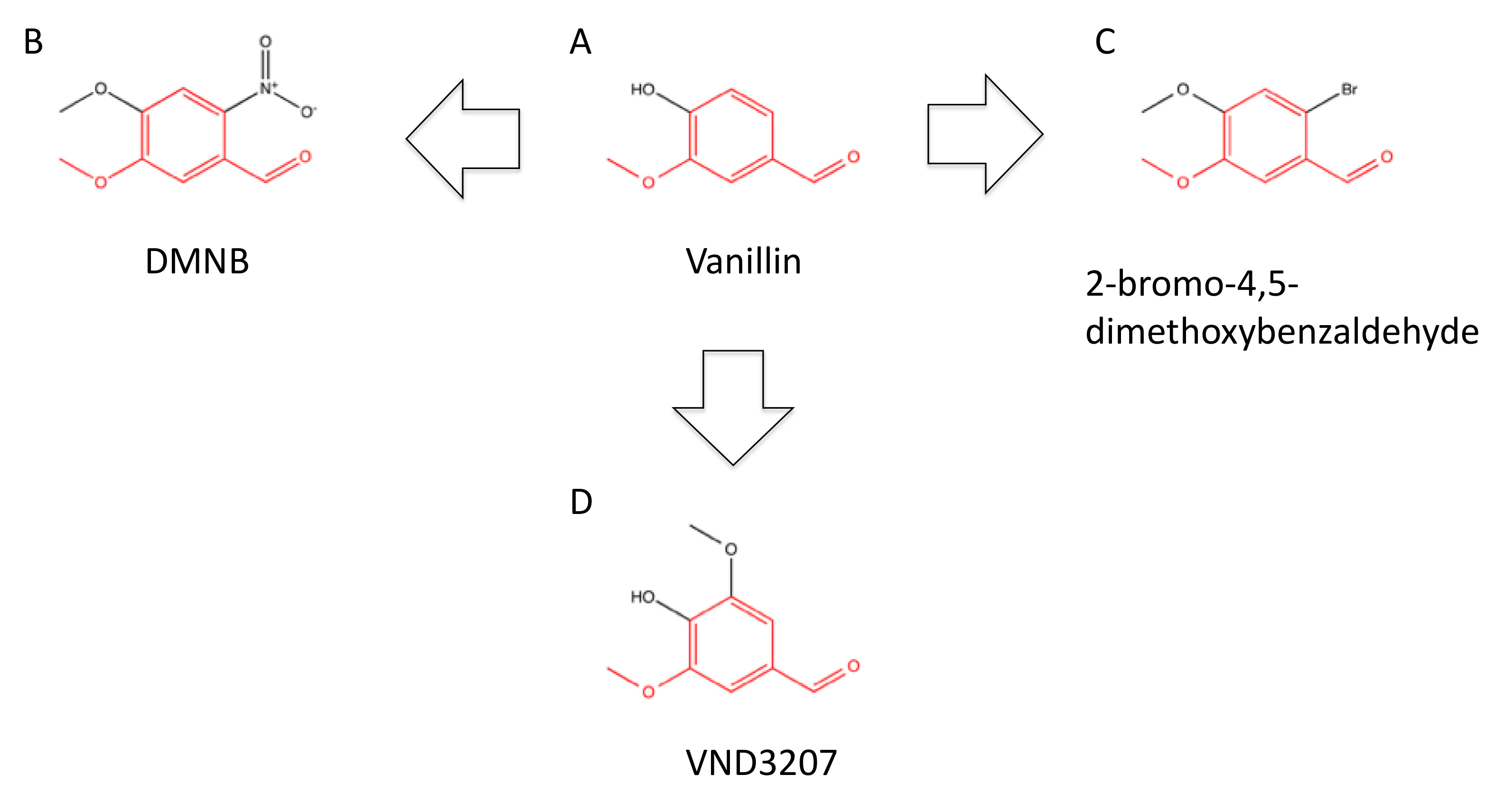
3.5. Vanillin-Based Inhibitors
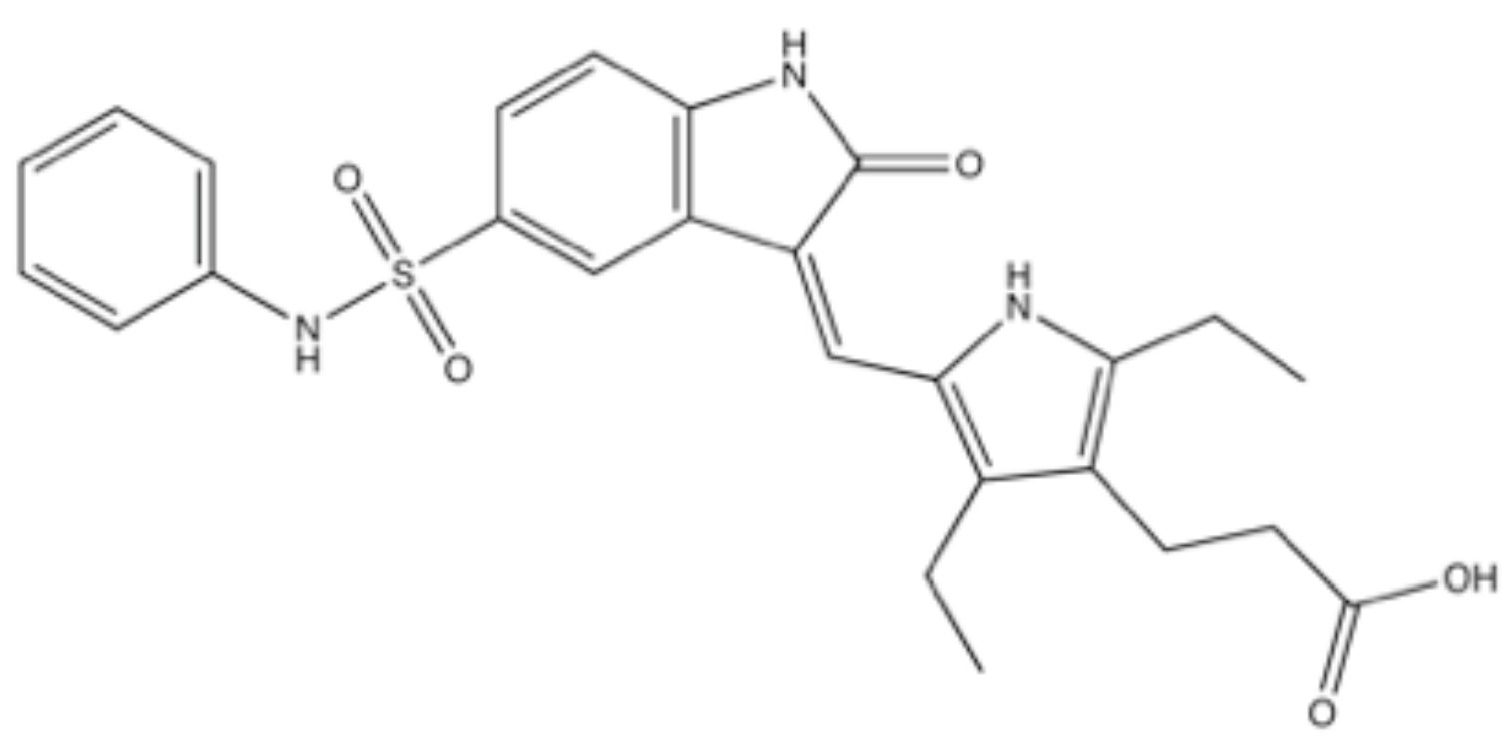
3.6. SU11752
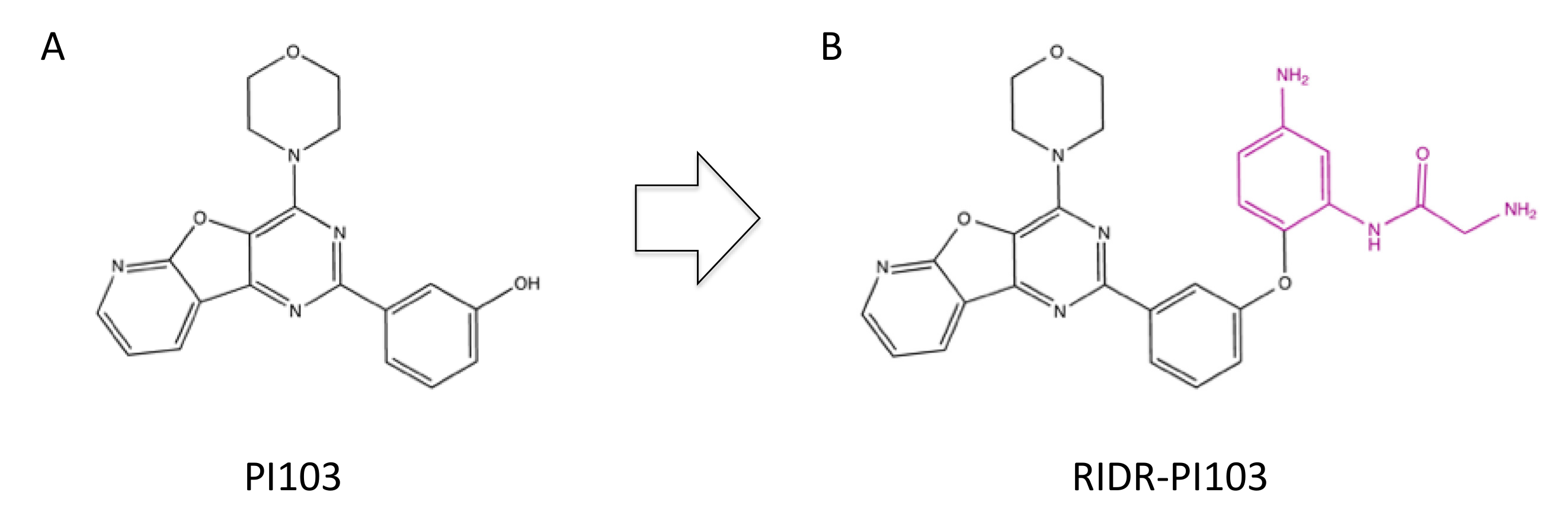
3.7. PI103
4. Development and Evolution of DNA-PK Inhibitors–Pursuit for Clinical Availability
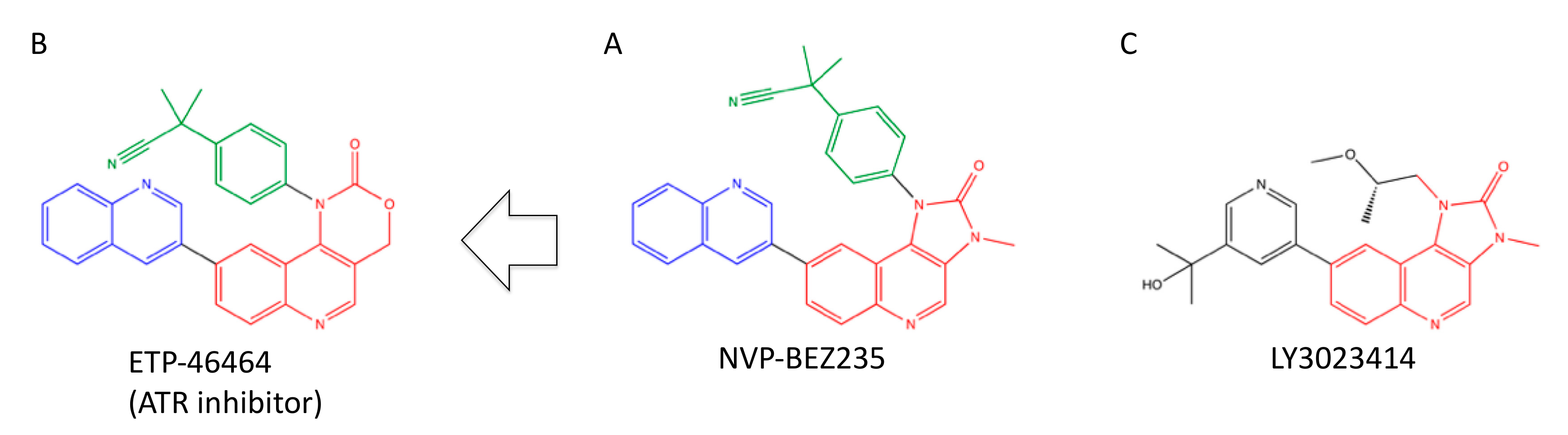
4.1. NVP-BEZ235 (Dactolicib)
4.2. LY3023414 (Samotolisib)
4.3. CC-115
4.4. VX-984 (M9831)
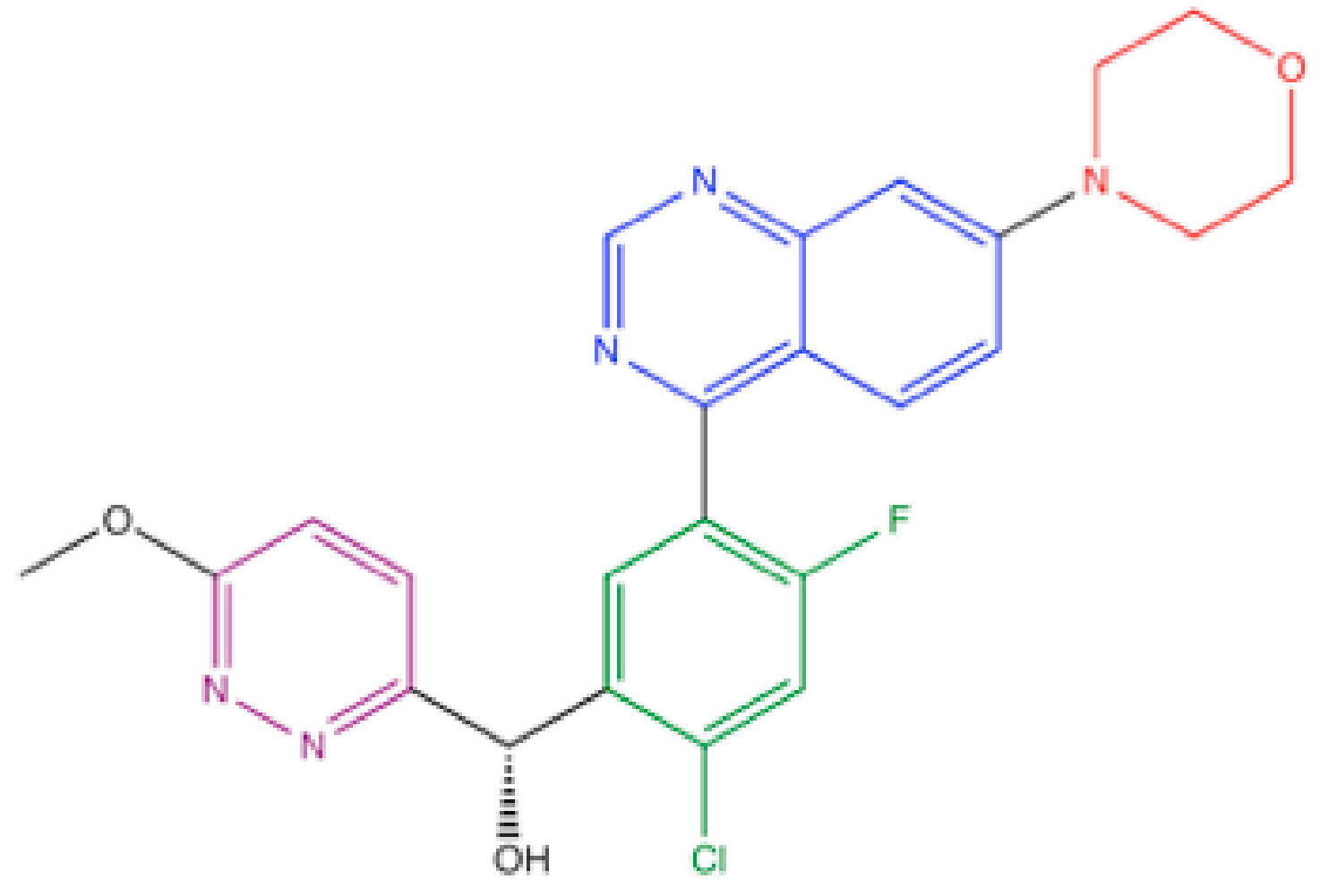
4.5. M3814 (Peposertib, Nedisertib)
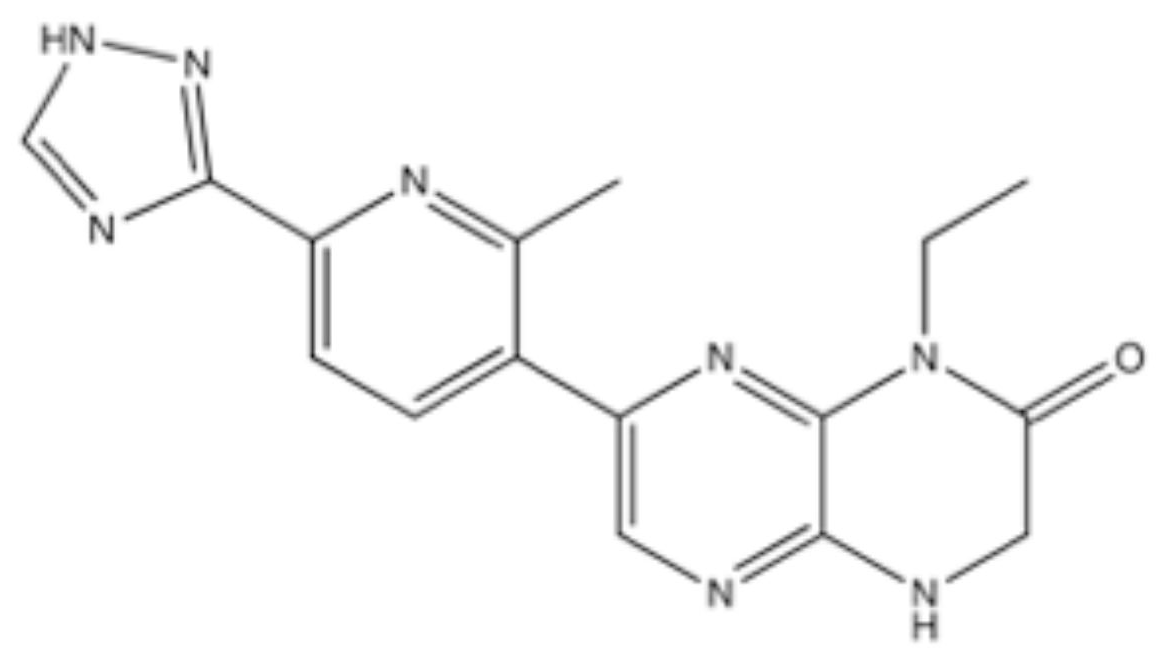
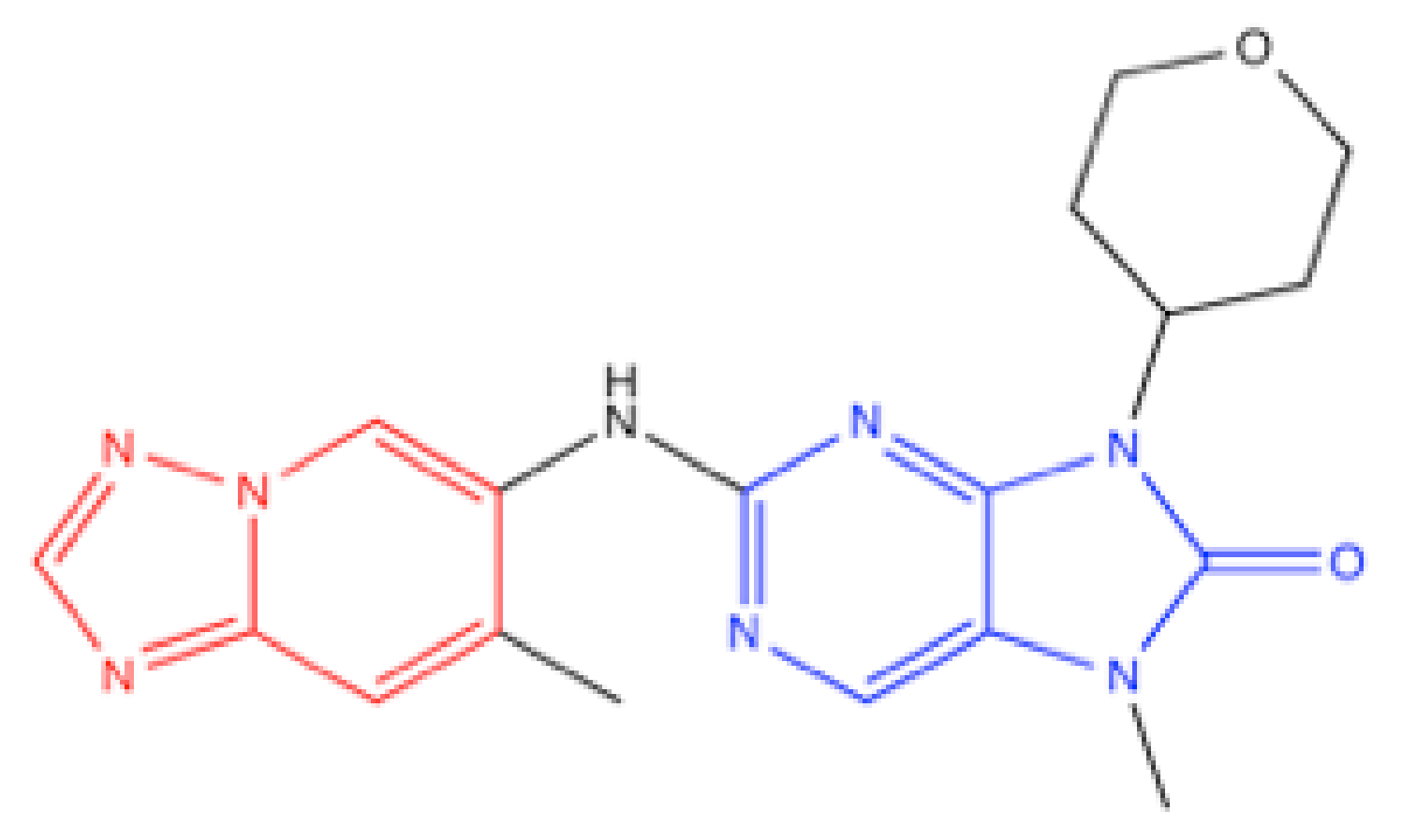
4.6. AZD7648
5. Summary and Future Directions
Funding
Acknowledgments
Conflicts of Interest
References
- United Nations Scientific Committee on the Effects of Atomic Radiation. Report of the United Nations Scientific Committee on the Effects of Atomic Radiation 2010 Fifty-Seventh Session, Includes Scientific Report: Summary of Low-Dose Radiation Effects on Health; United Nations Scientific Committee on the Effects of Atomic Radiation: Vienna, Austria, 2010. [Google Scholar]
- Zhao, B.; Rothenberg, E.; Ramsden, D.A.; Lieber, M.R. The molecular basis and disease relevance of non-homologous DNA end joining. Nat. Rev. Mol. Cell Biol. 2020, 21, 765–781. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, D.; Furst, A.; Meaburn, K.; Lezaja, A.; Wen, Y.; Altmeyer, M.; Reina-San-Martin, B.; Soutoglou, E. Activation of homologous recombination in G1 preserves centromeric integrity. Nature 2021, 600, 748–753. [Google Scholar] [CrossRef]
- Dvir, A.; Stein, L.Y.; Calore, B.L.; Dynan, W.S. Purification and characterization of a template-associated protein kinase that phosphorylates RNA polymerase II. J. Biol. Chem. 1993, 268, 10440–10447. [Google Scholar] [CrossRef]
- Gottlieb, T.M.; Jackson, S.P. The DNA-dependent protein kinase: Requirement for DNA ends and association with Ku antigen. Cell 1993, 72, 131–142. [Google Scholar] [CrossRef]
- Hartley, K.; Gell, D.; Smith, C.; Zhang, H.; Divecha, N.; Connelly, M.; Admon, A.; Lees-Miller, S.; Anderson, C.; Jackson, S. DNA-dependent protein kinase catalytic subunit: A relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell 1995, 82, 849–856. [Google Scholar] [CrossRef]
- Savitsky, K.; Bar-Shira, A.; Gilad, S.; Rotman, G.; Ziv, Y.; Vanagaite, L.; Tagle, D.A.; Smith, S.; Uziel, T.; Sfez, S.; et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 1995, 268, 1749–1753. [Google Scholar] [CrossRef] [PubMed]
- Bentley, N.J.; Holtzman, D.A.; Flaggs, G.; Keegan, K.S.; DeMaggio, A.; Ford, J.C.; Hoekstra, M.; Carr, A.M. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 1996, 15, 6641–6651. [Google Scholar] [CrossRef]
- Cimprich, K.A.; Shin, T.B.; Keith, C.T.; Schleiber, S.L. cDNA cloning and gene mapping of a candidate human cell cycle checkpoint protein. Proc. Natl. Acad. Sci. USA 1996, 93, 2850–2855. [Google Scholar] [CrossRef]
- Girard, P.M.; Riballo, E.; Begg, A.C.; Waugh, A.; Jeggo, P.A. Nbs1 promotes ATM dependent phosphorylation events including those required for G1/S arrest. Oncogene 2002, 21, 4191–4199. [Google Scholar] [CrossRef]
- Uziel, T.; Lerenthal, Y.; Moyal, L.; Andegeko, Y.; Mittelman, L.; Shiloh, Y. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 2003, 22, 5612–5621. [Google Scholar] [CrossRef]
- Zou, L.; Elledge, S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003, 300, 1542–1548. [Google Scholar] [CrossRef]
- Brown, E.J.; Albers, M.W.; Shin, T.B.; Ichikawa, K.; Keith, C.T.; Lane, W.S.; Schreiber, S.L. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 1994, 369, 756–758. [Google Scholar] [CrossRef]
- Sabatini, D.M.; Erdjument-Bromage, H.; Lui, M.; Tempst, P.; Snyder, S.H. RAFT1: A mammalian protein that binds to FKBP12 in a Rapamycin-dependent fashion and is homologous to yeast TORs. Cell 1994, 78, 35–43. [Google Scholar] [CrossRef]
- Denning, G.; Jamieson, L.; Maquat, L.E.; Thompson, E.A.; Fields, A.P. Cloning of a novel phosphatidylinositol kinase-related kinase: Characterization of the human SMG-1 RNA surveillance protein. J. Biol. Chem. 2001, 276, 22709–22714. [Google Scholar] [CrossRef]
- Yamashita, A.; Ohnishi, T.; Kashima, I.; Taya, Y.; Ohno, S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001, 15, 2215–2228. [Google Scholar] [CrossRef]
- McMahon, S.B.; Van Buskirk, H.A.; Dugan, K.A.; Copeland, T.D.; Cole, M.D. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 1998, 94, 363–374. [Google Scholar] [CrossRef]
- Alemi, F.; Sadigh, A.R.; Malakoti, F.; Elhaei, Y.; Ghaffari, S.H.; Maleki, M.; Asemi, Z.; Yousefi, B.; Targhazeh, N.; Majidinia, M. Molecular mechanisms involved in DNA repair in human cancers: An overview of PI3k/Akt signaling and PIKKs crosstalk. J. Cell Phisiol. 2022, 237, 313–328. [Google Scholar] [CrossRef]
- Huang, T.T.; Lampert, E.J.; Coots, C.; Lee, J.M. Targeting the PI3K pathway and DNA damage response as a therapeutic strategy in ovarian cancer. Cancer Treat. Rev. 2020, 86, 102021. [Google Scholar] [CrossRef]
- Wanigasooriya, K.; Tyler, R.; Barros-Silva, J.D.; Sinha, Y.; Ismail, T.; Beggs, A.D. Radiosensitising Cancer Using Phosphatidylinositol-3-Kinase (PI3K), Protein Kinase B (AKT) or Mammalian Target of Rapamycin (mTOR) Inhibitors. Cancers (Basel) 2020, 12, 1278. [Google Scholar] [CrossRef]
- Gewandter, J.S.; Bambara, R.A.; O’Reilly, M.A. The RNA surveillance protein SMG1 activates p53 in response to DNA double-strand breaks but not exogenously oxidized mRNA. Cell Cycle 2011, 10, 2561–2567. [Google Scholar] [CrossRef]
- Gubanova, E.; Issaeva, N.; Gokturk, C.; Djureinovic, T.; Helleday, T. SMG-1 suppresses CDK2 and tumor growth by regulating both the p53 and Cdc25A signaling pathways. Cell Cycle 2013, 12, 3770–3780. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Mi, J.; Deng, Y.; Deng, Z.; Long, D.; Liu, Z. DNMT1 Enhances the Radiosensitivity of HPV-Positive Head and Neck Squamous Cell Carcinomas via Downregulating SMG1. Onco. Targets Ther. 2020, 13, 4201–4211. [Google Scholar] [CrossRef] [PubMed]
- Long, D.; Xu, L.; Deng, Z.; Guo, D.; Zhang, Y.; Liu, Z.; Zhang, C. HPV16 E6 enhances the radiosensitivity in HPV-positive human head and neck squamous cell carcinoma by regulating the miR-27a-3p/SMG1 axis. Infect. Agent Cancer 2021, 16, 56. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.H.; Wei, M.F.; Wang, C.W.; Lee, H.W.; Pan, S.L.; Gao, M.; Kuo, S.H.; Cheng, A.L.; Teng, C.M. Dual phosphoinositide 3-kinase/mammalian target of rapamycin inhibitor is an effective radiosensitizer for colorectal cancer. Cancer Lett. 2015, 357, 582–590. [Google Scholar] [CrossRef]
- Del Peso, L.; González-García, M.; Page, C.; Herrera, R.; Nuñez, G. Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science 1997, 278, 687–689. [Google Scholar] [CrossRef]
- Datta, S.R.; Dudek, H.; Tao, X.; Masters, S.; Fu, H.; Gotoh, Y.; Greenberg, M.E. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 1997, 91, 231–241. [Google Scholar] [CrossRef]
- Zhou, B.P.; Liao, Y.; Xia, W.; Zou, Y.; Spohn, B.; Hung, M.C. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat. Cell Biol. 2001, 3, 973–982. [Google Scholar] [CrossRef]
- Oeck, S.; Al-Refae, K.; Riffkin, H.; Wiel, G.; Handrick, R.; Klein, D.; Iliakis, G.; Jendrossek, V. Activating Akt1 mutations alter DNA double strand break repair and radiosensitivity. Sci. Rep. 2017, 7, 42700. [Google Scholar] [CrossRef]
- Mueck, K.; Rebholz, S.; Harati, M.D.; Rodemann, H.P.; Toulany, M. Akt1 Stimulates Homologous Recombination Repair of DNA Double-Strand Breaks in a Rad51-Dependent Manner. Int. J. Mol. Sci. 2017, 18, 2473. [Google Scholar] [CrossRef]
- Bozulic, L.; Surucu, B.; Hynx, D.; Hemmings, B.A. PKBalpha/Akt1 acts downstream of DNA-PK in the DNA double-strand break response and promotes survival. Mol. Cell 2008, 30, 203–213. [Google Scholar] [CrossRef]
- Liu, L.; Dai, X.; Yin, S.; Liu, P.; Hill, E.G.; Wei, W.; Gan, W. DNA-PK promotes activation of the survival kinase AKT in response to DNA damage through an mTORC2-ECT2 pathway. Sci. Signal 2022, 15, eabh2290. [Google Scholar] [CrossRef]
- Yaneva, M.; Wen, J.; Ayala, A.; Cook, R. cDNA-derived amino acid sequence of the 86-kDa subunit of the Ku antigen. J. Biol. Chem. 1989, 264, 13407–13411. [Google Scholar] [CrossRef]
- Reeves, W.H.; Sthoeger, Z.M. Molecular cloning of cDNA encoding the p70 (Ku) lupus autoantigen. J. Biol. Chem. 1989, 264, 5047–5052. [Google Scholar] [CrossRef]
- Mimori, T.; Hardin, J.A. Mechanism of interaction between Ku protein and DNA. J. Biol. Chem. 1986, 261, 10375–10379. [Google Scholar] [CrossRef]
- Falck, J.; Coates, J.; Jackson, S.P. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 2005, 434, 605–611. [Google Scholar] [CrossRef]
- Walker, J.R.; Corpina, R.A.; Goldberg, J. Structure of the Ku heterodimer bound to DNA and its implication for double-strand break repair. Nature 2001, 412, 607–614. [Google Scholar] [CrossRef]
- Chen, X.; Xu, X.; Chen, Y.; Cheung, J.C.; Wang, H.; Jiang, J.; de Val, N.; Fox, T.; Gellert, M.; Yang, W. Structure of an activated DNA-PK and its implications for NHEJ. Mol. Cell 2021, 81, 801–810. [Google Scholar] [CrossRef]
- Liang, S.; Thomas, S.E.; Chaplin, A.K.; Hardwick, S.W.; Chirgadze, D.Y.; Blundell, T.L. Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs. Nature 2022, 601, 643–648. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Asa, A.D.D.C.; Modak, C.; Shimada, M. DNA-dependent protein kinase catalytic subunit: The sensor for DNA double-strand breaks structurally and functionally related to Ataxia telangiectasia mutated. Genes 2021, 12, 1143. [Google Scholar]
- Taccioli, G.E.; Gottlieb, T.M.; Blunt, T.; Priestley, A.; Demengeot, J.; Mizuta, R.; Lehmann, A.R.; Alt, F.W.; Jackson, S.P.; Jeggo, P.A. Ku80: Product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science 1994, 265, 1442–1445. [Google Scholar] [CrossRef]
- Smider, V.; Rathmell, W.K.; Lieber, M.R.; Chu, G. Restoration of X-ray resistance and V(D)J recombination in mutant cells by Ku cDNA. Science 1994, 266, 288–291. [Google Scholar] [CrossRef]
- Blunt, T.; Finnie, N.J.; Taccioli, G.E.; Smith, G.C.; Demengeot, J.; Gottlieb, T.M.; Mizuta, R.; Varghese, A.J.; Alt, F.W.; Jeggo, P.A. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell 1995, 80, 813–823. [Google Scholar] [CrossRef]
- Kirchgessner, C.U.; Patil, C.K.; Evans, J.W.; Cuomo, C.A.; Fried, L.M.; Carter, T.; Oettinger, M.A.; Brown, J.M. DNA-dependent kinase (p350) as a candidate gene for the murine SCID defect. Science 1995, 267, 1178–1183. [Google Scholar] [CrossRef]
- Peterson, S.R.; Kurimasa, A.; Oshimura, M.; Dynan, W.S.; Bradbury, E.M.; Chen, D.J. Loss of the catalytic subunit of the DNA-dependent protein kinase in DNA double-strand-break-repair mutant mammalian cells. Proc. Natl. Acad. Sci. USA 1995, 92, 3171–3174. [Google Scholar] [CrossRef]
- Ochi, T.; Blackford, A.N.; Coates, J.; Jhujh, S.; Mehmood, S.; Tamura, N.; Travers, J.; Wu, Q.; Draviam, V.M.; Robinson, C.V.; et al. DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science 2015, 347, 185–188. [Google Scholar] [CrossRef]
- Xing, M.; Yang, M.; Huo, W.; Feng, F.; Wei, L.; Jiang, W.; Ning, S.; Yan, Z.; Li, W.; Wang, Q.; et al. Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway. Nat. Commun. 2015, 6, 6233. [Google Scholar] [CrossRef]
- Craxton, A.; Somers, J.; Munnur, D.; Jukes-Jones, R.; Cain, K.; Malewicz, M. XLS (c9orf142) is a new component of mammalian DNA double-stranded break repair. Cell Death Diff. 2015, 22, 890–897. [Google Scholar] [CrossRef]
- Moshous, D.; Callebaut, I.; de Chasseval, R.; Corneo, B.; Cavazzana-Calvo, M.; Le Deist, F.; Tezcan, I.; Sanal, O.; Bertrand, Y.; Philippe, N.; et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 2001, 105, 177–186. [Google Scholar] [CrossRef]
- Ma, Y.; Pannicke, U.; Schwarz, K.; Lieber, M.R. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 2002, 108, 781–794. [Google Scholar] [CrossRef]
- Li, Z.; Otevrel, T.; Gao, Y.; Cheng, H.; Seed, B.; Stamato, T.; Taccioli, G.E.; Alt, F.W. The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell 1995, 83, 1079–1089. [Google Scholar] [CrossRef]
- Critchlow, S.; Bowater, R.; Jackson, S. Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr. Biol. 1997, 7, 588–598. [Google Scholar] [CrossRef]
- Grawunder, U.; Wilm, M.; Wu, X.; Kulesza, P.; Wilson, T.; Mann, M.; Leiber, M.R. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature 1997, 388, 492–495. [Google Scholar] [PubMed]
- Buck, D.; Malivert, L.; de Chasseval, R.; Barraud, A.; Fondanèche, M.C.; Sanal, O.; Plebani, A.; Stéphan, J.L.; Hufnagel, M.; le Deist, F.; et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 2006, 124, 287–299. [Google Scholar] [CrossRef] [PubMed]
- Ahnesorg, P.; Smith, P.; Jackson, S.P. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell 2006, 124, 301–313. [Google Scholar] [CrossRef]
- Hammel, M.; Yu, Y.; Fang, S.; Lees-Miller, S.P.; Tainer, J.A. XLF regulates filament architecture of the XRCC4·ligase IV complex. Structure 2010, 18, 1431–1442. [Google Scholar] [CrossRef]
- Kienker, L.J.; Shin, E.K.; Meek, K. Both V(D)J recombination and radioresistance require DNA-PK kinase activity, though minimal levels suffice for V(D)J recombination. Nucleic Acids Res. 2000, 28, 2752–2761. [Google Scholar] [CrossRef]
- Kurimasa, A.; Kumano, S.; Boubnov, N.; Story, M.; Tung, C.; Peterson, S.; Chen, D.J. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol. Cell Biol. 1999, 19, 3877–3884. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Sharma, M.K. DNA-dependent protein kinase in DNA damage response: Three decades and beyond. J. Radiat. Cancer Res. 2020, 11, 123–134. [Google Scholar] [CrossRef]
- Sehnal, D.; Bittrich, S.; Deshpande, M.; Svobodová, R.; Berka, K.; Bazgier, V.; Velankar, S.; Burley, S.K.; Koča, J.; Rose, A.S. Mol* Viewer: Modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acids Res. 2021, 49, 431–437. [Google Scholar] [CrossRef]
- Take, Y.; Kumano, M.; Hamano, Y.; Fukatsu, H.; Teraoka, H.; Nishimura, S.; Okumura, A. OK-1035, a selective inhibitor of DNA-dependent protein kinase. Biochem. Biophys. Res. Commun. 1995, 215, 41–47. [Google Scholar] [CrossRef]
- Stockley, M.; Clegg, W.; Fontana, G.; Golding, B.T.; Martin, N.; Rigoreau, L.J.; Smith, G.C.; Griffin, R.J. Synthesis, crystal structure determination, and biological properties of the DNA-dependent protein kinase (DNA-PK) inhibitor 3-cyano-6-hydrazonomethyl-5-(4-pyridyl)prid-[1H]-2-one (OK-1035). Bioorg. Med. Chem. Lett. 2001, 11, 2837–2841. [Google Scholar] [CrossRef]
- Kruszweski, M.; Wojewódzka, M.; Iwaneńko, T.; Szumiel, I.; Okuyama, A. Differential inhibitory effect of OK-1035 on DNA repair in L5178Y murine lymphoma sublines with functional or defective repair of double strand breaks. Mutat. Res. 1998, 409, 31–36. [Google Scholar] [CrossRef]
- Take, Y.; Kumano, M.; Teraoka, H.; Nishimura, S.; Okuyama, A. DNA-dependent protein kinase inhibitor (OK-1035) suppresses p21 expression in HCT116 cells containing wild-type p53 induced by adriamycin. Biochem. Biophys. Res. Commun. 1996, 221, 207–212. [Google Scholar] [CrossRef]
- Yano, H.; Nakanishi, S.; Kimura, K.; Hanai, N.; Saitoh, Y.; Fukui, Y.; Nonomura, Y.; Matsuda, Y. Inhibition of histamine secretion by wortmannin through the blockade of phosphatidylinositol 3-kinase in RBL-2H3 cells. J. Biol. Chem. 1993, 268, 25846–25856. [Google Scholar] [CrossRef]
- Banin, S.; Moyal, L.; Shieh, S.-Y.; Taya, Y.; Anderson, C.W.; Chessa, L.; Smorodinsky, N.I.; Prives, C.; Reiss, Y.; Shiloh, Y.; et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 1998, 281, 1674–1677. [Google Scholar] [CrossRef]
- Izzard, R.A.; Jackson, S.P.; Smith, G.C.M. Competitive and noncompetitive inhibition of the DNA-dependent protein kinase. Cancer Res. 1999, 59, 2581–2586. [Google Scholar]
- Cano, C.; Saravanan, K.; Bailey, C.; Bardos, J.; Curtin, N.J.; Frigerio, M.; Golding, B.T.; Hardcastle, I.R.; Hummersone, M.G.; Menear, K.A.; et al. 1-substituted (Dibenzo[b,d]thiophen-4-yl)-2-morpholino-4H-chromen-4-ones endowed with dual DNA-PK/PI3-K inhibitory activity. J. Med. Chem. 2013, 56, 6386–6401. [Google Scholar] [CrossRef]
- Veuger, S.J.; Curtin, N.J.; Richardson, C.J.; Smith, G.C.M.; Durkacz, B.W. Radiosensitization and DNA Repair Inhibition by the Combined Use of Novel Inhibitors of DNA-dependent Protein Kinase and Poly(ADP-Ribose) Polymerase-1. Cancer Res. 2003, 63, 6008–6015. [Google Scholar]
- Leahy, J.J.J.; Golding, B.T.; Griffin, R.J.; Hardcastle, I.R.; Richardson, C.; Rigoreau, L.; Smith, G.C.M. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg. Med. Chem. Lett. 2004, 14, 6083–6087. [Google Scholar] [CrossRef]
- Fok, J.H.L.; Ramos-Montoya, A.; Vazquez-Chantada, M.; Wijnhoven, P.W.G.; Follia, V.; James, N.; Farrington, P.M.; Karmokar, A.; Willis, S.E.; Cairns, J.; et al. AZD7648 is a potent and selective DNA-PK inhibitor that enhances radiation, chemotherapy and olaparib activity. Nat. Commun. 2019, 10, 5065. [Google Scholar] [CrossRef]
- Morrison, R.; Al-Rawi, J.M.; Jennings, I.G.; Thompson, P.E.; Angove, M.J. Synthesis, structure elucidation, DNA-PK and PI3K and anti-cancer activity of 8- and 6-aryl-substituted-1-3-benzoxazines. Eur. J. Med. Chem. 2016, 110, 326–339. [Google Scholar] [CrossRef] [PubMed]
- Willoughby, C.E.; Jiang, Y.; Thomas, H.D.; Willmore, E.; Kyle, S.; Wittner, A.; Phillips, N.; Zhao, Y.; Tudhope, S.J.; Prendergast, L.; et al. Selective DNA-PKcs inhibition extends the therapeutic index of localized radiotherapy and chemotherapy. J. Clin. Investig. 2020, 130, 258–271. [Google Scholar] [CrossRef] [PubMed]
- Kashishian, A.; Douangpanya, H.; Clark, D.; Schlachter, S.T.; Eary, C.T.; Schiro, J.G.; Huang, H.; Burgess, L.E.; Kesicki, E.A.; Halbrook, J. DNA-dependent protein kinase inhibitors as drug candidates for the treatment of cancer. Mol. Cancer Ther. 2003, 2, 1257–1264. [Google Scholar] [PubMed]
- Knight, Z.A.; Chiang, G.G.; Alaimo, P.J.; Kenski, D.M.; Ho, C.B.; Coan, K.; Abraham, R.T.; Shokat, K.M. Isoform-specific phosphoinositide 3-kinase inhibitors from an arylmorpholine scaffold. Bioorg. Med. Chem. Lett. 2004, 12, 4749–4759. [Google Scholar] [CrossRef] [PubMed]
- Durant, S.; Karran, P. Vanillins—A novel family of DNA-PK inhibitors. Nucleic Acids Res. 2003, 31, 5501–5512. [Google Scholar] [CrossRef] [PubMed]
- Ismail, I.H.; Marternsson, S.; Moshinsky, D.; Rice, A.; Tang, C.; Howlett, A.; McMahon, G.; Hammarste, O. SU11752 inhibits the DNA-dependent protein kinase and DNA double-strand break repair resulting in ionizing radiation sensitization. Oncogene 2004, 23, 873–882. [Google Scholar] [CrossRef]
- Fan, Q.W.; Knight, Z.A.; Goldenberg, D.D.; Yu, W.; Mostov, K.E.; Stokoe, D.; Shokat, K.M.; Weiss, W.A. A dual PI3 kinase/mTOR inhibitor reveals emergent efficacy in glioma. Cancer Cell 2006, 9, 341–349. [Google Scholar] [CrossRef]
- Kong, D.; Yaguchi, S.; Yamori, T. Effect of ZSTK474, a novel phosphatidylinositol 3-kinase inhibitor, on DNA-dependent protein kinase. Biol. Pharm. Bull. 2009, 32, 297–300. [Google Scholar] [CrossRef]
- Toledo, L.I.; Murga, M.; Zur, R.; Soria, R.; Rodriguez, A.; Martinez, S.; Oyarzabal, J.; Pastor, J.; Bischoff, J.R.; Fernandez-Capetillo, O. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat. Struct. Mol. Biol. 2011, 18, 721–727. [Google Scholar] [CrossRef]
- Smith, M.C.; Mader, M.M.; Cook, J.A.; Iversen, P.; Ajamie, R.; Perkins, E.; Bloem, L.; Yip, Y.Y.; Barda, D.A.; Waid, P.P.; et al. Characterization of LY3023414, a Novel PI3K/mTOR Dual Inhibitor Eliciting Transient Target Modulation to Impede Tumor Growth. Mol. Cancer Ther. 2016, 15, 2344–2356. [Google Scholar] [CrossRef]
- Mortensen, D.S.; Perrin-Ninkovic, S.M.; Shevlin, G.; Elsner, J.; Zhao, J.; Whitefield, B.; Tehrani, L.; Sapienza, J.; Riggs, J.R.; Parnes, J.S.; et al. Optimization of a Series of Triazole Containing Mammalian Target of Rapamycin (mTOR) Kinase Inhibitors and the Discovery of CC-115. J. Med. Chem. 2015, 58, 5599–5608. [Google Scholar] [CrossRef]
- Sun, Q.; Guo, Y.; Liu, X.; Czauderna, F.; Carr, M.I.; Zenke, F.T.; Blaukat, A.; Vassilev, L.T. Therapeutic Implications of p53 Status on Cancer Cell Fate Following Exposure to Ionizing Radiation and the DNA-PK Inhibitor M3814. Mol. Cancer Res. 2019, 17, 2457–2468. [Google Scholar] [CrossRef]
- Sarkaria, J.N.; Tibbetts, R.S.; Busby, E.C.; Kennedy, A.P.; Hill, D.E.; Abraham, R.T. Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res. 1998, 58, 4375–4382. [Google Scholar]
- Rosenzweig, K.E.; Youmell, M.B.; Palayoor, S.T.; Price, B.D. Radiosensitization of human tumor cells by the phosphatidylinositol3-kinase inhibitors wortmannin and LY294002 correlates with inhibition of DNA-dependent protein kinase and prolonged G2-M delay. Clin. Cancer Res. 1997, 3, 1149–1156. [Google Scholar]
- Okayasu, R.; Suetomi, K.; Ullrich, R.L. Wortmannin inhibits repair of DNA double-strand breaks in irradiated normal human cells. Radiat. Res. 1998, 149, 440–445. [Google Scholar] [CrossRef]
- Vlahos, C.J.; Matter, W.F.; Hui, K.Y.; Brown, R.F. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002). J. Biol. Chem. 1994, 269, 5241–5248. [Google Scholar] [CrossRef]
- Hollick, J.J.; Golding, B.T.; Hardcastle, I.R.; Martin, N.; Richardson, C.; Rigoreau, L.J.; Smith, G.C.; Griffin, R.J. 2,6-disubstituted pyran-4-one and thiopyran-4-one inhibitors of DNA-Dependent protein kinase (DNA-PK). Bioorg. Med. Chem. Lett. 2003, 13, 3083–3086. [Google Scholar] [CrossRef]
- Willmore, E.; de Caux, S.; Sunter, N.J.; Tilby, M.J.; Jackson, G.H.; Austin, C.A.; Durkacz, B.W. A novel DNA-dependent protein kinase inhibitor, NU7026, potentiates the cytotoxicity of topoisomerase II poisons used in the treatment of leukemia. Blood 2004, 103, 4659–4665. [Google Scholar] [CrossRef]
- Nutley, B.P.; Smith, N.F.; Hayes, A.; Kelland, L.R.; Brunton, L.; Golding, B.T.; Smith, G.C.; Martin, N.M.; Workman, P.; Raynaud, F.I. Preclinical pharmacokinetics and metabolism of a novel prototype DNA-PK inhibitor NU7026. Br. J. Cancer 2005, 93, 1011–1018. [Google Scholar] [CrossRef]
- Zhen, Y.F.; Li, S.T.; Zhu, Y.R.; Wang, X.D.; Zhou, X.Z.; Zhu, L.Q. Identification of DNA-PKcs as a primary resistance factor of salinomycin in osteosarcoma cells. Oncotarget 2016, 7, 79417–79427. [Google Scholar] [CrossRef]
- Cheng, L.; Liu, Y.Y.; Lu, P.H.; Peng, Y.; Yuan, Q.; Gu, X.S.; Jin, Y.; Chen, M.B.; Bai, X.M. Identification of DNA-PKcs as a primary resistance factor of TIC10 in hepatocellular carcinoma cells. Oncotarget 2017, 8, 28385–28394. [Google Scholar] [CrossRef]
- Yang, L.; Yang, X.; Tang, Y.; Zhang, D.; Zhu, L.; Wang, S.; Wang, B.; Ma, T. Inhibition of DNA-PK activity sensitizes A549 cells to X-ray irradiation by inducing the ATM-dependent DNA damage response. Mol. Med. Rep. 2018, 17, 7545–7552. [Google Scholar] [CrossRef]
- Hardcastle, I.R.; Cockcroft, X.; Curtin, N.J.; El-Murr, M.D.; Leahy, J.J.J.; Stockley, M.; Golding, B.T.; Rigoreau, L.; Richardson, C.; Smith, G.C.M.; et al. Discovery of potent chromen-4-one inhibitors of the DNA-dependent protein kinase (DNA-PK) using a small-molecule library approach. J. Med. Chem. 2005, 48, 7829–7846. [Google Scholar] [CrossRef]
- Zhao, Y.; Thomas, H.D.; Matey, M.A.; Cowell, I.G.; Rihardson, C.J.; Griffin, R.J.; Calvert, A.H.; Newell, D.R.; Smith, G.C.M.; Curtin, N.J. Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441. Cancer Res. 2006, 66, 5354–5362. [Google Scholar] [CrossRef]
- Yu, L.; Shang, Z.F.; Hsu, F.M.; Zhang, Z.; Tumati, V.; Lin, Y.F.; Chen, B.P.; Saha, D. NSCLC cells demonstrate differential mode of cell death in response to the combined treatment of radiation and a DNA-PKcs inhibitor. Oncotarget 2015, 6, 3848–3860. [Google Scholar] [CrossRef] [PubMed]
- Munck, J.M.; Batey, M.A.; Zhao, Y.; Jenkins, H.; Richardson, C.J.; Cano, C.; Tavecchio, M.; Barbeau, J.; Bardos, J.; Cornell, L.; et al. Chemosensitization of cancer cells by KU-0060648, a dual inhibitor of DNA-PK and PI-3K. Mol. Cancer Ther. 2012, 11, 1789–1798. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.B.; Zhou, Z.T.; Yang, L.; Wei, M.X.; Tang, M.; Ruan, T.Y.; Xu, J.Y.; Zhou, X.Z.; Chen, G.; Lu, P.H. KU-0060648 inhibits hepatocellular carcinoma cells through DNA-PKcs-dependent and DNA-PKcs-independent mechanisms. Oncotarget 2016, 7, 17047–17059. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Zhao, Z.; Qu, Y.; Zhang, M.; Wang, H.; Zhang, Z.; Zhou, W.; Fan, X.; Yu, C.; Zhan, Q.; et al. Targeting hyperactivated DNA-PKcs by KU0060648 inhibits glioma progression and enhances temozolomide therapy via suppression of AKT signaling. Oncotarget 2016, 7, 55555–55571. [Google Scholar] [CrossRef] [PubMed]
- Chao, O.S.; Goodman, O.B., Jr. DNA-PKc inhibition overcomes taxane resistance by promoting taxane-induced DNA damage in prostate cancer cells. Prostate 2021, 81, 1032–1048. [Google Scholar] [CrossRef] [PubMed]
- Hickson, I.; Zhao, Y.; Richardoson, C.J.; Green, S.J.; Martin, N.M.B.; Orr, A.I.; Reaper, P.M.; Jackson, S.P.; Curtin, N.J.; Smith, G.C.M. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004, 64, 9152–9159. [Google Scholar] [CrossRef] [PubMed]
- Golding, S.E.; Rosenberg, E.; Valerie, N.; Hussaini, I.; Frigerio, M.; Cockcroft, X.F.; Chong, W.Y.; Hummersone, M.; Rigoreau, L.; Menear, K.A.; et al. Improved ATM kinase inhibitor KU-60019 radiosensitizes glioma cells, compromises insulin, AKT and ERK prosurvival signaling, and inhibits migration and invasion. Mol. Cancer Ther. 2009, 8, 2894–2902. [Google Scholar] [CrossRef]
- Sturgeon, C.M.; Knight, Z.A.; Shokat, K.M.; Roberge, M. Effect of combined DNA repair inhibition and G2 checkpoint inhibition on cell cycle progression after DNA damage. Mol. Cancer Ther. 2006, 5, 885–892. [Google Scholar] [CrossRef][Green Version]
- Westhoff, M.A.; Kandenwein, J.A.; Karl, S.; Vellanki, S.H.; Braun, V.; Eramo, A.; Antoniadis, G.; Debatin, K.M.; Fulda, S. The pyridinylfuranopyrimidine inhibitor, PI-103, chemosensitizes glioblastoma cells for apoptosis by inhibiting DNA repair. Oncogene 2009, 28, 3586–3596. [Google Scholar] [CrossRef]
- Mukherjee, B.; Tomimatsu, N.; Amancherla, K.; Camacho, C.V.; Pichamoorthy, N.; Burma, S. The dual PI3K/mTOR inhibitor NVP-BEZ235 is a potent inhibitor of ATM- and DNAPKcs-mediated DNA damage responses. Neoplasia 2012, 14, 34–43. [Google Scholar] [CrossRef]
- Gil del Alcazar, C.R.; Hardebeck, M.C.; Mukherjee, B.; Tomimatsu, N.; Gao, X.; Yan, J.; Xie, X.J.; Bachoo, R.; Li, L.; Habib, A.A.; et al. Inhibition of DNA double-strand break repair by the dual PI3K/mTOR inhibitor NVP-BEZ235 as a strategy for radiosensitization of glioblastoma. Clin. Cancer Res. 2014, 20, 1235–1248. [Google Scholar] [CrossRef]
- Bürkel, F.; Jost, T.; Hecht, M.; Heinzerling, L.; Fietkau, R.; Distel, L. Dual mTOR/DNA-PK Inhibitor CC-115 Induces Cell Death in Melanoma Cells and Has Radiosensitizing Potential. Int. J. Mol. Sci. 2020, 21, 9321. [Google Scholar] [CrossRef]
- Timme, C.R.; Rath, B.H.; O’Neill, J.W.; Camphausen, K.; Tofilon, P.J. The DNA-PK Inhibitor VX-984 Enhances the Radiosensitivity of Glioblastoma Cells Grown In Vitro and as Orthotopic Xenografts. Mol. Cancer Ther. 2018, 17, 1207–1216. [Google Scholar] [CrossRef]
- Carr, M.I.; Zimmermann, A.; Chiu, L.Y.; Zenke, F.T.; Blaukat, A.; Vassilev, L.T. DNA-PK Inhibitor, M3814, as a New Combination Partner of Mylotarg in the Treatment of Acute Myeloid Leukemia. Front Oncol. 2020, 10, 127. [Google Scholar] [CrossRef]
- Zenke, F.T.; Zimmermann, A.; Sirrenberg, C.; Dahmen, H.; Kirkin, V.; Pehl, U.; Grombacher, T.; Wilm, C.; Fuchss, T.; Amendt, C.; et al. Pharmacologic Inhibitor of DNA-PK, M3814, Potentiates Radiotherapy and Regresses Human Tumors in Mouse Models. Mol. Cancer Ther. 2020, 19, 1091–1101. [Google Scholar] [CrossRef]
- Haines, E.; Nishida, Y.; Carr, M.I.; Montoya, R.H.; Ostermann, L.B.; Zhang, W.; Zenke, F.T.; Blaukat, A.; Andreeff, M.; Vassilev, L.T. DNA-PK inhibitor peposertib enhances p53-dependent cytotoxicity of DNA double-strand break inducing therapy in acute leukemia. Sci. Rep. 2021, 11, 12148. [Google Scholar] [CrossRef]
- Smithson, M.; Irwin, R.K.; Williams, G.; McLeod, M.C.; Choi, E.K.; Ganguly, A.; Pepple, A.; Cho, C.S.; Willey, C.D.; Leopold, J.; et al. Inhibition of DNA-PK may improve response to neoadjuvant chemoradiotherapy in rectal cancer. Neoplasia 2022, 25, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Chen, S.; Wei, Y.; Wei, X. DNA-PK inhibition by M3814 enhances chemosensitivity in non-small cell lung cancer. Acta. Pharm. Sin. B 2021, 11, 3935–3949. [Google Scholar] [CrossRef] [PubMed]
- Wise, H.C.; Iyer, G.V.; Moore, K.; Temkin, S.M.; Gordon, S.; Aghajanian, C.; Grisham, R.N. Activity of M3814, an Oral DNA-PK Inhibitor, In Combination with Topoisomerase II Inhibitors in Ovarian Cancer Models. Sci. Rep. 2019, 9, 18882. [Google Scholar] [CrossRef] [PubMed]
- Gordhandas, S.B.; Manning-Geist, B.; Henson, C.; Iyer, G.; Gardner, G.J.; Sonoda, Y.; Moore, K.N.; Aghajanian, C.; Chui, M.H.; Grisham, R.N. Pre-clinical activity of the oral DNA-PK inhibitor, peposertib (M3814), combined with radiation in xenograft models of cervical cancer. Sci. Rep. 2022, 12, 974. [Google Scholar] [CrossRef]
- Carr, M.I.; Chiu, L.Y.; Guo, Y.; Xu, C.; Lazorchak, A.S.; Yu, H.; Qin, G.; Qi, J.; Marelli, B.; Lan, Y.; et al. DNA-PK Inhibitor Peposertib Amplifies Radiation-Induced Inflammatory Micronucleation and Enhances TGFbeta/PD-L1 Targeted Cancer Immunotherapy. Mol. Cancer Res. 2022; online ahead of print. [Google Scholar] [CrossRef]
- Nakamura, K.; Karmokar, A.; Farrington, P.M.; James, N.H.; Ramos-Montoya, A.; Bickerton, S.J.; Hughes, G.D.; Illidge, T.M.; Cadogan, E.B.; Davies, B.R.; et al. Inhibition of DNA-PK with AZD7648 Sensitizes Tumor Cells to Radiotherapy and Induces Type I IFN-Dependent Durable Tumor Control. Clin. Cancer Res. 2021, 27, 4353–4366. [Google Scholar] [CrossRef]
- Hong, C.R.; Buckley, C.D.; Wong, W.W.; Anekal, P.V.; Dickson, B.D.; Bogle, G.; Hicks, K.O.; Hay, M.P.; Wilson, W.R. Radiosensitisation of SCCVII tumours and normal tissues in mice by the DNA-dependent protein kinase inhibitor AZD7648. Radiother. Oncol. 2022, 166, 162–170. [Google Scholar] [CrossRef]
- Anastasia, A.; Dellavedova, G.; Ramos-Montoya, A.; James, N.H.; Chiorino, G.; Russo, M.; Baakza, H.; Wilson, J.; Ghilardi, C.; Cadogan, E.B.; et al. The DNA-PK inhibitor AZD7648 sensitizes patient derived ovarian cancer xenografts to pegylated liposomal doxorubicin and olaparib preventing abdominal metastases. Mol. Cancer Ther. 2022; online ahead of print. [Google Scholar] [CrossRef]
- Shinohara, E.T.; Geng, L.; Tan, J.; Chen, H.; Shir, Y.; Edwards, E.; Halbrook, J.; Kesicki, E.A.; Kashishian, A.; Hallahan, D.E. DNA-dependent protein kinase is a molecular target for the development of noncytotoxic radiation-sensitizing drugs. Cancer Res. 2005, 65, 4987–4992. [Google Scholar] [CrossRef]
- Wong, W.W.; Jackson, R.K.; Liew, L.P.; Dickson, B.D.; Cheng, G.J.; Lipert, B.; Gu, Y.; Hunter, F.W.; Wilson, W.R.; Hay, M.P. Hypoxia-selective radiosensitisation by SN38023, a bioreductive prodrug of DNA-dependent protein kinase inhibitor IC87361. Biochem. Pharmacol. 2019, 169, 113641. [Google Scholar] [CrossRef]
- Zheng, H.; Chen, Z.W.; Wang, L.; Wang, S.Y.; Yan, Y.Q.; Wu, K.; Xu, Q.Z.; Zhang, S.M.; Zhou, P.K. Radioprotection of 4-hydroxy-3,5-dimethoxybenzaldehyde (VND3207) in culture cells is associated with minimizing DNA damage and activating Akt. Eur. J. Pharm. Sci. 2008, 33, 52–59. [Google Scholar] [CrossRef]
- Li, M.; Lang, Y.; Gu, M.M.; Shi, J.; Chen, B.P.C.; Yu, L.; Zhou, P.K.; Shang, Z.F. Vanillin derivative VND3207 activates DNA-PKcs conferring protection against radiation-induced intestinal epithelial cells injury in vitro and in vivo. Toxicol. Appl. Pharmacol. 2020, 387, 114855. [Google Scholar] [CrossRef]
- Hayakawa, M.; Kaizawa, H.; Moritomo, H.; Koizumi, T.; Ohishi, T.; Yamano, M.; Okada, M.; Ohta, M.; Tsukamoto, S.; Raynaud, F.I.; et al. Synthesis and biological evaluation of pyrido [3’,2’:4,5]furo [3,2-d]pyrimidine derivatives as novel PI3 kinase p110alpha inhibitors. Bioorg. Med. Chem. Lett. 2007, 17, 2438–2442. [Google Scholar] [CrossRef]
- Raynaud, F.I.; Eccles, S.; Clarke, P.A.; Hayes, A.; Nutley, B.; Alix, S.; Henley, A.; Di-Stefano, F.; Ahmad, Z.; Guillard, S.; et al. Pharmacologic characterization of a potent inhibitor of class I phosphatidylinositide 3-kinases. Cancer Res. 2007, 67, 5840–5850. [Google Scholar] [CrossRef]
- Zhu, H.; Mishra, R.; Yuan, L.; Salam, S.F.A.; Liu, J.; Gray, G.; Sterling, A.D.; Wunderlich, M.; Landero-Figueroa, J.; Garrett, J.T.; et al. Oxidative cyclization-induced activation of a phosphoinositide 3-kinase inhibitor for enhanced selectivity of cancer chemotherapeutics. Chem. Med. Chem. 2019, 14, 1933–1939. [Google Scholar] [CrossRef]
- Mishra, R.; Yuan, L.; Patel, H.; Karve, A.S.; Zhu, H.; White, A.; Alanazi, S.; Desai, P.; Merino, E.J.; Garrett, J.T. Phosphoinositide 3-Kinase (PI3K) Reactive Oxygen Species (ROS)-Activated Prodrug in Combination with Anthracycline Impairs PI3K Signaling, Increases DNA Damage Response and Reduces Breast Cancer Cell Growth. Int. J. Mol. Sci. 2021, 22, 2088. [Google Scholar] [CrossRef]
- Maira, S.M.; Stauffer, F.; Brueggen, J.; Furet, P.; Schnell, C.; Fritsch, C.; Brachmann, S.; Chene, P.; De Pover, A.; Schoemaker, K.; et al. Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol. Cancer Ther. 2008, 7, 1851–1863. [Google Scholar] [CrossRef]
- Seront, E.; Rottey, S.; Filleul, B.; Glorieux, P.; Goeminne, J.C.; Verschaeve, V.; Vandenbulcke, J.M.; Sautois, B.; Boegner, P.; Gillain, A.; et al. Phase II study of dual phosphoinositol-3-kinase (PI3K) and mammalian target of rapamycin (mTOR) inhibitor BEZ235 in patients with locally advanced or metastatic transitional cell carcinoma. BJU Int. 2016, 118, 408–415. [Google Scholar] [CrossRef]
- Fazio, N.; Buzzoni, R.; Baudin, E.; Antonuzzo, L.; Hubner, R.A.; Lahner, H.; de Herder, W.W.; Raderer, M.; Teulé, A.; Capdevila, J.; et al. A Phase II Study of BEZ235 in Patients with Everolimus-resistant, Advanced Pancreatic Neuroendocrine Tumours. Anticancer Res. 2016, 36, 713–719. [Google Scholar]
- Salazar, R.; Garcia-Carbonero, R.; Libutti, S.K.; Hendifar, A.E.; Custodio, A.; Guimbaud, R.; Lombard-Bohas, C.; Ricci, S.; Klümpen, H.J.; Capdevila, J.; et al. Phase II Study of BEZ235 versus Everolimus in Patients with Mammalian Target of Rapamycin Inhibitor-Naïve Advanced Pancreatic Neuroendocrine Tumors. Oncologist 2018, 23, 766–790. [Google Scholar] [CrossRef]
- Bendell, J.C.; Varghese, A.M.; Hyman, D.M.; Bauer, T.M.; Pant, S.; Callies, S.; Lin, J.; Martinez, R.; Wickremsinhe, E.; Fink, A.; et al. A First-in-Human Phase 1 Study of LY3023414, an Oral PI3K/mTOR Dual Inhibitor, in Patients with Advanced Cancer. Clin. Cancer Res. 2018, 24, 3253–3262. [Google Scholar] [CrossRef]
- Kondo, S.; Tajimi, M.; Funai, T.; Inoue, K.; Asou, H.; Ranka, V.K.; Wacheck, V.; Doi, T. Phase 1 dose-escalation study of a novel oral PI3K/mTOR dual inhibitor, LY3023414, in patients with cancer. Investig. New Drugs 2020, 38, 1836–1845. [Google Scholar] [CrossRef]
- Zauderer, M.G.; Alley, E.W.; Bendell, J.; Capelletto, E.; Bauer, T.M.; Callies, S.; Szpurka, A.M.; Kang, S.; Willard, M.D.; Wacheck, V.; et al. Phase 1 cohort expansion study of LY3023414, a dual PI3K/mTOR inhibitor, in patients with advanced mesothelioma. Investig. New Drugs 2021, 39, 1081–1088. [Google Scholar] [CrossRef]
- Rubinstein, M.M.; Hyman, D.M.; Caird, I.; Won, H.; Soldan, K.; Seier, K.; Iasonos, A.; Tew, W.P.; O’Cearbhaill, R.E.; Grisham, R.N.; et al. Phase 2 study of LY3023414 in patients with advanced endometrial cancer harboring activating mutations in the PI3K pathway. Cancer 2020, 126, 1274–1282. [Google Scholar] [CrossRef]
- Azaro, A.; Massard, C.; Tap, W.D.; Cassier, P.A.; Merchan, J.; Italiano, A.; Anderson, B.; Yuen, E.; Yu, D.; Oakley, G., 3rd; et al. A phase 1b study of the Notch inhibitor crenigacestat (LY3039478) in combination with other anticancer target agents (taladegib, LY3023414, or abemaciclib) in patients with advanced or metastatic solid tumors. Investig. New Drugs 2021, 39, 1089–1098. [Google Scholar] [CrossRef]
- Tsuji, T.; Sapinoso, L.M.; Tran, T.; Gaffney, B.; Wong, L.; Sankar, S.; Raymon, H.K.; Mortensen, D.S.; Xu, S. CC-115, a dual inhibitor of mTOR kinase and DNA-PK, blocks DNA damage repair pathways and selectively inhibits ATM-deficient cell growth in vitro. Oncotarget 2017, 8, 74688–74702. [Google Scholar] [CrossRef]
- Zheng, B.; Sun, X.; Chen, X.F.; Chen, Z.; Zhu, W.L.; Zhu, H.; Gu, D.H. Dual inhibition of DNA-PKcs and mTOR by CC-115 potently inhibits human renal cell carcinoma cell growth. Aging (Albany N. Y.) 2020, 12, 20445–20456. [Google Scholar] [CrossRef]
- Thijssen, R.; Ter Burg, J.; Garrick, B.; van Bochove, G.G.; Brown, J.R.; Fernandes, S.M.; Rodríguez, M.S.; Michot, J.M.; Hallek, M.; Eichhorst, B.; et al. Dual TORK/DNA-PK inhibition blocks critical signaling pathways in chronic lymphocytic leukemia. Blood 2016, 128, 574–583. [Google Scholar] [CrossRef]
- Munster, P.; Mita, M.; Mahipal, A.; Nemunaitis, J.; Massard, C.; Mikkelsen, T.; Cruz, C.; Paz-Ares, L.; Hidalgo, M.; Rathkopf, D.; et al. First-In-Human Phase I Study Of A Dual mTOR Kinase And DNA-PK Inhibitor (CC-115) In Advanced Malignancy. Cancer Manag. Res. 2019, 11, 10463–10476. [Google Scholar] [CrossRef]
- Khan, A.J.; Misenko, S.M.; Thandoni, A.; Schiff, D.; Jhawar, S.R.; Bunting, S.F.; Haffty, B.G. VX-984 is a selective inhibitor of non-homologous end joining, with possible preferential activity in transformed cells. Oncotarget 2018, 9, 25833–25841. [Google Scholar] [CrossRef]
- Wu, Z.X.; Peng, Z.; Yang, Y.; Wang, J.Q.; Teng, Q.X.; Lei, Z.N.; Fu, Y.G.; Patel, K.; Liu, L.; Lin, L.; et al. M3814, a DNA-PK Inhibitor, Modulates ABCG2-Mediated Multidrug Resistance in Lung Cancer Cells. Front. Oncol. 2020, 10, 674. [Google Scholar] [CrossRef]
- Van Bussel, M.T.J.; Awada, A.; de Jonge, M.J.A.; Mau-Sørensen, M.; Nielsen, D.; Schöffski, P.; Verheul, H.M.W.; Sarholz, B.; Berghoff, K.; El Bawab, S.; et al. A first-in-man phase 1 study of the DNA-dependent protein kinase inhibitor peposertib (formerly M3814) in patients with advanced solid tumours. Br. J. Cancer 2021, 124, 728–735. [Google Scholar] [CrossRef]
- Goldberg, F.W.; Finlay, M.R.V.; Ting, A.K.T.; Beattie, D.; Lamont, G.M.; Fallan, C.; Wrigley, G.L.; Schimpl, M.; Howard, M.R.; Williamson, B.; et al. The Discovery of 7-Methyl-2-[(7-methyl [1,2,4]triazolo [1,5-a]pyridin-6-yl)amino]-9-(tetrahydro-2H-pyran-4-yl)-7,9-dihydro-8H-purin-8-one (AZD7648), a Potent and Selective DNA-Dependent Protein Kinase (DNA-PK) Inhibitor. J. Med. Chem. 2020, 63, 3461–3471. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.G.; Sarkaria, J.N.; Kaufmann, S.H. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proc. Natl. Acad. Sci. USA 2011, 108, 3406–3411. [Google Scholar] [CrossRef] [PubMed]
- Williamson, C.T.; Kubota, E.; Hamill, J.D.; Klimowicz, A.; Ye, R.; Muzik, H.; Dean, M.; Tu, L.; Gilley, D.; Magliocco, A.M.; et al. Enhanced cytotoxicity of PARP inhibition in mantle cell lymphoma harbouring mutations in both ATM and p53. EMBO Mol. Med. 2012, 4, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Van Os, N.J.; Roeleveld, N.; Weemaes, C.M.; Jongmans, M.C.; Janssens, G.O.; Taylor, A.M.; Hoogerbrugge, N.; Willemsen, M.A. Health risks for ataxia-telangiectasia mutated heterozygotes: A systematic review, meta-analysis and evidence-based guideline. Clin. Genet. 2016, 90, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Jerzak, K.J.; Mancuso, T.; Eisen, A. Ataxia-telangiectasia gene (ATM) mutation heterozygosity in breast cancer: A narrative review. Curr. Oncol. 2018, 25, 176–180. [Google Scholar] [CrossRef] [PubMed]



| Substrates | Function | Substrates | Function |
|---|---|---|---|
| [DNA Repair and Damage Signaling] | [Transcription] | ||
| (NHEJ) | RNA polymerase II | Transcription (general) | |
| DNA-dependent protein kinase catalytic subunit (DNA-PKcs) | DNA-PK complex | ||
| TATA box-binding protein (TBP) | |||
| Ku autoantigen 80kDa subunit (Ku80) | p53 | Transcription (specific) | |
| Specificity protein 1 (Sp1) | |||
| Ku autoantigen 70 kDa subunit (Ku70) | c-Jun | ||
| c-Fos | |||
| DNA ligase IV (LIG4) | Ligation complex | c-Myc | |
| X-ray repair cross-complementing group 4 (XRCC4) | Octamer-binding factor 1 (Oct-1) | ||
| XRCC4-like factor (XLF) | Serum response factor (SRF) | ||
| Artemis | Nuclease | [RNA metabolism] | |
| Polynucleotide kinase phosphatase (PNKP) | Kinase, phosphatase | Nuclear DNA helicase II (NDHII) | Transcription and RNA processing |
| Werner syndrome protein (WRN) | Helicase, nuclease | Heterogeneous nuclear ribonucleoprotein A1 (hnRNP-A1) | RNA splicing |
| (Other DNA repair and damage signaling pathways) | |||
| Ataxia telangiectasia mutated (ATM) | Protein kinase; HR and cell cycle checkpoint | Heterogeneous nuclear ribonucleoprotein U (hnRNP-U) | |
| Replication protein A 2 (RPA2) | Single-stranded DNA binding; HR and DNA replication | Fused in sarcoma (FUS) | RNA binding |
| Poly(ADP-ribose) polymerase 1 (PARP1) | Single-strand break repair | [Signaling] | |
| Excision repair cross complementing 1 (ERCC1) | Nuclease component; nucleotide excision repair | Akt1 | Protein kinase |
| Akt2 | Protein kinase | ||
| [DNA replication] | Sty1/Spc1-interacting protein 1 (Sin1) | Protein kinase regulator | |
| DNA ligase I (LIG1) | Ligation | ||
| Minichromosome maintenance 3 (MCM3) | Initiation of replication | [Organelle, cytoskeleton] | |
| [Nucleosome and chromatin structure] | Golgi phosphoprotein 3 (GOLPH3) | Linking Golgi membrane to cytoskeleton | |
| Histone H2AX | Core histone component; recruitment of DSB repair proteins | Vimentin | Intermediate filament |
| Histone H1 | Linker histone | Tau | Microtubule regulation |
| High mobility group 1 (HMG1) | Maintenance and regulation of chromatin structure | [Protein maintenance] | |
| High mobility group 2 (HMG2) | Heat shock protein 90 alpha (HSP90a) | Protein chaperone | |
| C1D | Valosin-containing protein (VCP) | AAA+ ATPase | |
| Topoisomerase I | Regulation of topological status of DNA | [Metabolism] | |
| Topoisomerase II | Fumarate hydratase (FH) | Production of L-malate from fumarate; regulation of NHEJ | |
| Nuclear orphan receptor 4A2 (NR4A2) | Chromatin regulation; regulation of NHEJ | ||
| Pituitary tumor-transforming gene (PTTG) | Regulation of chromosome segregation | ||
| Name of Inhibitor | IC50 (nM) | Ref. | |||||||
|---|---|---|---|---|---|---|---|---|---|
| DNA-PK | ATM | ATR | mTOR | PI3Kα | PI3Kβ | PI3Kγ | PI3Kδ | ||
| OK-1035 | 8000 | [61] | |||||||
| 100,000 | [62] | ||||||||
| Wortmannin | 16 | 150 | [65] | ||||||
| 120 | [66] | ||||||||
| 260 | 300 | 4400 | 2500 | 3 | [67] | ||||
| LY294002 | 6000 | [67] | |||||||
| 1400 | >10,000 | >10,000 | 2800 | 300 | 270 | 3020 | 220 | [68] | |
| NU7026 | 230 | >100,000 | >100,000 | 6400 | 13,000 | [69] | |||
| NU7441 | 14 | >100,000 | >100,000 | 1700 | 5000 | [70] | |||
| 40 | >10,000 | >10,000 | 2400 | 130 | 16 | 220 | 30 | [68] | |
| 185 | >3100 | >30,000 | 1800 | 7800 | [71] | ||||
| KU-0060648 | 5 | >10,000 | >10,000 | 10,000 | 4 | 0.5 | 590 | <0.1 | [68] |
| 55 | >30,000 | >30,000 | 150 | 200 | [71] | ||||
| LTURM34 | 34 | >10,000 | 5,800 | >10,000 | 8500 | [72] | |||
| NU5455 | 8.2 | >10,000 | >10,000 | 4058 | 1870 | 9320 | >10,000 | 276 | [73] |
| IC60211 | 400 | [74] | |||||||
| IC86621 | 120 | 1400 | 135 | 880 | 1000 | [74] | |||
| IC87361 | 34 | 3800 | 1700 | 800 | 7900 | [74] | |||
| AMA37 | 270 | >100,000 | >100,000 | >100,000 | 32,000 | 3700 | ~100,000 | 22,000 | [75] |
| Vanillin | 1,500,000 | [76] | |||||||
| DMNB | 15,000 | [76] | |||||||
| 2-bromo-4,5-dimethoxybenzaldehyde | 30,000 | [76] | |||||||
| SU11752 | 130 | 1100 | [77] | ||||||
| PI103 | 14 | 2 | 3 | 15 | 3 | [78] | |||
| 7.5 | 8 | 7 | 15 | 172 | [79] | ||||
| NVP-BEZ235 | 1.7 | 7 | 72 | 6 | 38 | [79] | |||
| 5 | 7 | 21 | 2 | 2 | [80] | ||||
| LY3023414 | 4.24 | 165 | 6.07 | 77.6 | 23.8 | 38 | [81] | ||
| CC-115 | 13 | 21 | 852 | [82] | |||||
| VX-984 | 115 | >30,000 | >30,000 | >30,000 | >20,000 | >30,000 | 7100 | >30,000 | [71] |
| M3814 | 0.6–20 | 10,000 | 2800 | >10,000 | 330 | 250 | >1000 | 95 | [83] |
| 43 | >30,000 | >30,000 | 550 | 800 | 170 | 1590 | 350 | [71] | |
| AZD7648 1 | 91.3 | 17,930 | >29,770 | >30,000 | >8030 | >30,000 | 1370 | >30,000 | [71] |
| Name of Inhibitor | In Cellulo Sensitizing Effect | In Vivo Sensitizing Effect | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Radiation | Chemotherapeutic Drug | Radiation | Chemotherapeutic Drug | |||||||
| μM | Ref. | μM | Drug | Ref. | mg/kg 1 | Ref. | mg/kg 1 | Drug | Ref. | |
| NU7026 | 10 | [69] | 10 | idarubicin, daunorubicin, doxorubicin, etoposide, amsacrine (mAMSA), mitroxantrone | [89] | 25, i.p. | [91] | 50, i.p. | salinomycin | [92] |
| 50, i.p. | TIC10 3 | [93] | ||||||||
| NU7441 | 0.5 | [95] | 0.5 | etoposide | [95] | 25, i.p. | [96] | 10, i.p. | etoposide | [95] |
| KU-0060648 | 0.1 | [68] | 1 | etoposide, doxorubicin | [97] | 10, i.p. | etoposide | [97] | ||
| 1-10 | temozolomide | [99] | 10, 50, i.p. | temozolomide | [98] | |||||
| LTURM34 | 3 | docetaxel | [100] | |||||||
| NU5455 | 1 | [73] | 1 | etoposide, doxorubicin | [73] | 30, p.o. | [73] | 100, p.o. | etoposide | [73] |
| 30, p.o. | doxorubicin | [73] | ||||||||
| IC86621 | 50 | [74] | 400, s.c. | [74] | ||||||
| IC87361 | 7 | [74] | 75 2, i.p. | [74] | ||||||
| AMA37 | 20 | [103] | ||||||||
| Vanillin | 100, 300 | [76] | 100 | cisplatin | [76] | |||||
| DMNB | 15 | cisplatin | [76] | |||||||
| SU11752 | 50 | [77] | ||||||||
| PI103 | 0.06–1 | [104] | 0.06–1 | doxorubicin, etoposide, temozolomide | [104] | |||||
| NVP-BEZ235 | 0.1 | [105] | 50, 75, p.o. | [106] | ||||||
| LY3023414 | 15, p.o. | rapamicin, cisplatin+gemcitabin | [81] | |||||||
| CC-115 | 1 | [107] | ||||||||
| VX-984 | 0.1–0.5 | [108] | 50, p.o. | [108] | ||||||
| M3814 | 1 | [83] | 0.3–0.9 | calichiamicin | [109] | 5–50, p.o. | [110] | 100, p.o. | Mylotarg | [110] |
| 0.111–1 | [110] | 0.3 | daunorubicin | [111] | 50, p.o. | [112] | 50, i.g. | paclitaxel, etoposide | [113] | |
| 0.5–15 | [112] | 5 | paclitaxel, etoposide | [113] | 50, p.o. | PLD 4 | [114] | |||
| 50, p.o. | IR + 5-FU | [115] | ||||||||
| 50, p.o. | IR + bintrafusp alpha | [116] | ||||||||
| AZD7648 | 0.1, 1 | [71,117,118] | 0.1 | doxorubicin | [71] | 50, 100, p.o. | [71] | 37.5, 75, p.o. | doxorubicin, olaparib | [71] |
| 75, p.o. | [117,118] | 100, p.o. | PLD, olaparib | [119] | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matsumoto, Y. Development and Evolution of DNA-Dependent Protein Kinase Inhibitors toward Cancer Therapy. Int. J. Mol. Sci. 2022, 23, 4264. https://doi.org/10.3390/ijms23084264
Matsumoto Y. Development and Evolution of DNA-Dependent Protein Kinase Inhibitors toward Cancer Therapy. International Journal of Molecular Sciences. 2022; 23(8):4264. https://doi.org/10.3390/ijms23084264
Chicago/Turabian StyleMatsumoto, Yoshihisa. 2022. "Development and Evolution of DNA-Dependent Protein Kinase Inhibitors toward Cancer Therapy" International Journal of Molecular Sciences 23, no. 8: 4264. https://doi.org/10.3390/ijms23084264
APA StyleMatsumoto, Y. (2022). Development and Evolution of DNA-Dependent Protein Kinase Inhibitors toward Cancer Therapy. International Journal of Molecular Sciences, 23(8), 4264. https://doi.org/10.3390/ijms23084264





