Nocturnal Acidification: A Coordinating Cue in the Euprymna scolopes–Vibrio fischeri Symbiosis
Abstract
1. Introduction
2. Discussion
2.1. The Sepiolid Squid–Vibrio fischeri Mutualism
2.2. Bioluminescence in the Light Organ
2.3. Luminescence Is Required for Maintenance of Successful Colonization
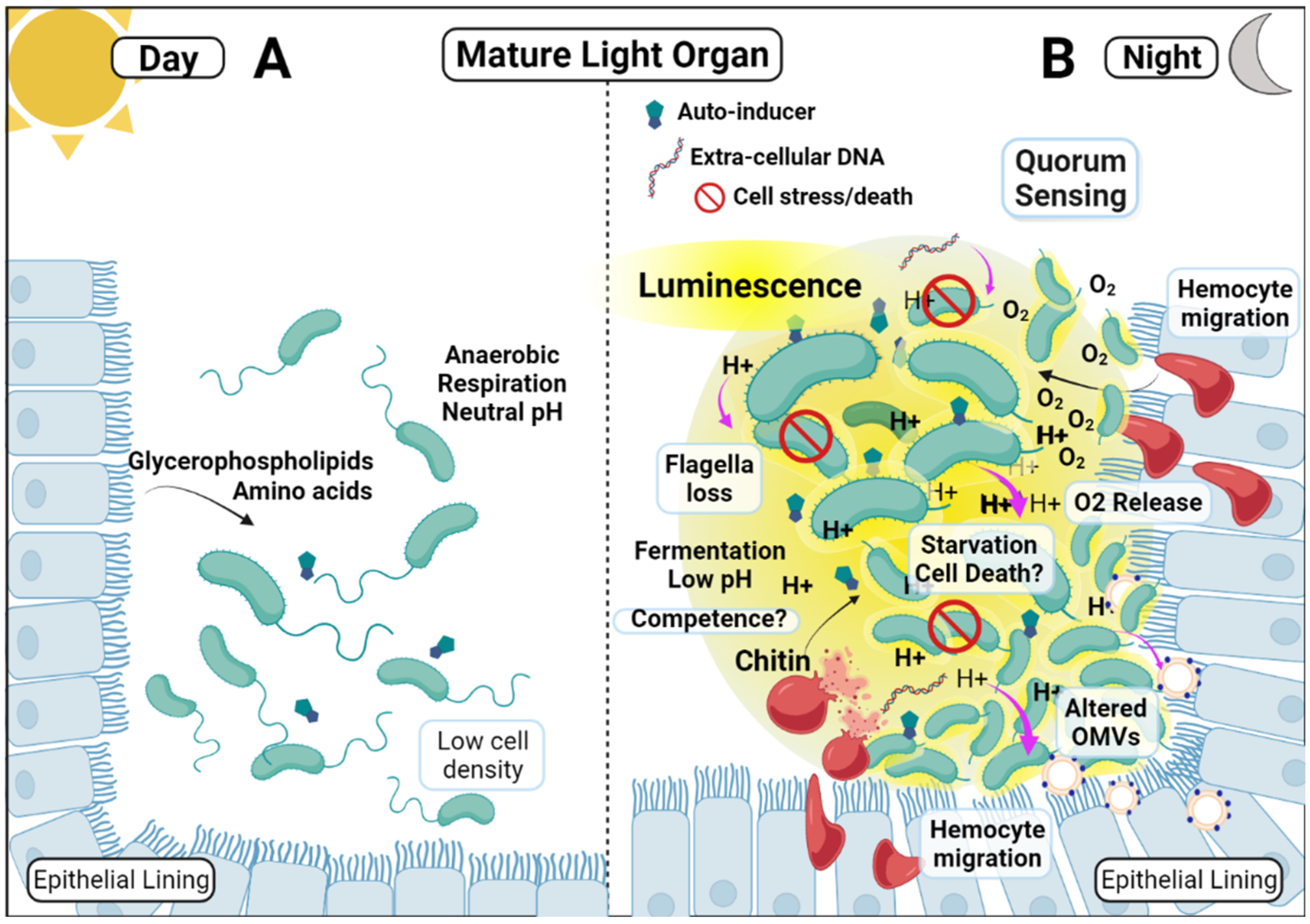
2.4. Light Organ Acidification
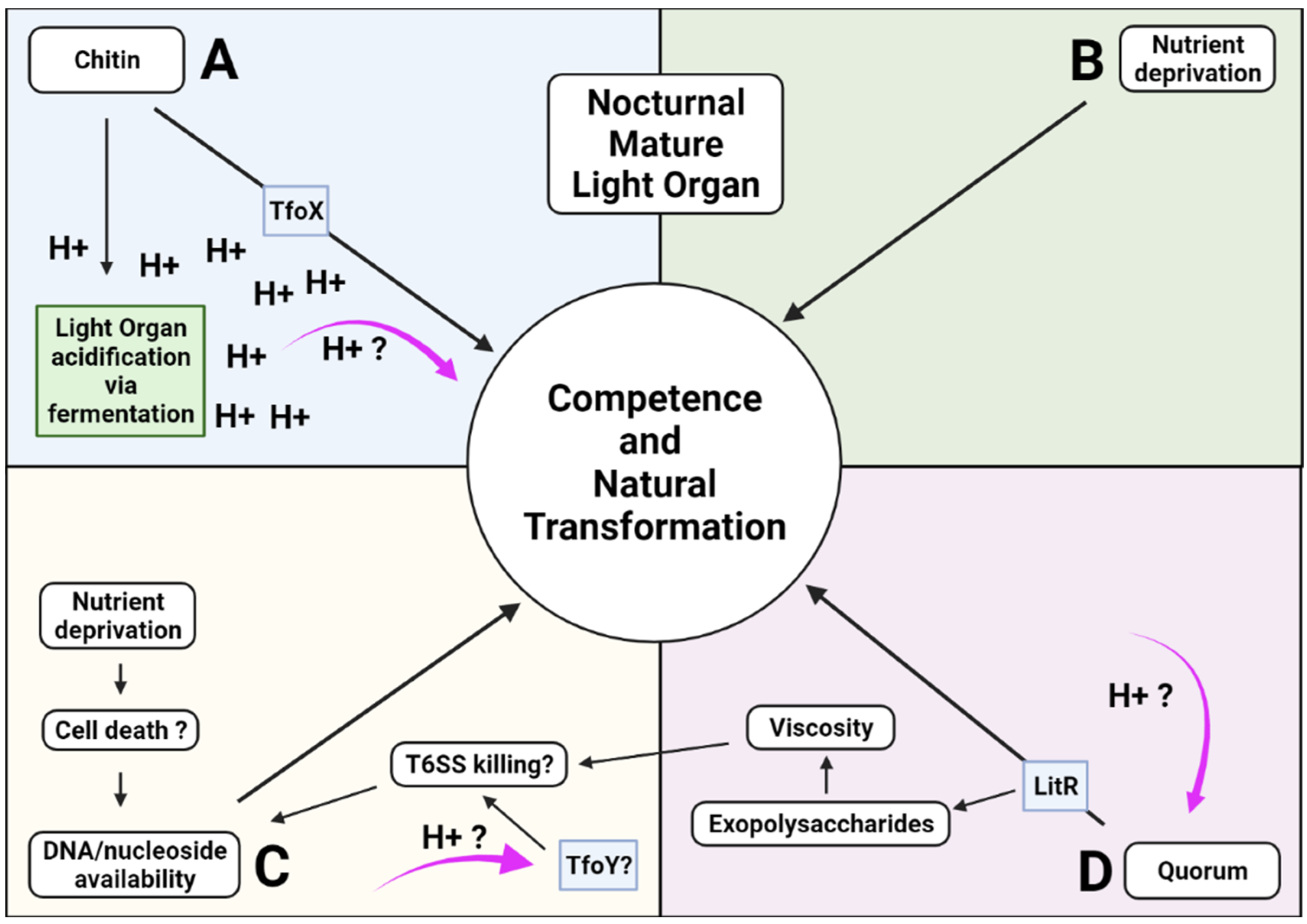
2.5. Nocturnal Acidification and the Induction of Competence and Natural Transformation
2.6. Competence and Natural Transformation in Vibrio fischeri
2.7. Acidification of the Light Organ Acts as a Cue
3. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chow, J.; Lee, S.M.; Shen, Y.; Khosravi, A.; Mazmanian, S.K. Host–Bacterial Symbiosis in Health and Disease. Adv. Immunol. 2010, 107, 243–274. [Google Scholar] [CrossRef] [PubMed]
- Neish, A.S. Microbes in Gastrointestinal Health and Disease. Gastroenterology 2009, 136, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Gordon, H.A.; Pesti, L. The gnotobiotic animal as a tool in the study of host microbial relationships. Bacteriol. Rev. 1971, 35, 390–429. [Google Scholar] [CrossRef] [PubMed]
- Samuel, B.S.; Gordon, J.I. A humanized gnotobiotic mouse model of host archaeal-bacterial mutualism. Proc. Natl. Acad. Sci. USA 2006, 103, 10011–10016. [Google Scholar] [CrossRef] [PubMed]
- Koch, E.J.; McFall-Ngai, M. Model systems for the study of how symbiotic associations between animals and extracellular bacte-rial partners are established and maintained. Drug Discov. Today Dis. Models 2018, 28, 3–12. [Google Scholar] [CrossRef]
- Blockley, A.; Elliott, D.R.; Roberts, A.P.; Sweet, M.J. Symbiotic Microbes from Marine Invertebrates: Driving a New Era of Natural Product Drug Discovery. Diversity 2017, 9, 49. [Google Scholar] [CrossRef]
- Leal, M.C.; Sheridan, C.; Osinga, R.; Dionísio, G.; Rocha, R.J.M.; Silva, B.; Rosa, R.; Calado, R. Marine Microorganism-Invertebrate Assemblages: Perspectives to Solve the “Supply Problem” in the Initial Steps of Drug Discovery. Mar. Drugs 2014, 12, 3929–3952. [Google Scholar] [CrossRef]
- Tauber, A.I. Metchnikoff and the phagocytosis theory. Nat. Rev. Mol. Cell Biol. 2003, 4, 897–901. [Google Scholar] [CrossRef]
- Rohwer, F.; Seguritan, V.; Azam, F.; Knowlton, N. Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 2002, 243, 1–10. [Google Scholar] [CrossRef]
- Knowlton, N.; Rohwer, F. Multispecies Microbial Mutualisms on Coral Reefs: The Host as a Habitat. Am. Nat. 2003, 162, S51–S62. [Google Scholar] [CrossRef]
- Ainsworth, T.; Hoegh-Guldberg, O. Bacterial communities closely associated with coral tissues vary under experimental and natural reef conditions and thermal stress. Aquat. Biol. 2009, 4, 289–296. [Google Scholar] [CrossRef]
- Paul, V.J.; Ritson-Williams, R.; Sharp, K. Marine chemical ecology in benthic environments. Nat. Prod. Rep. 2011, 28, 345–388. [Google Scholar] [CrossRef]
- Nyholm, S.V.; McFall-Ngai, M.J. A lasting symbiosis: How the Hawaiian bobtail squid finds and keeps its bioluminescent bacterial partner. Nat. Rev. Genet. 2021, 19, 666–679. [Google Scholar] [CrossRef]
- Visick, K.L.; Stabb, E.V.; Ruby, E.G. A lasting symbiosis: How Vibrio fischeri finds a squid partner and persists within its natural host. Nat. Rev. Genet. 2021, 19, 654–665. [Google Scholar] [CrossRef]
- Wier, A.M.; Nyholm, S.V.; Mandel, M.J.; Massengo-Tiassé, R.P.; Schaefer, A.L.; Koroleva, I.; Splinter-BonDurant, S.; Brown, B.; Manzella, L.; Snir, E.; et al. Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc. Natl. Acad. Sci. USA 2010, 107, 2259–2264. [Google Scholar] [CrossRef]
- Ii, J.F.B.; Gyllborg, M.C.; Cronin, D.C.; Quillin, S.J.; Mallama, C.A.; Foxall, R.; Whistler, C.; Goodman, A.L.; Mandel, M.J. Global discovery of colonization determinants in the squid symbiont Vibrio fischeri. Proc. Natl. Acad. Sci. USA 2014, 111, 17284–17289. [Google Scholar] [CrossRef]
- Pérez-Montaño, F.; Del Cerro, P.; Jiménez-Guerrero, I.; López-Baena, F.J.; Cubo, M.T.; Hungria, M.; Megías, M.; Ollero, F.J. RNA-seq analysis of the Rhizobium tropici CIAT 899 transcriptome shows similarities in the activation patterns of symbiotic genes in the presence of apigenin and salt. BMC Genom. 2016, 17, 198. [Google Scholar] [CrossRef]
- Thompson, L.R.; Nikolakakis, K.; Pan, S.; Reed, J.; Knight, R.; Ruby, E.G. Transcriptional characterization of Vibrio fischeri during colonization of juvenile Euprymna scolopes. Environ. Microbiol. 2017, 19, 1845–1856. [Google Scholar] [CrossRef]
- Wang, Y.; Ruby, E.G. The roles of NO in microbial symbioses. Cell. Microbiol. 2011, 13, 518–526. [Google Scholar] [CrossRef]
- Moriano-Gutierrez, S.; Koch, E.J.; Bussan, H.; Romano, K.; Belcaid, M.; Rey, F.E.; Ruby, E.G.; McFall-Ngai, M.J. Critical symbiont signals drive both local and systemic changes in diel and developmental host gene expression. Proc. Natl. Acad. Sci. USA 2019, 116, 7990–7999. [Google Scholar] [CrossRef]
- Rader, B.A.; Nyholm, S.V. Host/microbe interactions revealed through “omics” in the symbiosis between the Hawaiian bobtail squid Euprymna scolopes and the bioluminescent bacterium Vibrio fischeri. Biol. Bull. 2012, 223, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Breakspear, A.; Liu, C.; Roy, S.; Stacey, N.; Rogers, C.; Trick, M.; Morieri, G.; Mysore, K.; Wen, J.; Oldroyd, G.E.; et al. The Root Hair “Infectome” of Medicago truncatula Uncovers Changes in Cell Cycle Genes and Reveals a Requirement for Auxin Signaling in Rhizobial Infection. Plant Cell 2014, 26, 4680–4701. [Google Scholar] [CrossRef] [PubMed]
- McFall-Ngai, M.J. The Importance of Microbes in Animal Development: Lessons from the Squid-Vibrio Symbiosis. Annu. Rev. Microbiol. 2014, 68, 177–194. [Google Scholar] [CrossRef] [PubMed]
- Chun, C.K.; Troll, J.V.; Koroleva, I.; Brown, B.; Manzella, L.; Snir, E.; Almabrazi, H.; Scheetz, T.E.; Bonaldo, M.D.F.; Casavant, T.L.; et al. Effects of colonization, luminescence, and autoinducer on host transcription during development of the squid-Vibrio association. Proc. Natl. Acad. Sci. USA 2008, 105, 11323–11328. [Google Scholar] [CrossRef]
- Kremer, N.; Philipp, E.E.; Carpentier, M.-C.; Brennan, C.; Kraemer, L.; Altura, M.A.; Augustin, R.; Häsler, R.; Heath-Heckman, E.A.; Peyer, S.M.; et al. Initial Symbiont Contact Orchestrates Host-Organ-wide Transcriptional Changes that Prime Tissue Colonization. Cell Host Microbe 2013, 14, 183–194. [Google Scholar] [CrossRef]
- Borges, R.M. Co-niche construction between hosts and symbionts: Ideas and evidence. J. Genet. 2017, 96, 483–489. [Google Scholar] [CrossRef]
- Gilbert, S.F. Developmental plasticity and developmental symbiosis: The return of Eco-Devo. Curr. Top. Dev. Biol. 2016, 116, 415–433. [Google Scholar]
- Ruby, E.G.; Lee, K.-H. The Vibrio fischeri-Euprymna scolopes Light Organ Association: Current Ecological Paradigms. Appl. Environ. Microbiol. 1998, 64, 805–812. [Google Scholar] [CrossRef]
- McFall-Ngai, M.J. The development of cooperative associations between animals and bacteria: Establishing détente between domains. Am. Zool. 1998, 38, 3–18. [Google Scholar] [CrossRef]
- Meighen, E.A.; Dunlap, P.V. Physiological, Biochemical and Genetic Control of Bacterial Bioluminescence. Adv. Microb. Physiol. 1993, 34, 1–67. [Google Scholar] [CrossRef]
- Visick, K.L.; Foster, J.; Doino, J.; McFall-Ngai, M.; Ruby, E.G. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J. Bacteriol. 2000, 182, 4578–4586. [Google Scholar] [CrossRef]
- Fuqua, C.; Winans, S.C.; Greenberg, E.P. Census and consensus in bacterial ecosystems: The LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 1996, 50, 727–751. [Google Scholar] [CrossRef]
- Eberhard, A.; Burlingame, A.L.; Eberhard, C.; Kenyon, G.L.; Nealson, K.H.; Oppenheimer, N.J. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 1981, 28, 2444–2449. [Google Scholar] [CrossRef]
- Kaplan, H.B.; Greenberg, E.P. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J. Bacteriol. 1985, 163, 1210–1214. [Google Scholar] [CrossRef]
- Dunlap, P.V.; Kuo, A. Cell-density modulation of the Vibrio fischeri luminescence system in the absence of autoinducer and LuxR protein. J. Bacteriol. 1992, 174, 2440–2448. [Google Scholar] [CrossRef]
- Gilson, L.; Kuo, A.; Dunlap, P.V. AinS and a new family of autoinducer synthesis proteins. J. Bacteriol. 1995, 177, 6946–6951. [Google Scholar] [CrossRef]
- Kuo, A.; Blough, N.V.; Dunlap, P.V. Multiple N-acyl-l-homoserine lactone autoinducers of luminescence in the marine symbiotic bacterium Vibrio fischeri. J. Bacteriol. 1994, 176, 7558–7565. [Google Scholar] [CrossRef]
- Visick, K.G.; Ruby, E.G. Construction and symbiotic competence of a luxA-deletion mutant of Vibrio fischeri. Gene 1996, 175, 89–94. [Google Scholar] [CrossRef]
- Ruby, E.G.; McFall-Ngai, M.J. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends Microbiol. 1999, 7, 414–419. [Google Scholar] [CrossRef]
- Thompson, F.L.; Iida, T.; Swings, J. Biodiversity of Vibrios. Microbiol. Mol. Biol. Rev. 2004, 68, 403–431. [Google Scholar] [CrossRef]
- Soto, W.; Nishiguchi, M.K. Microbial experimental evolution as a novel research approach in the Vibrionaceae and squid-Vibrio symbiosis. Front. Microbiol. 2014, 5, 593. [Google Scholar] [CrossRef]
- Krause, E.; Wichels, A.; Giménez, L.; Lunau, M.; Schilhabel, M.B.; Gerdts, G. Small Changes in pH Have Direct Effects on Marine Bacterial Community Composition: A Microcosm Approach. PLoS ONE 2012, 7, e47035. [Google Scholar] [CrossRef]
- Cohen, M.L.; Mashanova, E.V.; Jagannathan, S.V.; Soto, W. Adaptation to pH stress by Vibrio fischeri can affect its symbiosis with the Hawaiian bobtail squid (Euprymna scolopes). Microbiology 2020, 166, 262–277. [Google Scholar] [CrossRef]
- Jones, D. pH Tolerance and Luminescence in Vibrio fischeri. Undergraduate Honors Theses, William & Mary, Williamsburg, VA, USA, 2018. Available online: https://scholarworks.wm.edu/honorstheses/1182 (accessed on 1 March 2022).
- Nourabadi, N.; Nishiguchi, M.K. pH Adaptation Drives Diverse Phenotypes in a Beneficial Bacterium-Host Mutualism. Front. Ecol. Evol. 2021, 9, 611411. [Google Scholar] [CrossRef]
- Heath-Heckman, E.A.; McFall-Ngai, M.J. The occurrence of chitin in the hemocytes of invertebrates. Zoology 2011, 114, 191–198. [Google Scholar] [CrossRef]
- Wolfe, A.J. The Acetate Switch. Microbiol. Mol. Biol. Rev. 2005, 69, 12–50. [Google Scholar] [CrossRef]
- Graf, J.; Ruby, E.G. Host-derived amino acids support the proliferation of symbiotic bacteria. Proc. Natl. Acad. Sci. USA 1998, 95, 1818–1822. [Google Scholar] [CrossRef]
- Schwartzman, J.A.; Koch, E.; Heath-Heckman, E.A.C.; Zhou, L.; Kremer, N.; McFall-Ngai, M.J.; Ruby, E.G. The chemistry of negotiation: Rhythmic, glycan-driven acidification in a symbiotic conversation. Proc. Natl. Acad. Sci. USA 2014, 112, 566–571. [Google Scholar] [CrossRef]
- Rader, B.A.; Kremer, N.; Apicella, M.A.; Goldman, W.; McFall-Ngai, M.J. Modulation of Symbiont Lipid A Signaling by Host Alkaline Phosphatases in the Squid-Vibrio Symbiosis. mBio 2012, 3, e00093-12. [Google Scholar] [CrossRef]
- Royet, J.; Gupta, D.; Dziarski, R. Peptidoglycan recognition proteins: Modulators of the microbiome and inflammation. Nat. Rev. Immunol. 2011, 11, 837–851. [Google Scholar] [CrossRef]
- Bates, J.M.; Akerlund, J.; Mittge, E.; Guillemin, K. Intestinal Alkaline Phosphatase Detoxifies Lipopolysaccharide and Prevents Inflammation in Zebrafish in Response to the Gut Microbiota. Cell Host Microbe 2007, 2, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, S.F. A holobiont birth narrative: The epigenetic transmission of the human microbiome. Front. Genet. 2014, 5, 282. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.B.; Schwartzman, J.A.; Bennett, B.D.; McAnulty, S.J.; Knop, M.; Nyholm, S.V.; Ruby, E.G. Ambient pH Alters the Protein Content of Outer Membrane Vesicles, Driving Host Development in a Beneficial Symbiosis. J. Bacteriol. 2019, 201, e00319-19. [Google Scholar] [CrossRef] [PubMed]
- Robledo, M.; Peregrina, A.; Millán, V.; García-Tomsig, N.I.; Quesada, O.T.; Mateos, P.; Becker, A.; Jiménez-Zurdo, J.I. A conserved α-proteobacterial small RNA contributes to osmoadaptation and symbiotic efficiency of rhizobia on legume roots. Environ. Microbiol. 2017, 19, 2661–2680. [Google Scholar] [CrossRef]
- Koropatnick, T.A.; Engle, J.T.; Apicella, M.A.; Stabb, E.V.; Goldman, W.E.; McFall-Ngai, M.J. Microbial Factor-Mediated Development in a Host-Bacterial Mutualism. Science 2004, 306, 1186–1188. [Google Scholar] [CrossRef]
- Rakoff-Nahoum, S.; Paglino, J.; Eslami-Varzaneh, F.; Edberg, S.; Medzhitov, R. Recognition of Commensal Microflora by Toll-Like Receptors Is Required for Intestinal Homeostasis. Cell 2004, 118, 229–241. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Zeevi, D.; Levy, M.; Zilberman-Schapira, G.; Suez, J.; Tengeler, A.C.; Abramson, L.; Katz, M.N.; Korem, T.; Zmora, N.; et al. Trans kingdom Control of Microbiota Diurnal Oscillations Promotes Metabolic Homeostasis. Cell 2014, 159, 514–529. [Google Scholar] [CrossRef]
- Mukherji, A.; Kobiita, A.; Ye, T.; Chambon, P. Homeostasis in Intestinal Epithelium Is Orchestrated by the Circadian Clock and Microbiota Cues Transduced by TLRs. Cell 2013, 153, 812–827. [Google Scholar] [CrossRef]
- Clayton, A.L.; Enomoto, S.; Su, Y.; Dale, C. The regulation of antimicrobial peptide resistance in the transition to insect symbiosis. Mol. Microbiol. 2017, 103, 958–972. [Google Scholar] [CrossRef]
- Hood, G.; Karunakaran, R.; Downie, J.A.; Poole, P. MgtE From Rhizobium leguminosarum Is a Mg2+ Channel Essential for Growth at Low pH and N2 Fixation on Specific Plants. Mol. Plant-Microbe Interact. 2015, 28, 1281–1287. [Google Scholar] [CrossRef]
- Foster, J.S.; McFall-Ngai, M.J. Induction of apoptosis by cooperative bacteria in the morphogenesis of host epithelial tissues. Dev. Genes Evol. 1998, 208, 295–303. [Google Scholar] [CrossRef]
- Lyell, N.L.; Dunn, A.K.; Bose, J.L.; Stabb, E.V. Bright Mutants of Vibrio fischeri ES114 Reveal Conditions and Regulators That Control Bioluminescence and Expression of the lux Operon. J. Bacteriol. 2010, 192, 5103–5114. [Google Scholar] [CrossRef]
- Melkina, O.E.; Goryanin, I.; Bazhenov, S.V.; Manukhov, I.; Zavilgelsky, G.B. Comparative analysis of Aliivibrio logei luxR1 and luxR2 genes regulation in Escherichia coli cells. Arch. Microbiol. 2019, 201, 1415–1425. [Google Scholar] [CrossRef]
- Chaparian, R.; Tran, M.L.N.; Conrad, L.M.; Rusch, D.B.; Van Kessel, J.C. Global H-NS counter-silencing by LuxR activates quorum sensing gene expression. Nucleic Acids Res. 2019, 48, 171–183. [Google Scholar] [CrossRef]
- Lee, K.-H.; Ruby, E.G. Effect of the Squid Host on the Abundance and Distribution of Symbiotic Vibrio fischeri in Nature. Appl. Environ. Microbiol. 1994, 60, 1565–1571. [Google Scholar] [CrossRef]
- Bongrand, C.; Ruby, E.G. Achieving a multi-strain symbiosis: Strain behavior and infection dynamics. ISME J. 2018, 13, 698–706. [Google Scholar] [CrossRef]
- Jones, B.W.; Nishiguchi, M.K. Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda). Mar. Biol. 2004, 144, 1151–1155. [Google Scholar] [CrossRef]
- McFall-Ngai, M.J.; Ruby, E.G. Developmental biology in marine invertebrate symbioses. Curr. Opin. Microbiol. 2000, 3, 603–607. [Google Scholar] [CrossRef]
- Moriano-Gutierrez, S.; Bongrand, C.; Essock-Burns, T.; Wu, L.; McFall-Ngai, M.J.; Ruby, E.G. The noncoding small RNA SsrA is released by Vibrio fischeri and modulates critical host responses. PLoS Biol. 2020, 18, e3000934. [Google Scholar] [CrossRef]
- Schwartzman, J.A.; Ruby, E.G. Stress as a Normal Cue in the Symbiotic Environment. Trends Microbiol. 2016, 24, 414–424. [Google Scholar] [CrossRef]
- Kremer, N.; Schwartzman, J.; Augustin, R.; Zhou, L.; Ruby, E.G.; Hourdez, S.; McFall-Ngai, M.J. The dual nature of haemocyanin in the establishment and persistence of the squid–vibrio symbiosis. Proc. R. Soc. B Biol. Sci. 2014, 281, 20140504. [Google Scholar] [CrossRef] [PubMed]
- Seitz, P.; Blokesch, M. DNA-uptake machinery of naturally competent Vibrio cholerae. Proc. Natl. Acad. Sci. USA 2013, 110, 17987–17992. [Google Scholar] [CrossRef] [PubMed]
- Heath-Heckman, E.A.C.; Peyer, S.M.; Whistler, C.A.; Apicella, M.A.; Goldman, W.; McFall-Ngai, M.J. Bacterial Bioluminescence Regulates Expression of a Host Cryptochrome Gene in the Squid-Vibrio Symbiosis. mBio 2013, 4, e00167-13. [Google Scholar] [CrossRef] [PubMed]
- Bouskra, D.; Brézillon, C.; Bérard, M.; Werts, C.; Varona, R.; Boneca, I.G.; Eberl, G. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 2008, 456, 507–510. [Google Scholar] [CrossRef]
- Pollack-Berti, A.; Wollenberg, M.S.; Ruby, E.G. Natural transformation of Vibrio fischeri requires tfoX and tfoY. Environ. Microbiol. 2010, 12, 2302–2311. [Google Scholar] [CrossRef]
- Blokesch, M.; Schoolnik, G.K. The Extracellular Nuclease Dns and Its Role in Natural Transformation of Vibrio cholerae. J. Bacteriol. 2008, 190, 7232–7240. [Google Scholar] [CrossRef]
- Sun, Y.; Bernardy, E.E.; Hammer, B.K.; Miyashiro, T. Competence and natural transformation in vibrios. Mol. Microbiol. 2013, 89, 583–595. [Google Scholar] [CrossRef]
- Antonova, E.S.; Hammer, B.K. Quorum-sensing autoinducer molecules produced by members of a multispecies biofilm promote horizontal gene transfer to Vibrio cholerae. FEMS Microbiol. Lett. 2011, 322, 68–76. [Google Scholar] [CrossRef]
- Cohen, J.J.; Eichinger, S.J.; Witte, D.A.; Cook, C.J.; Fidopiastis, P.M.; Tepavčević, J.; Visick, K.L. Control of Competence in Vibrio fischeri. Appl. Environ. Microbiol. 2021, 87, e01962-20. [Google Scholar] [CrossRef]
- Ball, A.S.; Chaparian, R.R.; van Kessel, J.C. Quorum Sensing Gene Regulation by LuxR/HapR Master Regulators in Vibrios. J. Bacteriol. 2017, 199, e00105-17. [Google Scholar] [CrossRef]
- Suckow, G.; Seitz, P.; Blokesch, M. Quorum sensing contributes to natural transformation of Vibrio cholerae in a species-specific manner. J. Bacteriol. 2011, 193, 4914–4924. [Google Scholar] [CrossRef]
- Speare, L.; Woo, M.; Bultman, K.M.; Mandel, M.J.; Wollenberg, M.S.; Septer, A.N. Host-Like Conditions Are Required for T6SS-Mediated Competition among Vibrio fischeri Light Organ Symbionts. mSphere 2021, 6, e0128820. [Google Scholar] [CrossRef]
- Bennett, B.D.; Essock-Burns, T.; Ruby, E.G. HbtR, a Heterofunctional Homolog of the Virulence Regulator TcpP, Facilitates the Transition between Symbiotic and Planktonic Lifestyles in Vibrio fischeri. mBio 2020, 11, e01624-20. [Google Scholar] [CrossRef]
- Ruby, E.G.; Asato, L.M. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch. Microbiol. 1993, 159, 160–167. [Google Scholar] [CrossRef]
- Amy, P.S.; Pauling, C.; Morita, R.Y. Recovery from nutrient starvation by a marine Vibrio sp. Appl. Environ. Microbiol. 1983, 45, 1685–1690. [Google Scholar] [CrossRef]
- Baker, R.M.; Singleton, F.L.; Hood, M.A. Effects of nutrient deprivation on Vibrio cholerae. Appl. Environ. Microbiol. 1983, 46, 930–940. [Google Scholar] [CrossRef]
- Malmcrona-Friberg, K.; Goodman, A.; Kjelleberg, S. Chemotactic Responses of Marine Vibrio sp. Strain S14 (CCUG 15956) to Low-Molecular-Weight Substances under Starvation and Recovery Conditions. Appl. Environ. Microbiol. 1990, 56, 3699–3704. [Google Scholar] [CrossRef]
- Moreno-Gámez, S.; Sorg, R.A.; Domenech, A.; Kjos, M.; Weissing, F.J.; van Doorn, G.S.; Veening, J.-W. Quorum sensing integrates environmental cues, cell density and cell history to control bacterial competence. Nat. Commun. 2017, 8, 1–12. [Google Scholar] [CrossRef]
- Keller, L.; Surette, M.G. Communication in bacteria: An ecological and evolutionary perspective. Nat. Rev. Genet. 2006, 4, 249–258. [Google Scholar] [CrossRef]
- Cao, M.; Goodrich-Blair, H. Ready or Not: Microbial Adaptive Responses in Dynamic Symbiosis Environments. J. Bacteriol. 2017, 199, e00883-16. [Google Scholar] [CrossRef]
- Wilson, C.M.; Aggio, R.B.M.; O’Toole, P.W.; Villas-Boas, S.; Tannock, C.W. Transcriptional and metabolomic consequences of LuxS inactivation reveal a metabolic rather than quorum-sensing role for luxS in Lactobacillus reuteri 100-23. J. Bacteriol. 2012, 194, 1743–1746. [Google Scholar] [CrossRef]
- Aschtgen, M.S.; Brennan, C.A.; Nikolakakis, K.; Cohen, S.; McFall-Ngai, M.; Ruby, E.G. Insights into flagellar function and mecha-nism from the squid-Vibrio symbiosis. NPJ Biofilms Microbiomes 2019, 5, 32. [Google Scholar] [CrossRef]
- Millikan, D.S.; Ruby, E.G. Alterations in Vibrio fischeri Motility Correlate with a Delay in Symbiosis Initiation and Are Associated with Additional Symbiotic Colonization Defects. Appl. Environ. Microbiol. 2002, 68, 2519–2528. [Google Scholar] [CrossRef]
- Akerley, B.J.; Monack, D.M.; Falkow, S.; Miller, J.F. The bvgAS locus negatively controls motility and synthesis of flagella in Bordetella bronchiseptica. J. Bacteriol. 1992, 174, 980–990. [Google Scholar] [CrossRef]
- Lupp, C.; Ruby, E.G. Vibrio fischeri Uses Two Quorum-Sensing Systems for the Regulation of Early and Late Colonization Factors. J. Bacteriol. 2005, 187, 3620–3629. [Google Scholar] [CrossRef]
- Li, Y.; Yan, J.; Guo, X.; Wang, X.; Liu, F.; Cao, B. The global regulators ArcA and CytR collaboratively modulate Vibrio cholerae motility. BMC Microbiol. 2022, 22, 22. [Google Scholar] [CrossRef]
- Jones, B.W.; Nishiguchi, M.K. Differentially expressed genes reveal adaptations between free-living and symbiotic niches of Vibrio fischeri in a fully established mutualism. Can. J. Microbiol. 2006, 52, 1218–1227. [Google Scholar] [CrossRef][Green Version]
- Ferreira, J.L.; Gao, F.Z.; Rossmann, F.M.; Nans, A.; Brenzinger, S.; Hosseini, R.; Wilson, A.; Briegel, A.; Thormann, K.M.; Rosenthal, P.B.; et al. γ-proteobacteria eject their polar flagella under nutrient depletion, retaining flagellar motor relic structures. PLoS Biol. 2019, 17, e3000165. [Google Scholar] [CrossRef]
- Schwartzman, J.A.; Lynch, J.B.; Flores Ramos, S.; Zhou, L.; Apicella, M.A.; Yew, J.Y.; Ruby, E.G. Acidic pH promotes lipopolysaccharide modification and alters colonization in a bacteria-animal mutualism. Mol. Microbiol. 2019, 112, 1326–1338. [Google Scholar] [CrossRef]
- Nyholm, S.V.; Graf, J. Knowing your friends: Invertebrate innate immunity fosters beneficial bacterial symbioses. Nat. Rev. Genet. 2012, 10, 815–827. [Google Scholar] [CrossRef]
- McFall-Ngai, M.; Ruby, E. Getting the Message Out: The Many Modes of Host-Symbiont Communication during Early-Stage Establishment of the Squid-Vibrio Partnership. mSystems 2021, 6, e0086721. [Google Scholar] [CrossRef] [PubMed]
- Okada, Y.; Setoyama, H.; Matsumoto, S.; Imaoka, A.; Nanno, M.; Kawaguchi, M.; Umesaki, Y. Effects of fecal microorganisms and their chloroform-resistant variants derived from mice, rats, and humans on immunological and physiological characteristics of the intestines of ex-germfree mice. Infect. Immun. 1994, 62, 5442–5446. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pipes, B.L.; Nishiguchi, M.K. Nocturnal Acidification: A Coordinating Cue in the Euprymna scolopes–Vibrio fischeri Symbiosis. Int. J. Mol. Sci. 2022, 23, 3743. https://doi.org/10.3390/ijms23073743
Pipes BL, Nishiguchi MK. Nocturnal Acidification: A Coordinating Cue in the Euprymna scolopes–Vibrio fischeri Symbiosis. International Journal of Molecular Sciences. 2022; 23(7):3743. https://doi.org/10.3390/ijms23073743
Chicago/Turabian StylePipes, Brian L., and Michele K. Nishiguchi. 2022. "Nocturnal Acidification: A Coordinating Cue in the Euprymna scolopes–Vibrio fischeri Symbiosis" International Journal of Molecular Sciences 23, no. 7: 3743. https://doi.org/10.3390/ijms23073743
APA StylePipes, B. L., & Nishiguchi, M. K. (2022). Nocturnal Acidification: A Coordinating Cue in the Euprymna scolopes–Vibrio fischeri Symbiosis. International Journal of Molecular Sciences, 23(7), 3743. https://doi.org/10.3390/ijms23073743





