Atomic Force Microscopy (AFM) Applications in Arrhythmogenic Cardiomyopathy
Abstract
1. Introduction
2. Cell Mechanics and Mechanobiology
3. AFM—The Mechanical Machine
4. Force Spectroscopy with AFM
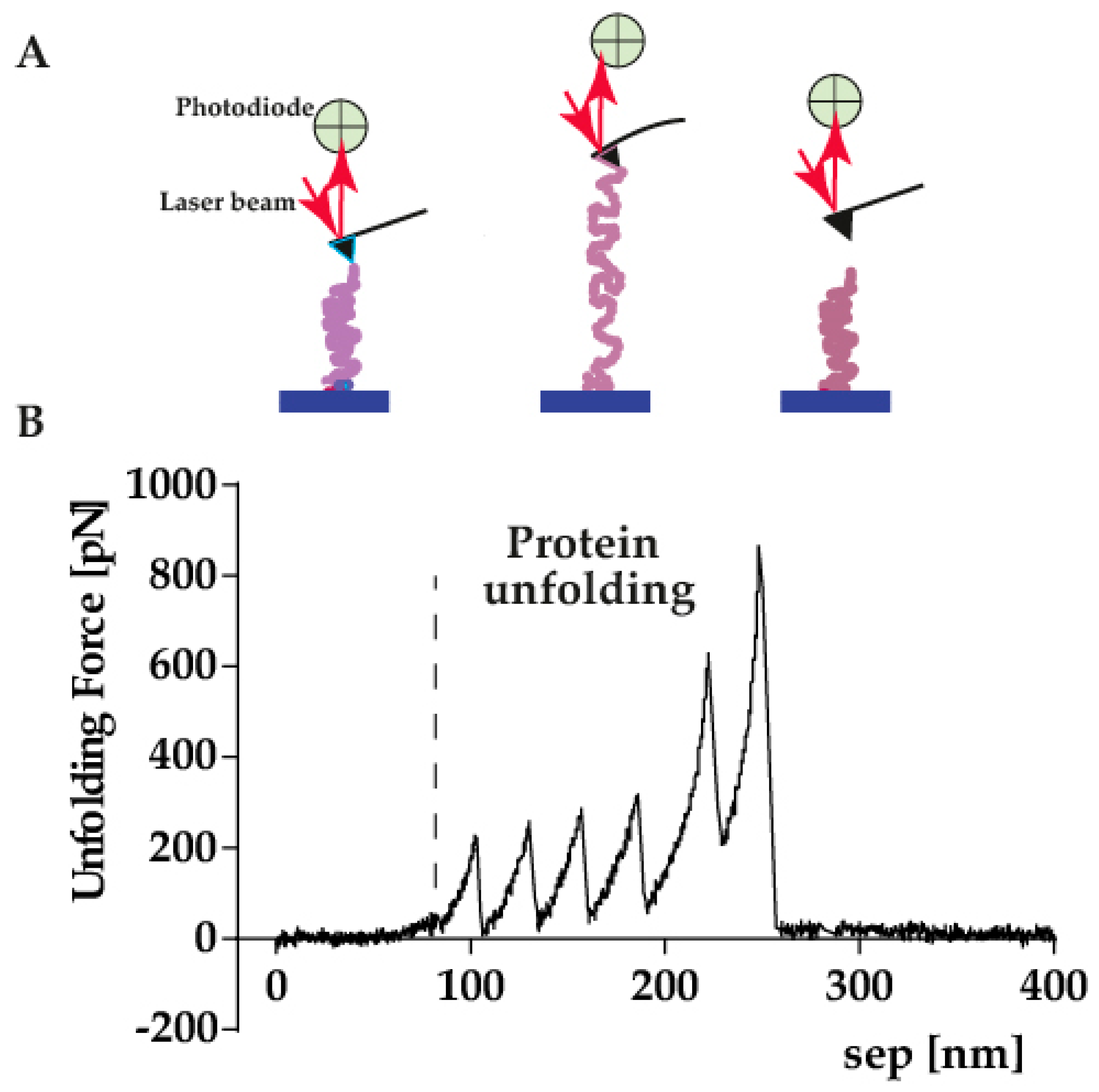
4.1. Single Molecule Force Spectroscopy
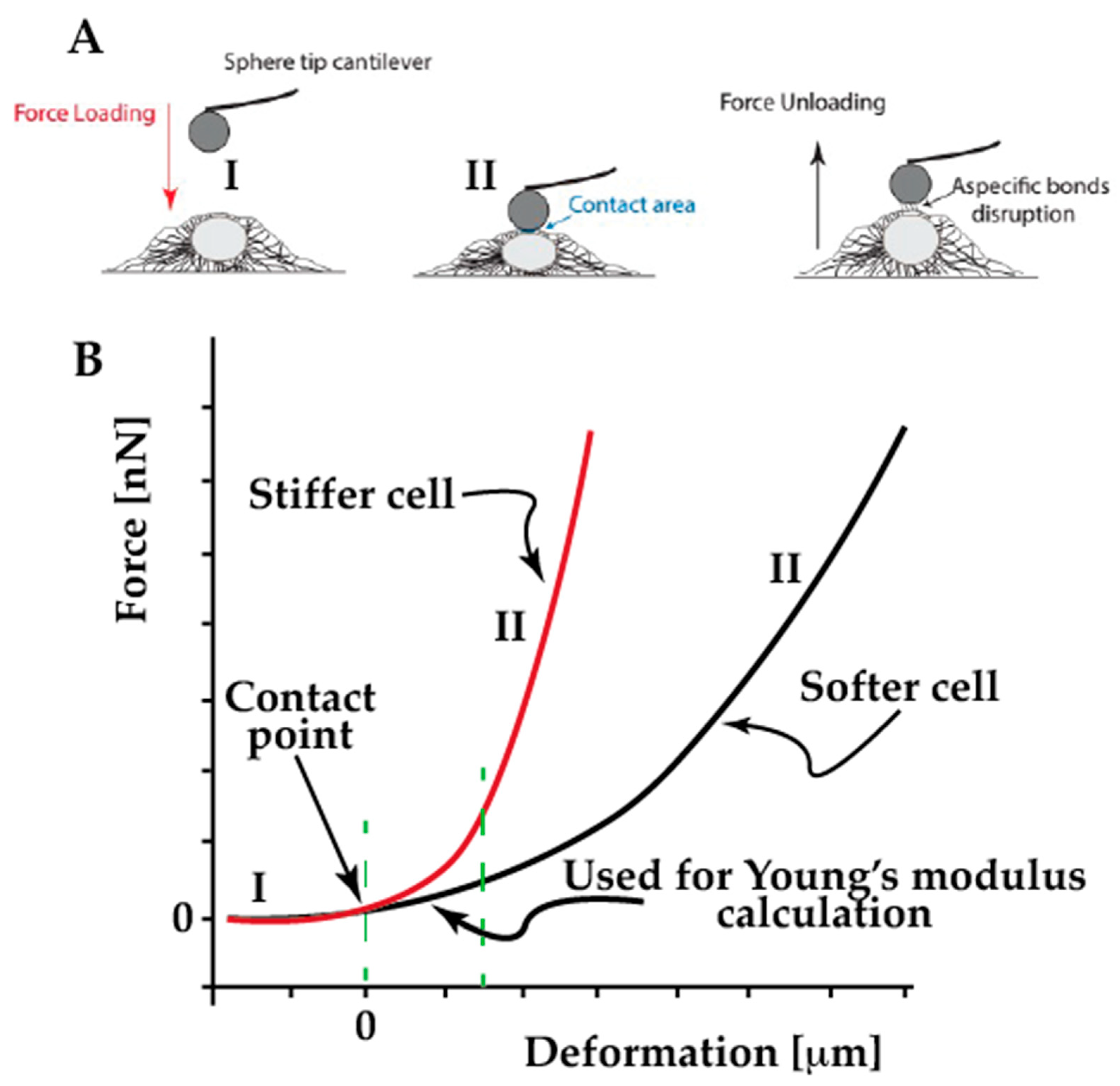
4.2. Single Cell Force Spectroscopy
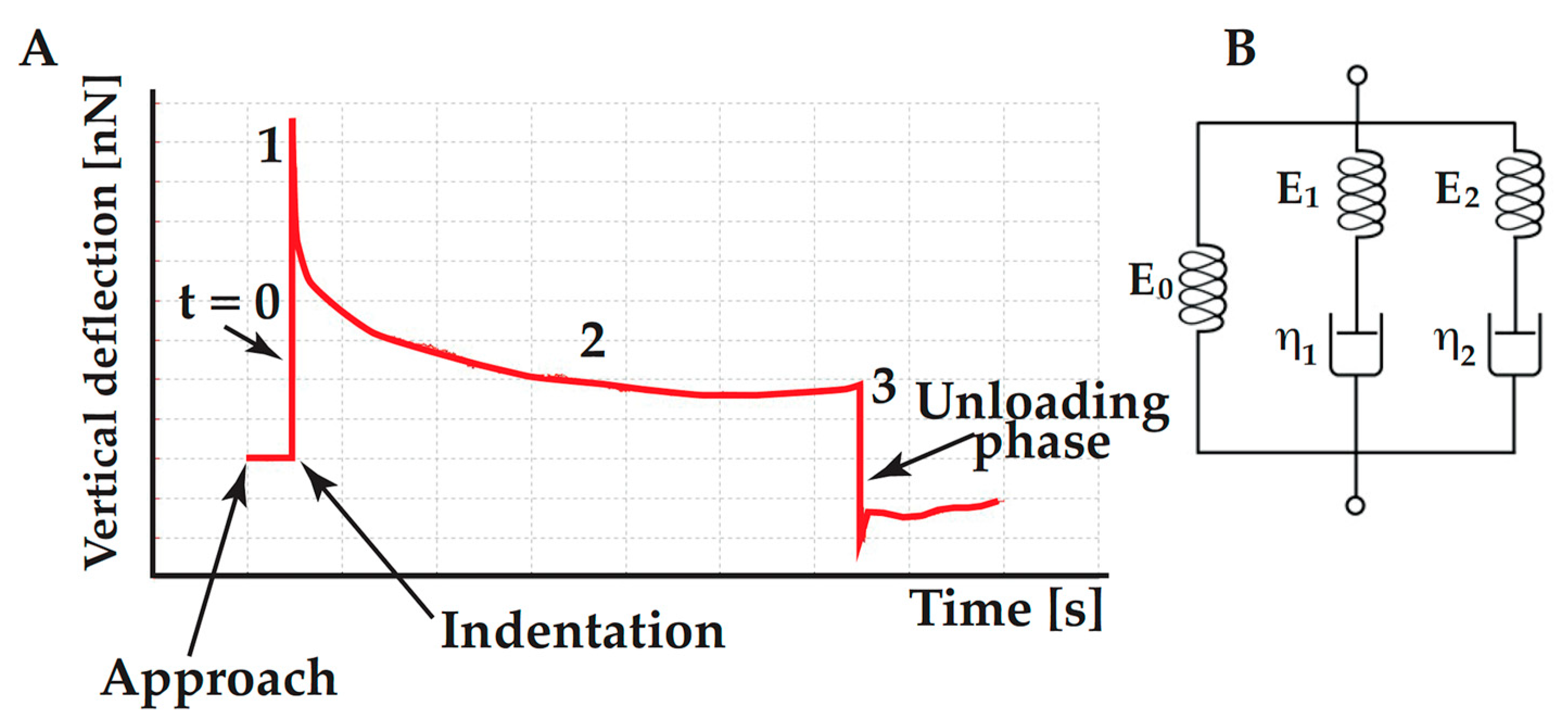
4.3. Assessing the Cell Viscoelasticity
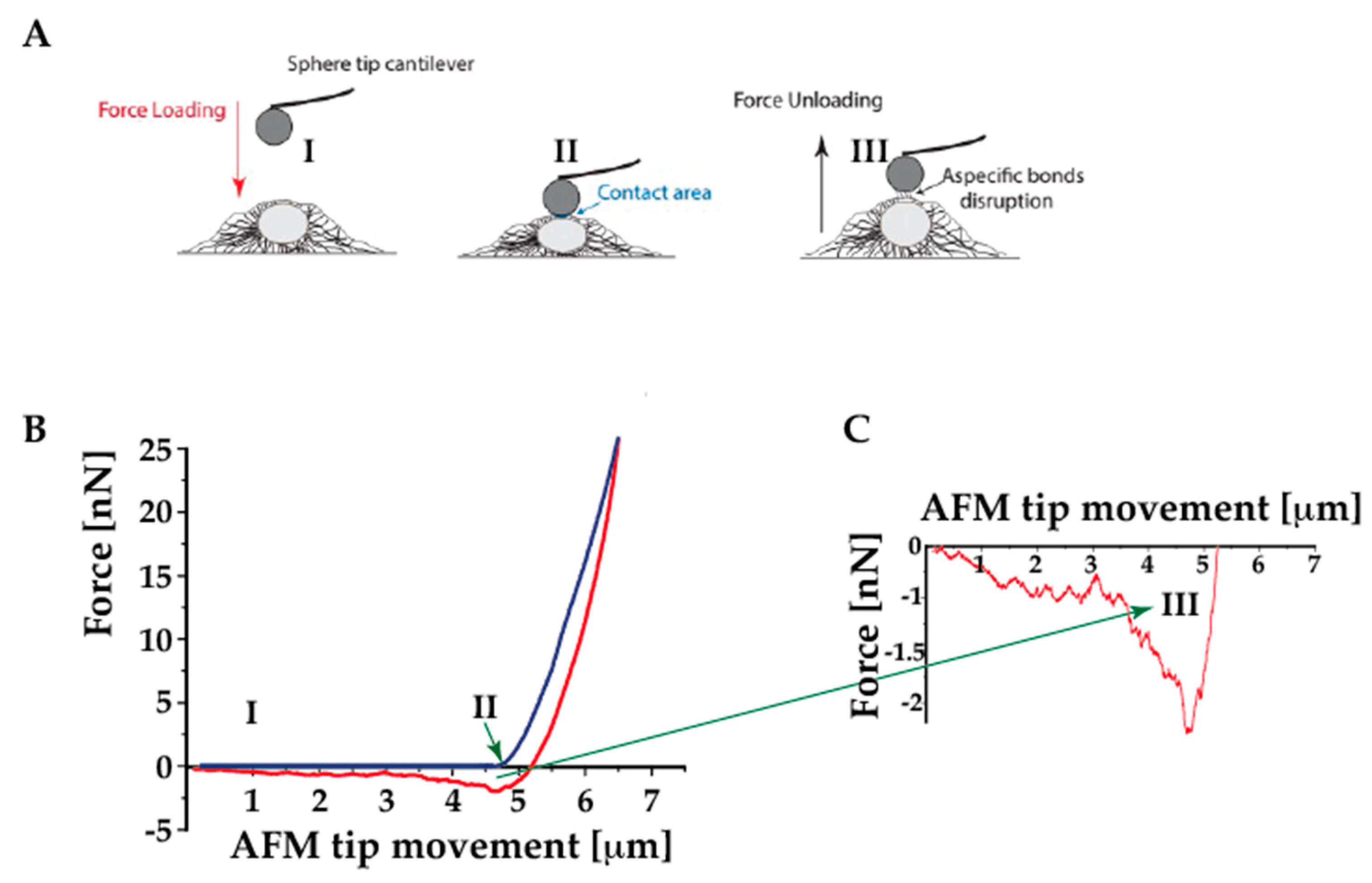
4.4. Assessing the Cell Adhesion Behavior
- (1)
- a cell on a substrate is “touched” by either a tipless cantilever or a cantilever-tip assembly
- (2)
- a cell adhered to the cantilever is brought in contact with either a substrate or another cell.
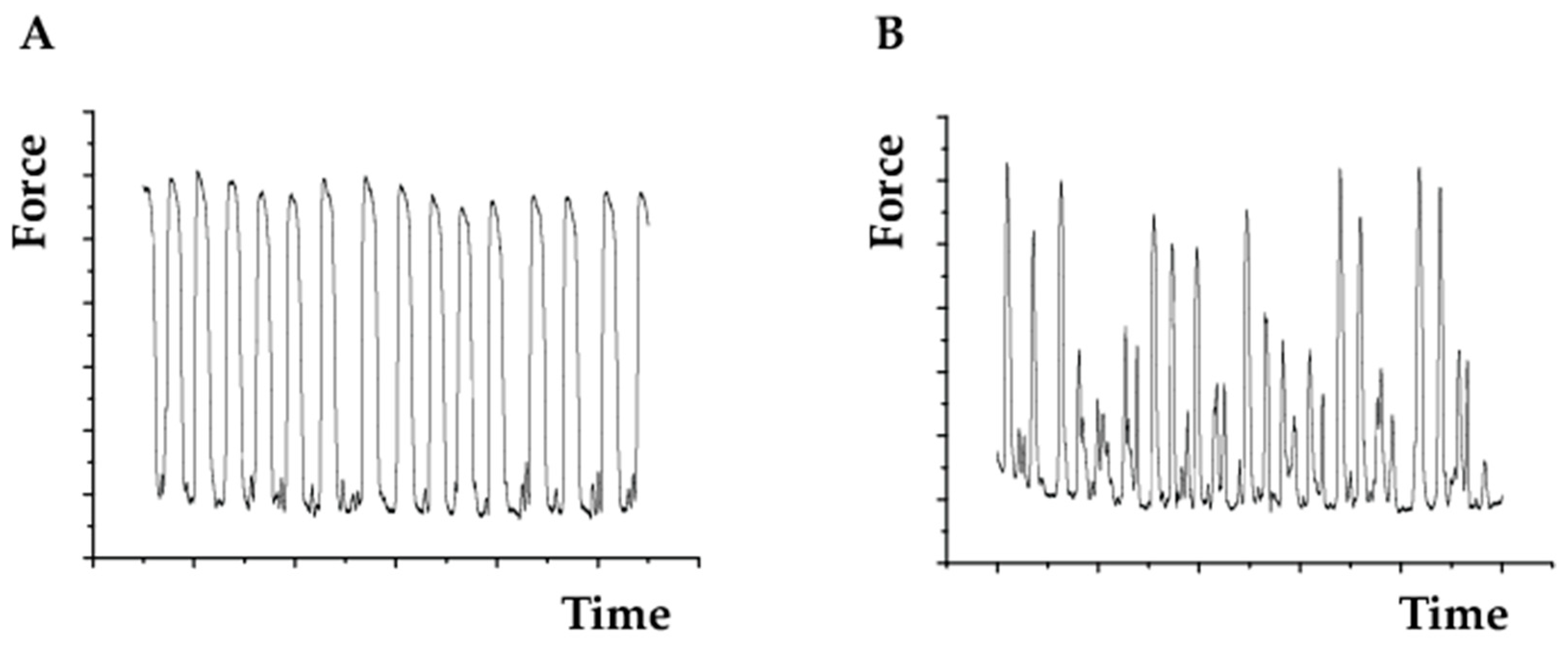
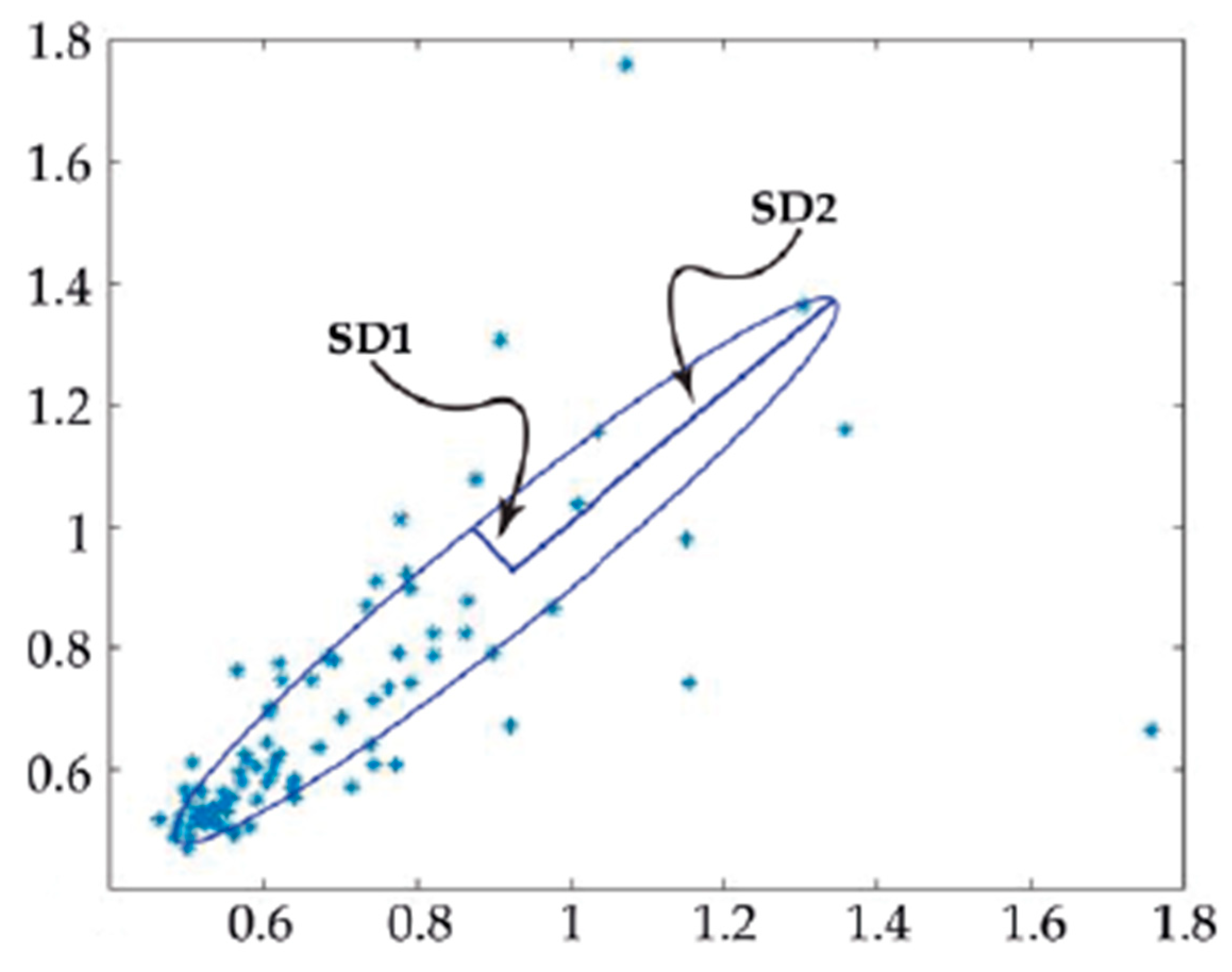
4.5. Assessing Cell Beating Force and Frequency
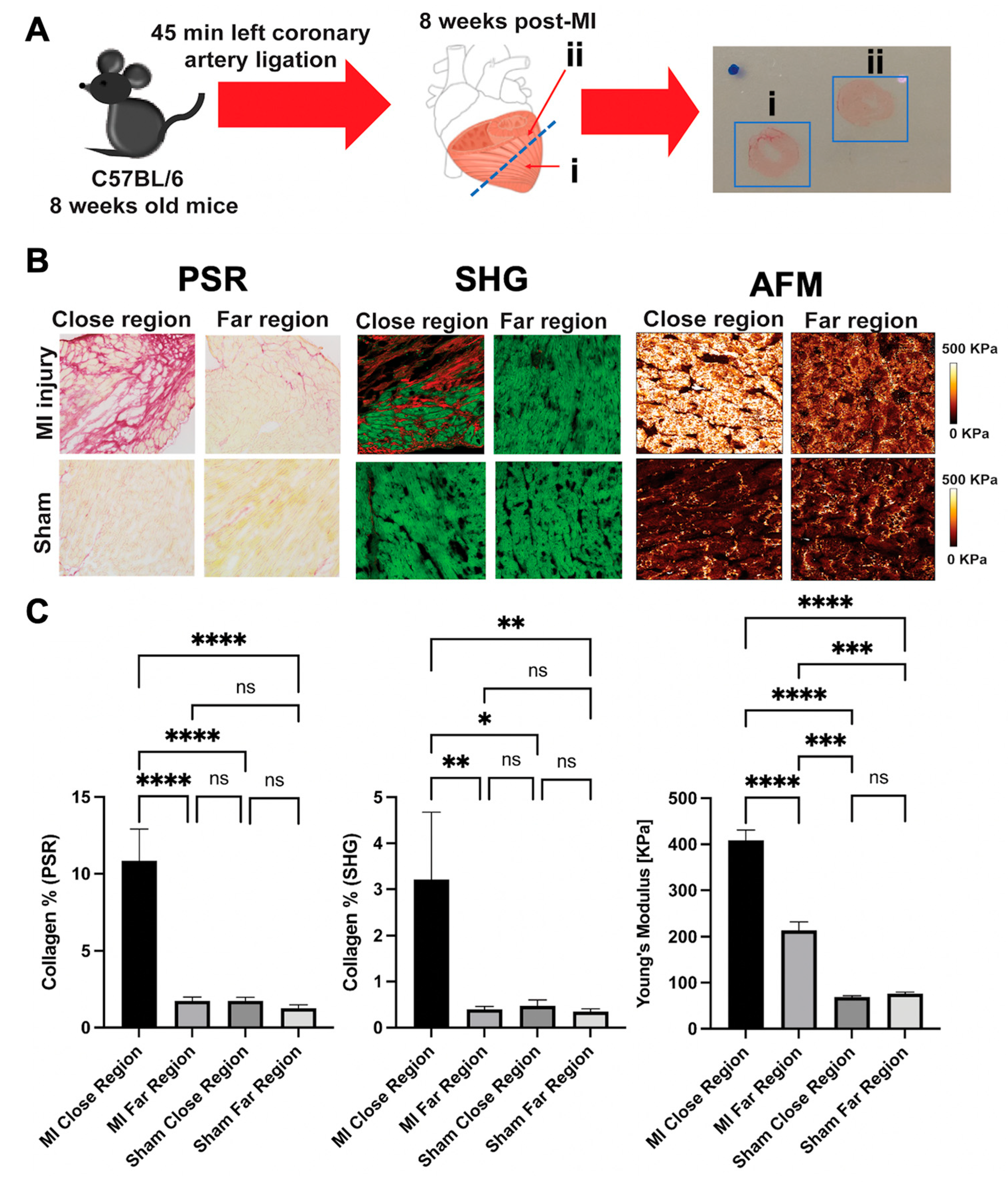
5. AFM for Cardiac Tissue Analysis
Tissue Scanning Methodologies
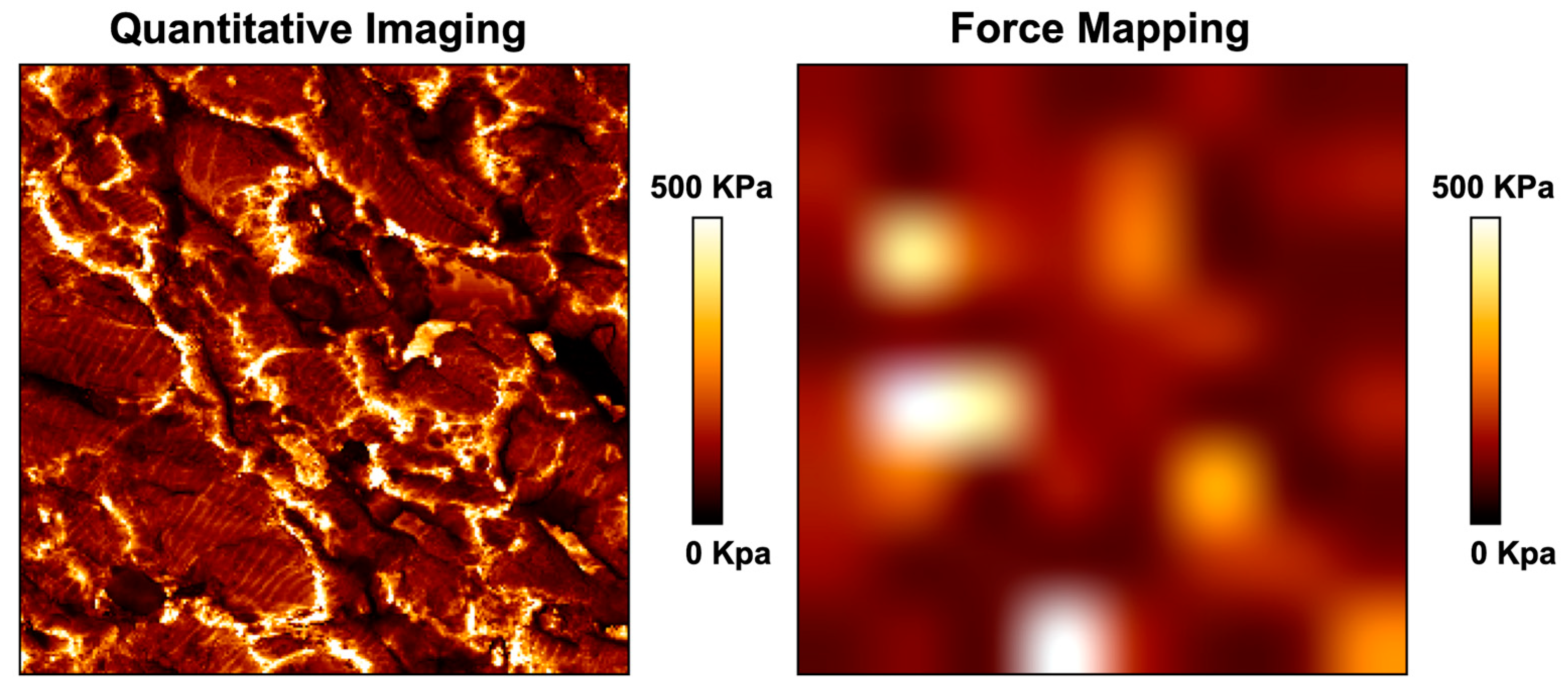
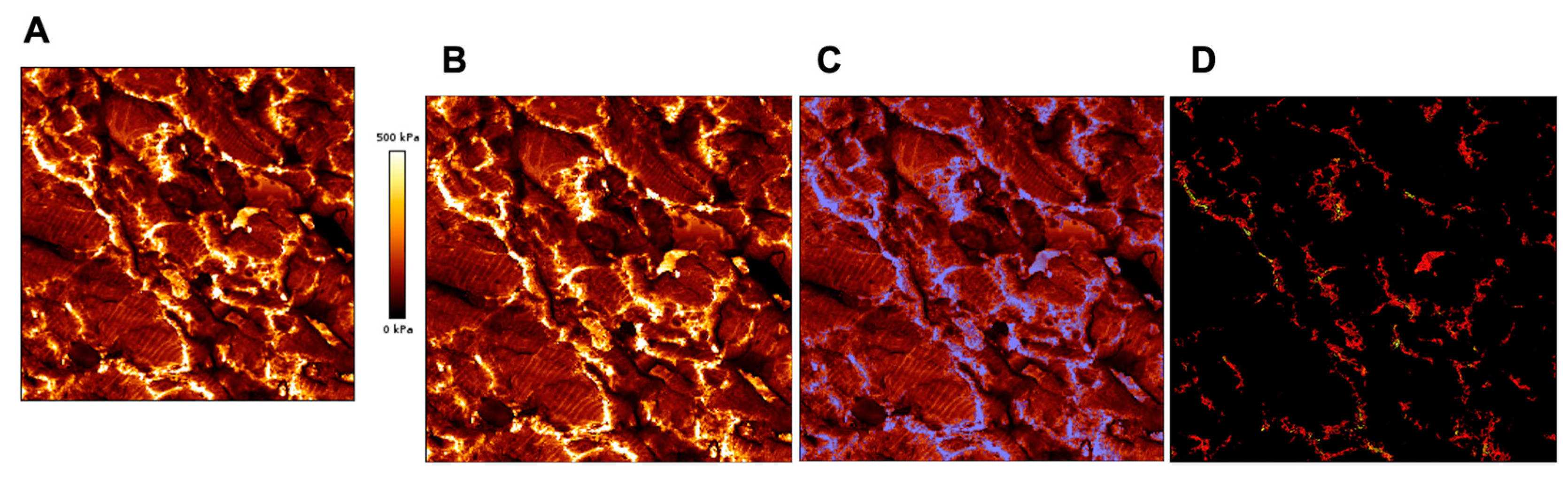
6. AFM as a Complementary Tool to Detect Fibrosis in Cardiac Tissue
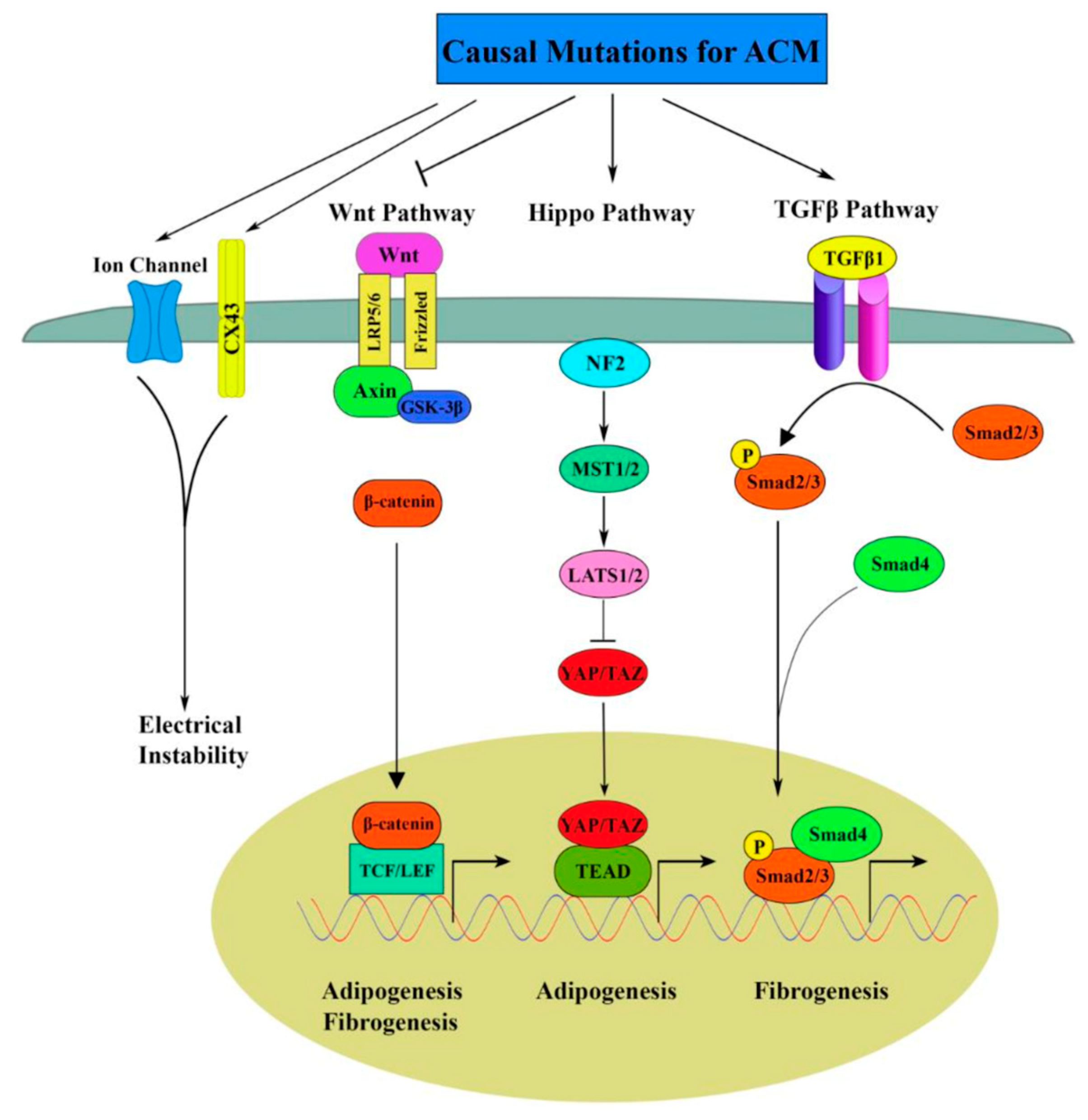
7. AFM and Arrhythmogenic Cardiomyopathy
8. Data Reliability: Variables to Be Considered
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Beffagna, G.; Zorzi, A.; Pilichou, K.; Perazzolo Marra, M.; Rigato, I.; Corrado, D.; Migliore, F.; Rampazzo, A.; Bauce, B.; Basso, C.; et al. Arrhythmogenic Cardiomyopathy. Eur. Heart J. 2020, 41, 4457–4462. [Google Scholar] [CrossRef] [PubMed]
- Thiene, G.; Basso, C. Arrhythmogenic right ventricular cardiomyopathy: An update. Cardiovasc. Pathol. 2001, 10, 109–117. [Google Scholar] [CrossRef]
- Corrado, D.; Basso, C.; Thiene, G. Letter by Corrado et al. regarding article, “sudden deaths in young competitive athletes: Analysis of 1866 deaths in the United States, 1980–2006”. Circulation 2009, 120, e143, author reply e144. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pilichou, K.; Remme, C.A.; Basso, C.; Campian, M.E.; Rizzo, S.; Barnett, P.; Scicluna, B.P.; Bauce, B.; van den Hoff, M.J.; de Bakker, J.M.; et al. Myocyte necrosis underlies progressive myocardial dystrophy in mouse dsg2-related arrhythmogenic right ventricular cardiomyopathy. J. Exp. Med. 2009, 206, 1787–1802. [Google Scholar] [CrossRef] [PubMed]
- Stevens, T.L.; Wallace, M.J.; Refaey, M.E.; Roberts, J.D.; Koenig, S.N.; Mohler, P.J. Arrhythmogenic Cardiomyopathy: Molecular Insights for Improved Therapeutic Design. J. Cardiovasc. Dev. Dis. 2020, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Groeneweg, J.A.; Bhonsale, A.; James, C.A.; te Riele, A.S.; Dooijes, D.; Tichnell, C.; Murray, B.; Wiesfeld, A.C.; Sawant, A.C.; Kassamali, B.; et al. Clinical Presentation, Long-Term Follow-Up, and Outcomes of 1001 Arrhythmogenic Right Ventricular Dysplasia/Cardiomyopathy Patients and Family Members. Circ. Cardiovasc. Genet. 2015, 8, 437–446. [Google Scholar] [CrossRef]
- van Lint, F.H.M.; Murray, B.; Tichnell, C.; Zwart, R.; Amat, N.; Lekanne Deprez, R.H.; Dittmann, S.; Stallmeyer, B.; Calkins, H.; van der Smagt, J.J.; et al. Arrhythmogenic Right Ventricular Cardiomyopathy-Associated Desmosomal Variants Are Rarely De Novo. Circ. Genom. Precis Med. 2019, 12, e002467. [Google Scholar] [CrossRef]
- Gandjbakhch, E.; Redheuil, A.; Pousset, F.; Charron, P.; Frank, R. Clinical Diagnosis, Imaging, and Genetics of Arrhythmogenic Right Ventricular Cardiomyopathy/Dysplasia: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2018, 72, 784–804. [Google Scholar] [CrossRef]
- Rampazzo, A.; Calore, M.; van Hengel, J.; van Roy, F. Intercalated discs and arrhythmogenic cardiomyopathy. Circ. Cardiovasc. Genet. 2014, 7, 930–940. [Google Scholar] [CrossRef]
- Calore, M.; Lorenzon, A.; De Bortoli, M.; Poloni, G.; Rampazzo, A. Arrhythmogenic cardiomyopathy: A disease of intercalated discs. Cell Tissue Res. 2015, 360, 491–500. [Google Scholar] [CrossRef]
- Baskin, B.; Skinner, J.R.; Sanatani, S.; Terespolsky, D.; Krahn, A.D.; Ray, P.N.; Scherer, S.W.; Hamilton, R.M. TMEM43 mutations associated with arrhythmogenic right ventricular cardiomyopathy in non-Newfoundland populations. Hum. Genet. 2013, 132, 1245–1252. [Google Scholar] [CrossRef] [PubMed]
- Mayosi, B.M.; Fish, M.; Shaboodien, G.; Mastantuono, E.; Kraus, S.; Wieland, T.; Kotta, M.C.; Chin, A.; Laing, N.; Ntusi, N.B.; et al. Identification of Cadherin 2. Circ. Cardiovasc. Genet. 2017, 10, e001605. [Google Scholar] [CrossRef] [PubMed]
- Quarta, G.; Syrris, P.; Ashworth, M.; Jenkins, S.; Zuborne Alapi, K.; Morgan, J.; Muir, A.; Pantazis, A.; McKenna, W.J.; Elliott, P.M. Mutations in the Lamin A/C gene mimic arrhythmogenic right ventricular cardiomyopathy. Eur. Heart J. 2012, 33, 1128–1136. [Google Scholar] [CrossRef] [PubMed]
- Captur, G.; Arbustini, E.; Bonne, G.; Syrris, P.; Mills, K.; Wahbi, K.; Mohiddin, S.A.; McKenna, W.J.; Pettit, S.; Ho, C.Y.; et al. Lamin and the heart. Heart 2018, 104, 468–479. [Google Scholar] [CrossRef]
- Mestroni, L.; Sbaizero, O. Arrhythmogenic Cardiomyopathy: Mechanotransduction Going Wrong. Circulation 2018, 137, 1611–1613. [Google Scholar] [CrossRef]
- van Hengel, J.; Calore, M.; Bauce, B.; Dazzo, E.; Mazzotti, E.; De Bortoli, M.; Lorenzon, A.; Li Mura, I.E.; Beffagna, G.; Rigato, I.; et al. Mutations in the area composita protein αT-catenin are associated with arrhythmogenic right ventricular cardiomyopathy. Eur. Heart J. 2013, 34, 201–210. [Google Scholar] [CrossRef]
- Roberts, J.D. TJP1 Mutations in Arrhythmogenic Cardiomyopathy. Circ. Genom. Precis Med. 2018, 11, e002337. [Google Scholar] [CrossRef]
- Thiene, G. The research venture in arrhythmogenic right ventricular cardiomyopathy: A paradigm of translational medicine. Eur. Heart J. 2015, 36, 837–846. [Google Scholar] [CrossRef]
- Karmouch, J.; Protonotarios, A.; Syrris, P. Genetic basis of arrhythmogenic cardiomyopathy. Curr. Opin. Cardiol. 2018, 33, 276–281. [Google Scholar] [CrossRef]
- Hoffman, B.D.; Grashoff, C.; Schwartz, M.A. Dynamic molecular processes mediate cellular mechanotransduction. Nature 2011, 475, 316–323. [Google Scholar] [CrossRef]
- Howard, J.; Grill, S.W.; Bois, J.S. Turing’s next steps: The mechanochemical basis of morphogenesis. Nat. Rev. Mol. Cell Biol. 2011, 12, 392–398. [Google Scholar] [CrossRef]
- Petridou, N.I.; Spiró, Z.; Heisenberg, C.P. Multiscale force sensing in development. Nat. Cell Biol. 2017, 19, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Crest, J.; Diz-Muñoz, A.; Chen, D.Y.; Fletcher, D.A.; Bilder, D. Organ sculpting by patterned extracellular matrix stiffness. eLife 2017, 6, e24958. [Google Scholar] [CrossRef] [PubMed]
- Brand, R.A. Biographical sketch: Julius Wolff, 1836–1902. Clin. Orthop. Relat. Res. 2010, 468, 1047–1049. [Google Scholar] [CrossRef] [PubMed]
- Paluch, E.K.; Nelson, C.M.; Biais, N.; Fabry, B.; Moeller, J.; Pruitt, B.L.; Wollnik, C.; Kudryasheva, G.; Rehfeldt, F.; Federle, W. Mechanotransduction: Use the force(s). BMC Biol. 2015, 13, 47. [Google Scholar] [CrossRef]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix elasticity directs stem cell lineage specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef]
- Liu, N.; Zhou, M.; Zhang, Q.; Yong, L.; Zhang, T.; Tian, T.; Ma, Q.; Lin, S.; Zhu, B.; Cai, X. Effect of substrate stiffness on proliferation and differentiation of periodontal ligament stem cells. Cell Prolif. 2018, 51, e12478. [Google Scholar] [CrossRef]
- Gilbert, P.M.; Havenstrite, K.L.; Magnusson, K.E.; Sacco, A.; Leonardi, N.A.; Kraft, P.; Nguyen, N.K.; Thrun, S.; Lutolf, M.P.; Blau, H.M. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science 2010, 329, 1078–1081. [Google Scholar] [CrossRef]
- Denais, C.; Lammerding, J. Nuclear mechanics in cancer. Adv. Exp. Med. Biol. 2014, 773, 435–470. [Google Scholar] [CrossRef]
- Kim, C.; Wong, J.; Wen, J.; Wang, S.; Wang, C.; Spiering, S.; Kan, N.G.; Forcales, S.; Puri, P.L.; Leone, T.C.; et al. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature 2013, 494, 105–110. [Google Scholar] [CrossRef]
- Binnig, G.; Quate, C.F.; Gerber, C. Atomic force microscope. Phys. Rev. Lett. 1986, 56, 930–933. [Google Scholar] [CrossRef] [PubMed]
- Benitez, R.; Toca-Herrera, J.L. Looking at cell mechanics with atomic force microscopy: Experiment and theory. Microsc. Res. Tech. 2014, 77, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Friedrichs, J.; Legate, K.R.; Schubert, R.; Bharadwaj, M.; Werner, C.; Müller, D.J.; Benoit, M. A practical guide to quantify cell adhesion using single-cell force spectroscopy. Methods 2013, 60, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Liu, Z.; Liu, H.; Nash, M.A. Next Generation Methods for Single-Molecule Force Spectroscopy on Polyproteins and Receptor-Ligand Complexes. Front. Mol. Biosci. 2020, 7, 85. [Google Scholar] [CrossRef]
- Gavara, N. A beginner’s guide to atomic force microscopy probing for cell mechanics. Microsc. Res. Tech. 2017, 80, 75–84. [Google Scholar] [CrossRef]
- Moreno-Cencerrado, A.; Iturri, J.; Pecorari, I.; Vivanco, M.D.M.; Sbaizero, O.; Toca-Herrera, J.L. Investigating cell-substrate and cell-cell interactions by means of single-cell-probe force spectroscopy. Microsc. Res. Tech. 2017, 80, 124–130. [Google Scholar] [CrossRef]
- Thomas, G.; Burnham, N.A.; Camesano, T.A.; Wen, Q. Measuring the mechanical properties of living cells using atomic force microscopy. J. Vis. Exp. 2013, 76, 50497. [Google Scholar] [CrossRef]
- Sirghi, L.; Ponti, J.; Broggi, F.; Rossi, F. Probing elasticity and adhesion of live cells by atomic force microscopy indentation. Eur. Biophys. J. 2008, 37, 935–945. [Google Scholar] [CrossRef]
- Kuznetsova, T.G.; Starodubtseva, M.N.; Yegorenkov, N.I.; Chizhik, S.A.; Zhdanov, R.I. Atomic force microscopy probing of cell elasticity. Micron 2007, 38, 824–833. [Google Scholar] [CrossRef]
- Brückner, B.R.; Janshoff, A. Elastic properties of epithelial cells probed by atomic force microscopy. Biochim. Biophys. Acta 2015, 1853, 3075–3082. [Google Scholar] [CrossRef]
- Chiang, M.Y.; Yangben, Y.; Lin, N.J.; Zhong, J.L.; Yang, L. Relationships among cell morphology, intrinsic cell stiffness and cell-substrate interactions. Biomaterials 2013, 34, 9754–9762. [Google Scholar] [CrossRef] [PubMed]
- Lekka, M.; Fornal, M.; Pyka-Fościak, G.; Lebed, K.; Wizner, B.; Grodzicki, T.; Styczeń, J. Erythrocyte stiffness probed using atomic force microscope. Biorheology 2005, 42, 307–317. [Google Scholar] [PubMed]
- Dulińska, I.; Targosz, M.; Strojny, W.; Lekka, M.; Czuba, P.; Balwierz, W.; Szymoński, M. Stiffness of normal and pathological erythrocytes studied by means of atomic force microscopy. J. Biochem. Biophys. Methods 2006, 66, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Benech, J.C.; Benech, N.; Zambrana, A.I.; Rauschert, I.; Bervejillo, V.; Oddone, N.; Damián, J.P. Diabetes increases stiffness of live cardiomyocytes measured by atomic force microscopy nanoindentation. Am. J. Physiol. Cell Physiol. 2014, 307, C910–C919. [Google Scholar] [CrossRef]
- Laurini, E.; Martinelli, V.; Lanzicher, T.; Puzzi, L.; Borin, D.; Chen, S.N.; Long, C.S.; Lee, P.; Mestroni, L.; Taylor, M.R.G.; et al. Biomechanical defects and rescue of cardiomyocytes expressing pathologic nuclear lamins. Cardiovasc. Res. 2018, 114, 846–857. [Google Scholar] [CrossRef]
- Puzzi, L.; Borin, D.; Martinelli, V.; Mestroni, L.; Kelsell, D.P.; Sbaizero, O. Cellular biomechanics impairment in keratinocytes is associated with a C-terminal truncated desmoplakin: An atomic force microscopy investigation. Micron 2018, 106, 27–33. [Google Scholar] [CrossRef]
- Jeffrey, D.A.; Da Silva, J.P.; Garcia, A.M.; Jiang, X.; Karimpour-Fard, A.; Toni, L.S.; Lanzicher, T.; Pena, B.; Miyano, C.A.; Nunley, K.; et al. Serum circulating proteins from pediatric patients with dilated cardiomyopathy cause pathologic remodeling and cardiomyocyte stiffness. JCI Insight 2021, 6, e148637. [Google Scholar] [CrossRef]
- Moreno-Flores, S.; Benitez, R.; Vivanco, M.; Toca-Herrera, J.L. Stress relaxation and creep on living cells with the atomic force microscope: A means to calculate elastic moduli and viscosities of cell components. Nanotechnology 2010, 21, 445101. [Google Scholar] [CrossRef]
- Khalili, A.A.; Ahmad, M.R. A Review of Cell Adhesion Studies for Biomedical and Biological Applications. Int. J. Mol. Sci. 2015, 16, 18149–18184. [Google Scholar] [CrossRef]
- Lachaize, V.; Pena, B.; Ciubotaru, C.; Cojoc, D.; Chen, S.N.; Taylor, M.R.G.; Mestroni, L.; Sbaizero, O. Compromised Biomechanical Properties, Cell-Cell Adhesion and Nanotubes Communication in Cardiac Fibroblasts Carrying the Lamin A/C D192G Mutation. Int. J. Mol. Sci. 2021, 22, 9193. [Google Scholar] [CrossRef]
- Pena, B.; Bosi, S.; Aguado, B.A.; Borin, D.; Farnsworth, N.L.; Dobrinskikh, E.; Rowland, T.J.; Martinelli, V.; Jeong, M.; Taylor, M.R.G.; et al. Injectable Carbon Nanotube-Functionalized Reverse Thermal Gel Promotes Cardiomyocytes Survival and Maturation. ACS Appl. Mater. Interfaces 2017, 9, 31645–31656. [Google Scholar] [CrossRef] [PubMed]
- Tulppo, M.P.; Mäkikallio, T.H.; Takala, T.E.; Seppänen, T.; Huikuri, H.V. Quantitative beat-to-beat analysis of heart rate dynamics during exercise. Am. J. Physiol. 1996, 271, H244–H252. [Google Scholar] [CrossRef] [PubMed]
- Ai, Z.; Fischer, A.; Spray, D.C.; Brown, A.M.; Fishman, G.I. Wnt-1 regulation of connexin43 in cardiac myocytes. J. Clin. Investig. 2000, 105, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Müller, D.J.; Dufrêne, Y.F. Atomic force microscopy: A nanoscopic window on the cell surface. Trends Cell Biol. 2011, 21, 461–469. [Google Scholar] [CrossRef]
- Chiou, Y.W.; Lin, H.K.; Tang, M.J.; Lin, H.H.; Yeh, M.L. The influence of physical and physiological cues on atomic force microscopy-based cell stiffness assessment. PLoS ONE 2013, 8, e77384. [Google Scholar] [CrossRef]
- Smolyakov, G.; Formosa-Dague, C.; Severac, C.; Duval, R.E.; Dague, E. High speed indentation measures by FV, QI and QNM introduce a new understanding of bionanomechanical experiments. Micron 2016, 85, 8–14. [Google Scholar] [CrossRef]
- Laklai, H.; Miroshnikova, Y.A.; Pickup, M.W.; Collisson, E.A.; Kim, G.E.; Barrett, A.S.; Hill, R.C.; Lakins, J.N.; Schlaepfer, D.D.; Mouw, J.K.; et al. Genotype tunes pancreatic ductal adenocarcinoma tissue tension to induce matricellular fibrosis and tumor progression. Nat. Med. 2016, 22, 497–505. [Google Scholar] [CrossRef]
- Graham, H.K.; Hodson, N.W.; Hoyland, J.A.; Millward-Sadler, S.J.; Garrod, D.; Scothern, A.; Griffiths, C.E.; Watson, R.E.; Cox, T.R.; Erler, J.T.; et al. Tissue section AFM: In situ ultrastructural imaging of native biomolecules. Matrix Biol. 2010, 29, 254–260. [Google Scholar] [CrossRef]
- Sewell-Loftin, M.K.; Brown, C.B.; Baldwin, H.S.; Merryman, W.D. A novel technique for quantifying mouse heart valve leaflet stiffness with atomic force microscopy. J. Heart Valve Dis. 2012, 21, 513–520. [Google Scholar]
- Collier, D.M.; Peterson, Z.J.; Blokhin, I.O.; Benson, C.J.; Snyder, P.M. Identification of extracellular domain residues required for epithelial Na+ channel activation by acidic pH. J. Biol. Chem. 2012, 287, 40907–40914. [Google Scholar] [CrossRef]
- Travers, J.G.; Wennersten, S.A.; Pena, B.; Bagchi, R.A.; Smith, H.E.; Hirsch, R.A.; Vanderlinden, L.A.; Lin, Y.H.; Dobrinskikh, E.; Demos-Davies, K.M.; et al. HDAC Inhibition Reverses Preexisting Diastolic Dysfunction and Blocks Covert Extracellular Matrix Remodeling. Circulation 2021, 143, 1874–1890. [Google Scholar] [CrossRef] [PubMed]
- Usukura, E.; Narita, A.; Yagi, A.; Sakai, N.; Uekusa, Y.; Imaoka, Y.; Ito, S.; Usukura, J. A Cryosectioning Technique for the Observation of Intracellular Structures and Immunocytochemistry of Tissues in Atomic Force Microscopy (AFM). Sci. Rep. 2017, 7, 6462. [Google Scholar] [CrossRef] [PubMed]
- Jacot, J.G.; Martin, J.C.; Hunt, D.L. Mechanobiology of cardiomyocyte development. J. Biomech. 2010, 43, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Berry, M.F.; Engler, A.J.; Woo, Y.J.; Pirolli, T.J.; Bish, L.T.; Jayasankar, V.; Morine, K.J.; Gardner, T.J.; Discher, D.E.; Sweeney, H.L. Mesenchymal stem cell injection after myocardial infarction improves myocardial compliance. Am. J. Physiol. Heart Circ. Physiol 2006, 290, H2196–H2203. [Google Scholar] [CrossRef]
- Shen, J.; Xie, Y.; Liu, Z.; Zhang, S.; Wang, Y.; Jia, L.; Cai, Z.; Ma, H.; Xiang, M. Increased myocardial stiffness activates cardiac microvascular endothelial cell via VEGF paracrine signaling in cardiac hypertrophy. J. Mol. Cell Cardiol. 2018, 122, 140–151. [Google Scholar] [CrossRef]
- Fu, X.; Khalil, H.; Kanisicak, O.; Boyer, J.G.; Vagnozzi, R.J.; Maliken, B.D.; Sargent, M.A.; Prasad, V.; Valiente-Alandi, I.; Blaxall, B.C.; et al. Specialized fibroblast differentiated states underlie scar formation in the infarcted mouse heart. J. Clin. Investig. 2018, 128, 2127–2143. [Google Scholar] [CrossRef]
- Yang, H.; Liu, Y.; Lu, X.-L.; Li, X.-H.; Zhang, H.-G. Transmembrane transport of the Gαq protein carboxyl terminus imitation polypeptide GCIP-27. Eur. J. Pharm. Sci. 2013, 49, 791–799. [Google Scholar] [CrossRef]
- Kliche, K.; Kuhn, M.; Hillebrand, U.; Ludwig, Y.; Stock, C.; Oberleithner, H. Direct aldosterone action on mouse cardiomyocytes detected with atomic force microscopy. Cell Physiol Biochem. 2006, 18, 265–274. [Google Scholar] [CrossRef]
- Dague, E.; Genet, G.; Lachaize, V.; Guilbeau-Frugier, C.; Fauconnier, J.; Mias, C.; Payré, B.; Chopinet, L.; Alsteens, D.; Kasas, S.; et al. Atomic force and electron microscopic-based study of sarcolemmal surface of living cardiomyocytes unveils unexpected mitochondrial shift in heart failure. J. Mol. Cell Cardiol. 2014, 74, 162–172. [Google Scholar] [CrossRef]
- Wang, L.; Chen, T.; Zhou, X.; Huang, Q.; Jin, C. Atomic force microscopy observation of lipopolysaccharide-induced cardiomyocyte cytoskeleton reorganization. Micron 2013, 51, 48–53. [Google Scholar] [CrossRef]
- Saneyasu, T.; Akhtar, R.; Sakai, T. Molecular Cues Guiding Matrix Stiffness in Liver Fibrosis. BioMed Res. Int. 2016, 2016, 2646212. [Google Scholar] [CrossRef] [PubMed]
- Urbanczyk, M.; Layland, S.L.; Schenke-Layland, K. The role of extracellular matrix in biomechanics and its impact on bioengineering of cells and 3D tissues. Matrix Biol. 2020, 85–86, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, M.; Wirrig, E.; Phelps, A.; Wessels, A. Extracellular Matrix and Heart Development. Birth Defects Res. Part A-Clin. Mol. Teratol. 2011, 91, 535–550. [Google Scholar] [CrossRef] [PubMed]
- Little, C.D.; Rongish, B.J. The Extracellular-Matrix during Heart Development. Experientia 1995, 51, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Coker, M.L.; Spinale, F.G. Myocardial extracellular matrix remodeling with the development of pacing induced congestive heart failure: Contributory mechanisms. Cardiovasc. Pathol. 1998, 7, 161–168. [Google Scholar] [CrossRef]
- Voorhees, A.P.; Han, H.C. Biomechanics of Cardiac Function. Compr. Physiol. 2015, 5, 1623–1644. [Google Scholar] [CrossRef]
- Avazmohammadi, R.; Soares, J.S.; Li, D.S.; Raut, S.S.; Gorman, R.C.; Sacks, M.S. A Contemporary Look at Biomechanical Models of Myocardium. Annu. Rev. Biomed. Eng. 2019, 21, 417–442. [Google Scholar] [CrossRef]
- Emig, R.; Knodt, W.; Krussig, M.J.; Zgierski-Johnston, C.M.; Gorka, O.; Gross, O.; Kohl, P.; Ravens, U.; Peyronnet, R. Piezo1 Channels Contribute to the Regulation of Human Atrial Fibroblast Mechanical Properties and Matrix Stiffness Sensing. Cells 2021, 10, 663. [Google Scholar] [CrossRef]
- Ranjit, S.; Dvornikov, A.; Stakic, M.; Hong, S.H.; Levi, M.; Evans, R.M.; Gratton, E. Imaging Fibrosis and Separating Collagens using Second Harmonic Generation and Phasor Approach to Fluorescence Lifetime Imaging. Sci. Rep. 2015, 5, 13378. [Google Scholar] [CrossRef]
- Hinderer, S.; Schenke-Layland, K. Cardiac fibrosis—A short review of causes and therapeutic strategies. Adv. Drug Deliv. Rev. 2019, 146, 77–82. [Google Scholar] [CrossRef]
- Chute, M.; Aujla, P.; Jana, S.; Kassiri, Z. The Non-Fibrillar Side of Fibrosis: Contribution of the Basement Membrane, Proteoglycans, and Glycoproteins to Myocardial Fibrosis. J. Cardiovasc. Dev. Dis. 2019, 6, 35. [Google Scholar] [CrossRef] [PubMed]
- Kong, P.; Christia, P.; Frangogiannis, N.G. The pathogenesis of cardiac fibrosis. Cell. Mol. Life Sci. 2014, 71, 549–574. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.C.; Pereira, C.; Fonseca, A.; Pinto-do-O, P.; Nascimento, D.S. Bearing My Heart: The Role of Extracellular Matrix on Cardiac Development, Homeostasis, and Injury Response. Front. Cell Dev. Biol. 2021, 8, 621644. [Google Scholar] [CrossRef] [PubMed]
- De Padilla, C.M.L.; Coenen, M.J.; Tovar, A.; De la Vega, R.E.; Evans, C.H.; Muller, S.A. Picrosirius Red Staining: Revisiting Its Application to the Qualitative and Quantitative Assessment of Collagen Type I and Type III in Tendon. J. Histochem. Cytochem. 2021, 69, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Matlack, K.H.; Kim, J.Y.; Jacobs, L.J.; Qu, J. Review of Second Harmonic Generation Measurement Techniques for Material State Determination in Metals. J. Nondestruct. Eval. 2015, 34, 273. [Google Scholar] [CrossRef]
- Austin, K.M.; Trembley, M.A.; Chandler, S.E.; Sanders, S.P.; Saffitz, J.E.; Abrams, D.J.; Pu, W.T. Molecular mechanisms of arrhythmogenic cardiomyopathy. Nat. Rev. Cardiol. 2019, 16, 519–537. [Google Scholar] [CrossRef]
- Pilichou, K.; Thiene, G.; Bauce, B.; Rigato, I.; Lazzarini, E.; Migliore, F.; Marra, M.P.; Rizzo, S.; Zorzi, A.; Daliento, L.; et al. Arrhythmogenic cardiomyopathy. Orphanet J. Rare Dis. 2016, 11, 33. [Google Scholar] [CrossRef]
- Gao, S.S.; Puthenvedu, D.; Lombardi, R.; Chen, S.N. Established and Emerging Mechanisms in the Pathogenesis of Arrhythmogenic Cardiomyopathy: A Multifaceted Disease. Int. J. Mol. Sci. 2020, 21, 6320. [Google Scholar] [CrossRef]
- Kirchner, F.; Schuetz, A.; Boldt, L.H.; Martens, K.; Dittmar, G.; Haverkamp, W.; Thierfelder, L.; Heinemann, U.; Gerull, B. Molecular Insights into Arrhythmogenic Right Ventricular Cardiomyopathy Caused by Plakophilin-2 Missense Mutations. Circ. -Cardiovasc. Genet. 2012, 5, 400–411. [Google Scholar] [CrossRef]
- Schlipp, A.; Schinner, C.; Spindler, V.; Vielmuth, F.; Gehmlich, K.; Syrris, P.; McKenna, W.J.; Dendorfer, A.; Hartlieb, E.; Waschke, J. Desmoglein-2 interaction is crucial for cardiomyocyte cohesion and function. Cardiovasc. Res. 2014, 104, 245–257. [Google Scholar] [CrossRef]
- Forleo, C.; Carmosino, M.; Resta, N.; Rampazzo, A.; Valecce, R.; Sorrentino, S.; Iacoviello, M.; Pisani, F.; Procino, G.; Gerbino, A.; et al. Clinical and Functional Characterization of a Novel Mutation in Lamin A/C Gene in a Multigenerational Family with Arrhythmogenic Cardiac Laminopathy. PLoS ONE 2015, 10, e0121723. [Google Scholar] [CrossRef]
- Fotiadis, D. Atomic force microscopy for the study of membrane proteins. Curr. Opin. Biotechnol. 2012, 23, 510–515. [Google Scholar] [CrossRef] [PubMed]
- Milting, H.; Klauke, B.; Christensen, A.H.; Musebeck, J.; Walhorn, V.; Grannemann, S.; Munnich, T.; Saric, T.; Rasmussen, T.B.; Jensen, H.K.; et al. The TMEM43 Newfoundland mutation p.S358L causing ARVC-5 was imported from Europe and increases the stiffness of the cell nucleus. Eur. Heart J. 2015, 36, 872–881. [Google Scholar] [CrossRef]
- Brun, F.; Gigli, M.; Graw, S.L.; Judge, D.P.; Merlo, M.; Murray, B.; Calkins, H.; Sinagra, G.; Taylor, M.R.G.; Mestroni, L.; et al. FLNC truncations cause arrhythmogenic right ventricular cardiomyopathy. J. Med. Genet. 2020, 57, 254–257. [Google Scholar] [CrossRef] [PubMed]
- Perkin, J.; Slater, R.; Del Favero, G.; Lanzicher, T.; Hidalgo, C.; Anderson, B.; Smith, J.E.; Sbaizero, O.; Labeit, S.; Granzier, H. Phosphorylating Titin’s Cardiac N2B Element by ERK2 or CaMKII delta Lowers the Single Molecule and Cardiac Muscle Force. Biophys. J. 2015, 109, 2592–2601. [Google Scholar] [CrossRef] [PubMed]
- Lanzicher, T.; Zhou, T.K.; Saripalli, C.; Keschrumrus, V.; Smith, J.E.; Mayans, O.; Sbaizero, O.; Granzier, H. Single-Molecule Force Spectroscopy on the N2A Element of Titin: Effects of Phosphorylation and CARP. Front. Physiol. 2020, 11, 173. [Google Scholar] [CrossRef]
- Lanzicher, T.; Martinelli, V.; Long, C.S.; Del Favero, G.; Puzzi, L.; Borelli, M.; Mestroni, L.; Taylor, M.R.G.; Sbaizero, O. AFM single-cell force spectroscopy links altered nuclear and cytoskeletal mechanics to defective cell adhesion in cardiac myocytes with a nuclear lamin mutation. Nucleus 2015, 6, 394–407. [Google Scholar] [CrossRef]
- Lanzicher, T.; Martinelli, V.; Puzzi, L.; Del Favero, G.; Codan, B.; Long, C.S.; Mestroni, L.; Taylor, M.R.G.; Sbaizero, O. The Cardiomyopathy Lamin A/C D192G Mutation Disrupts Whole-Cell Biomechanics in Cardiomyocytes as Measured by Atomic Force Microscopy Loading-Unloading Curve Analysis. Sci. Rep. 2015, 5, 13388. [Google Scholar] [CrossRef]
- Borin, D.; Pena, B.; Chen, S.N.; Long, C.S.; Taylor, M.R.G.; Mestroni, L.; Sbaizero, O. Altered microtubule structure, hemichannel localization and beating activity in cardiomyocytes expressing pathologic nuclear lamin A/C. Heliyon 2020, 6, e03175. [Google Scholar] [CrossRef]
- Borin, D.; Pena, B.; Taylor, M.R.G.; Mestroni, L.; Lapasin, R.; Sbaizero, O. Viscoelastic behavior of cardiomyocytes carrying LMNA mutations. Biorheology 2020, 57, 1–14. [Google Scholar] [CrossRef]
- Liu, J.W.; Sun, N.; Bruce, M.A.; Wu, J.C.; Butte, M.J. Atomic Force Mechanobiology of Pluripotent Stem Cell-Derived Cardiomyocytes. PLoS ONE 2012, 7, e0037559. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Yazawa, M.; Liu, J.W.; Han, L.; Sanchez-Freire, V.; Abilez, O.J.; Navarrete, E.G.; Hu, S.J.; Wang, L.; Lee, A.; et al. Patient-Specific Induced Pluripotent Stem Cells as a Model for Familial Dilated Cardiomyopathy. Sci. Transl. Med. 2012, 4, 130ra47. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.T.; Yu, D.; Lai, Y.C.; Lin, K.Y.; Liau, I. Characterization of the Mechanodynamic Response of Cardiomyocytes with Atomic Force Microscopy. Anal. Chem. 2013, 85, 1395–1400. [Google Scholar] [CrossRef] [PubMed]
| Quantitative Imaging | Force Spectroscopy |
|---|---|
Advantages
| Advantages
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peña, B.; Adbel-Hafiz, M.; Cavasin, M.; Mestroni, L.; Sbaizero, O. Atomic Force Microscopy (AFM) Applications in Arrhythmogenic Cardiomyopathy. Int. J. Mol. Sci. 2022, 23, 3700. https://doi.org/10.3390/ijms23073700
Peña B, Adbel-Hafiz M, Cavasin M, Mestroni L, Sbaizero O. Atomic Force Microscopy (AFM) Applications in Arrhythmogenic Cardiomyopathy. International Journal of Molecular Sciences. 2022; 23(7):3700. https://doi.org/10.3390/ijms23073700
Chicago/Turabian StylePeña, Brisa, Mostafa Adbel-Hafiz, Maria Cavasin, Luisa Mestroni, and Orfeo Sbaizero. 2022. "Atomic Force Microscopy (AFM) Applications in Arrhythmogenic Cardiomyopathy" International Journal of Molecular Sciences 23, no. 7: 3700. https://doi.org/10.3390/ijms23073700
APA StylePeña, B., Adbel-Hafiz, M., Cavasin, M., Mestroni, L., & Sbaizero, O. (2022). Atomic Force Microscopy (AFM) Applications in Arrhythmogenic Cardiomyopathy. International Journal of Molecular Sciences, 23(7), 3700. https://doi.org/10.3390/ijms23073700







