In Silico Characterisation of the Late Embryogenesis Abundant (LEA) Protein Families and Their Role in Desiccation Tolerance in Ramonda serbica Panc
Abstract
1. Introduction
2. Results
2.1. Identification and Classification of R. serbica LEAPs
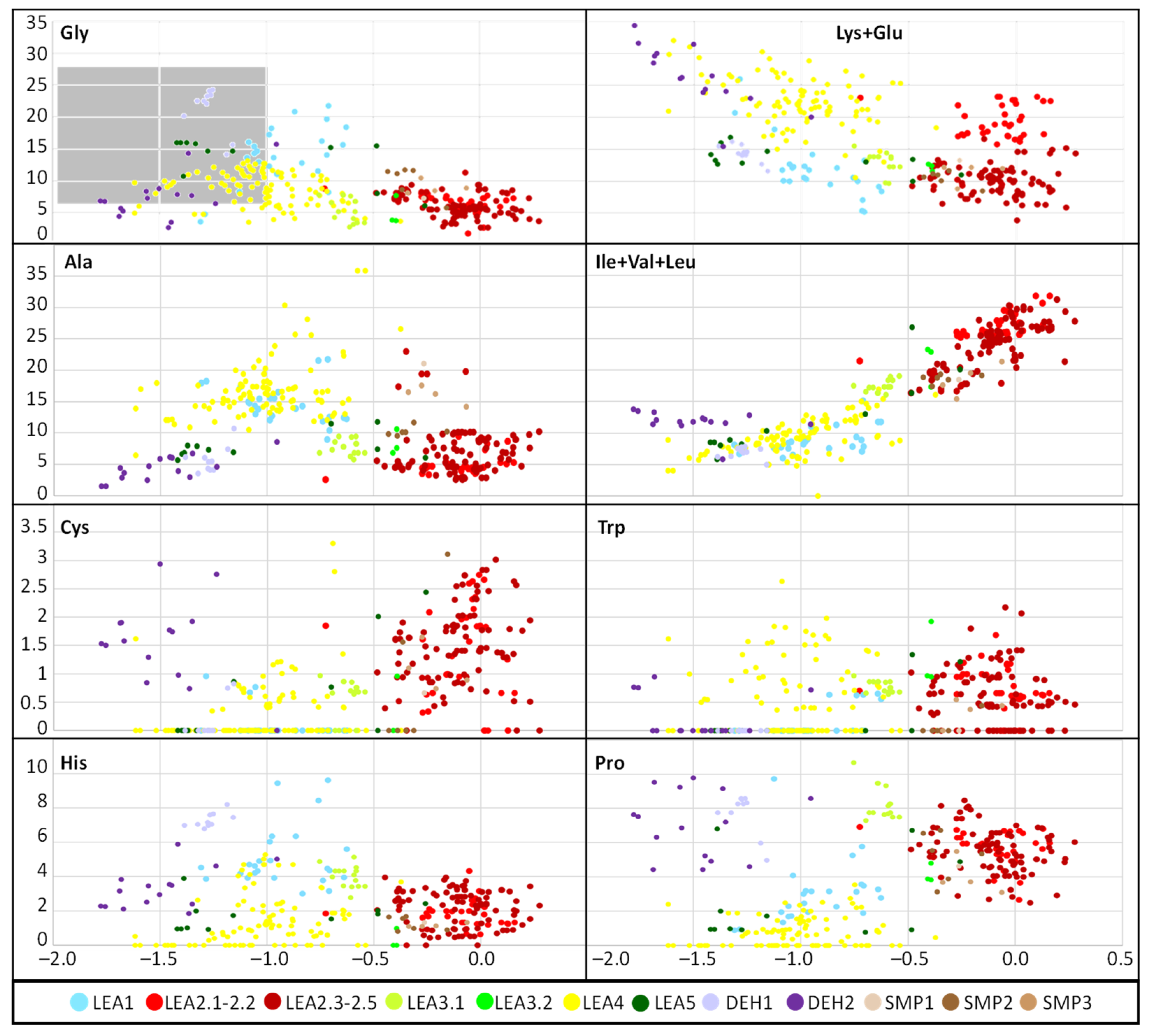
2.2. Physicochemical Analysis of R. serbica LEAPs
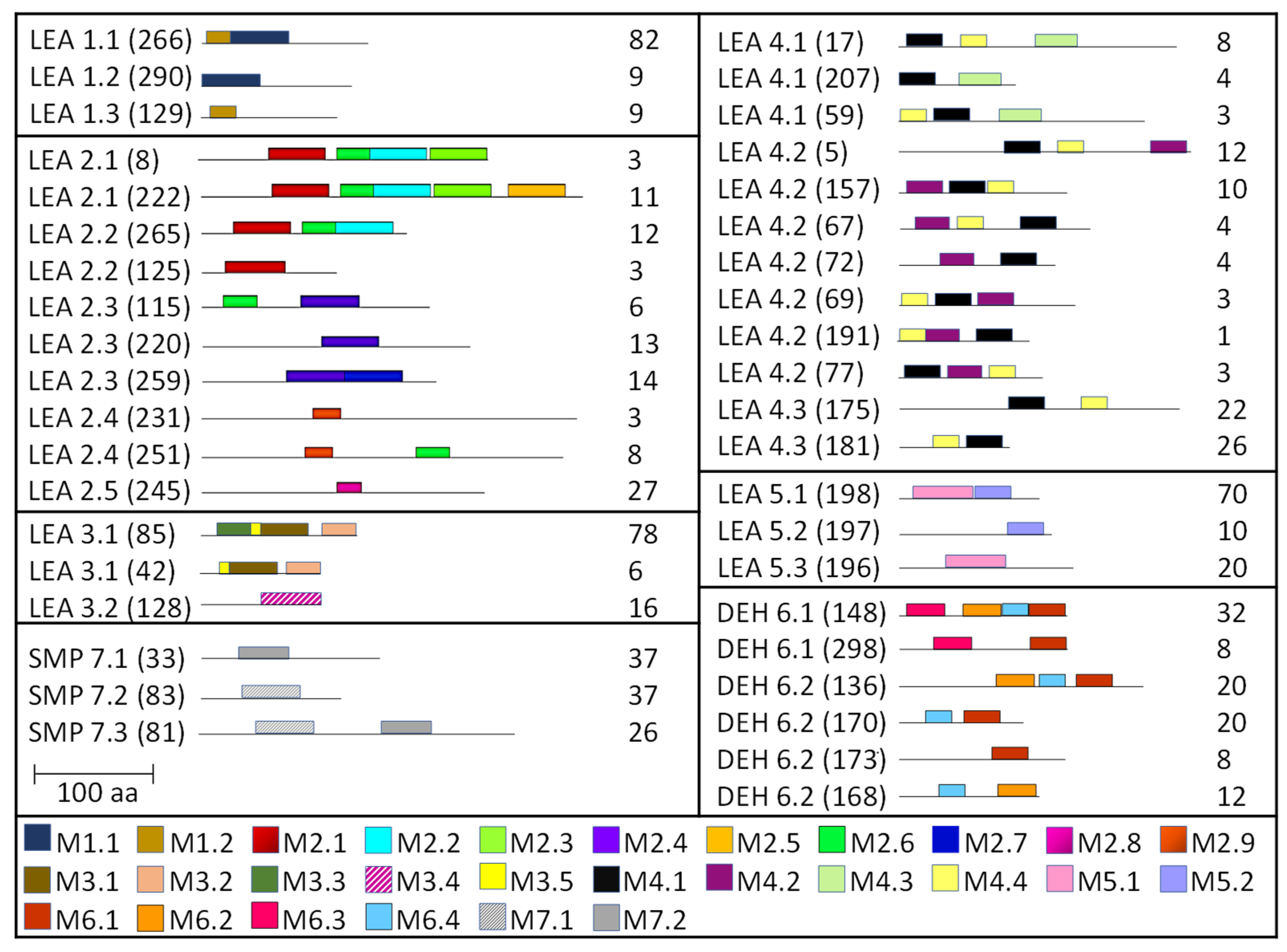
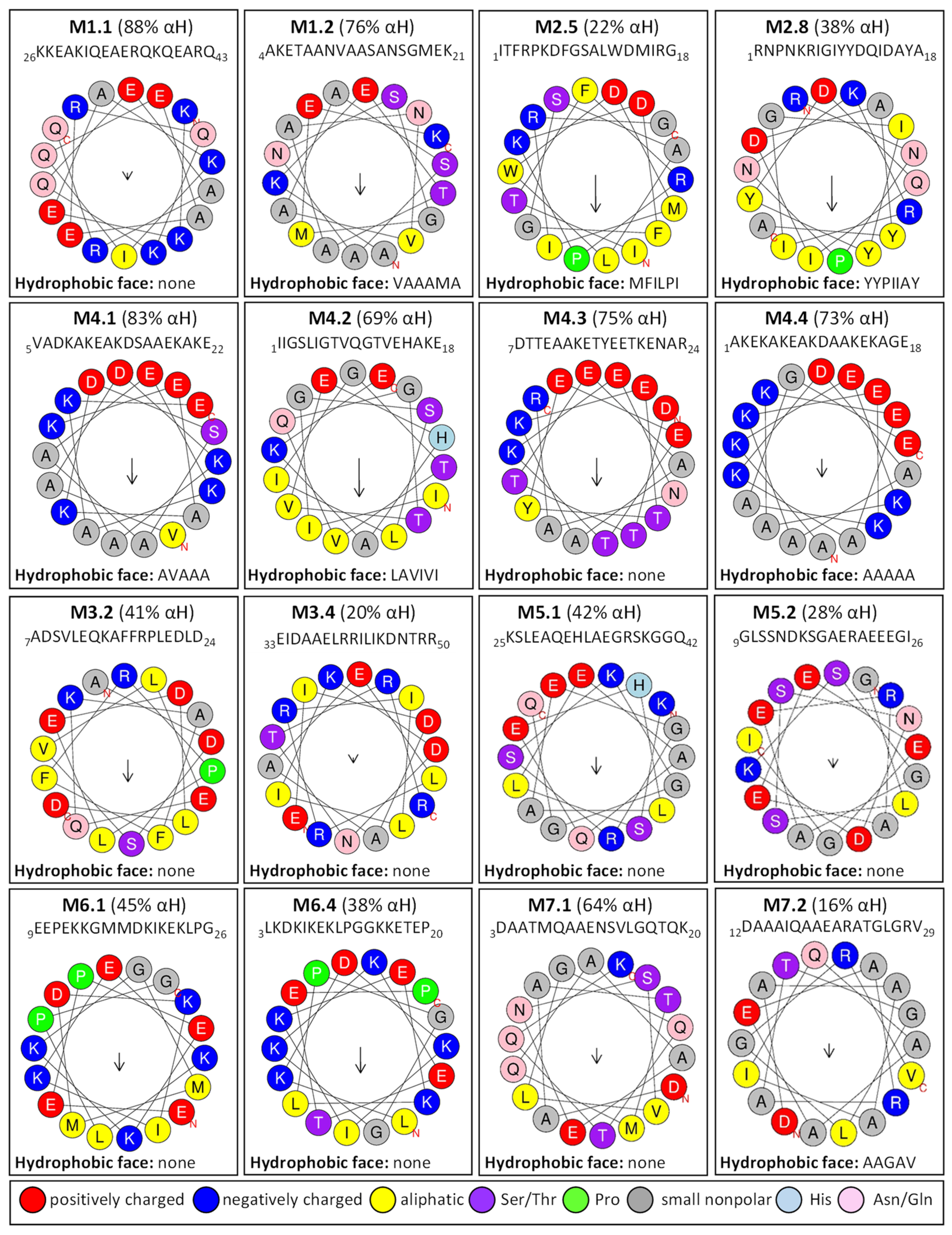
2.3. Homology Motifs Analyses of R. serbica LEAPs
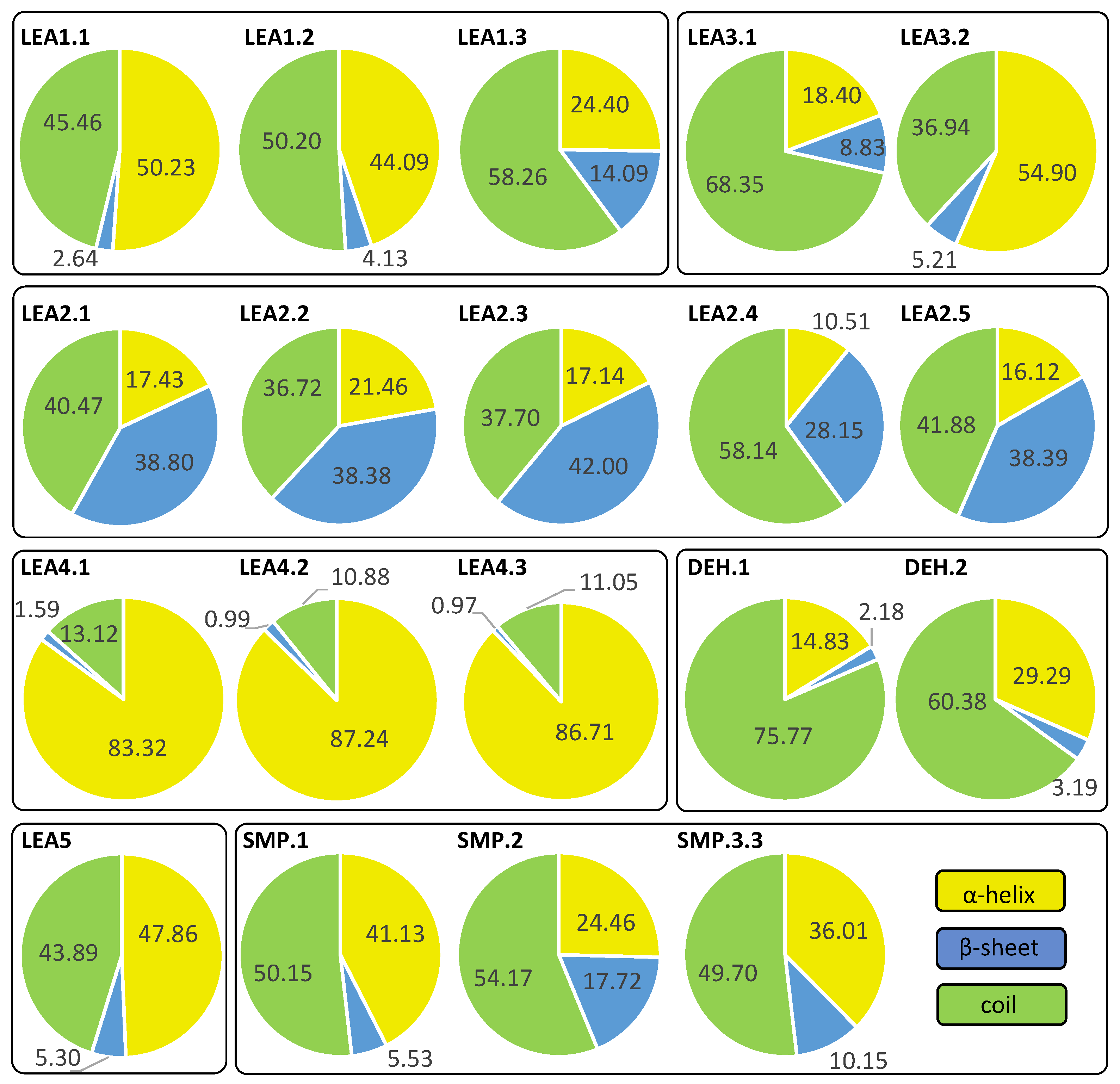
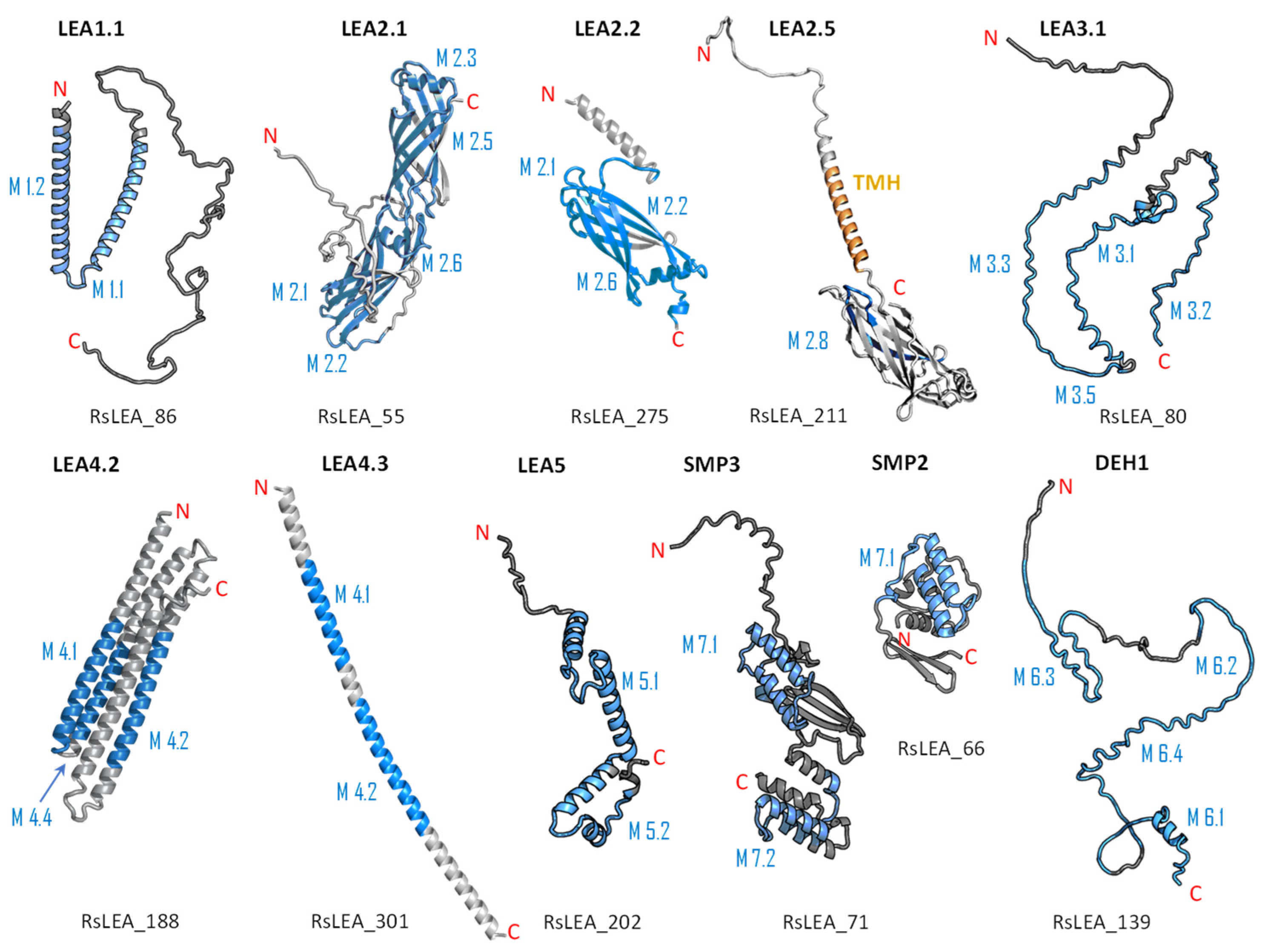
2.4. Structure and Disorder Prediction of R. serbica LEAPs
2.5. Calculated Hydroxyl Radical Scavenging Ability (HRSA) of R. serbica LEAPs
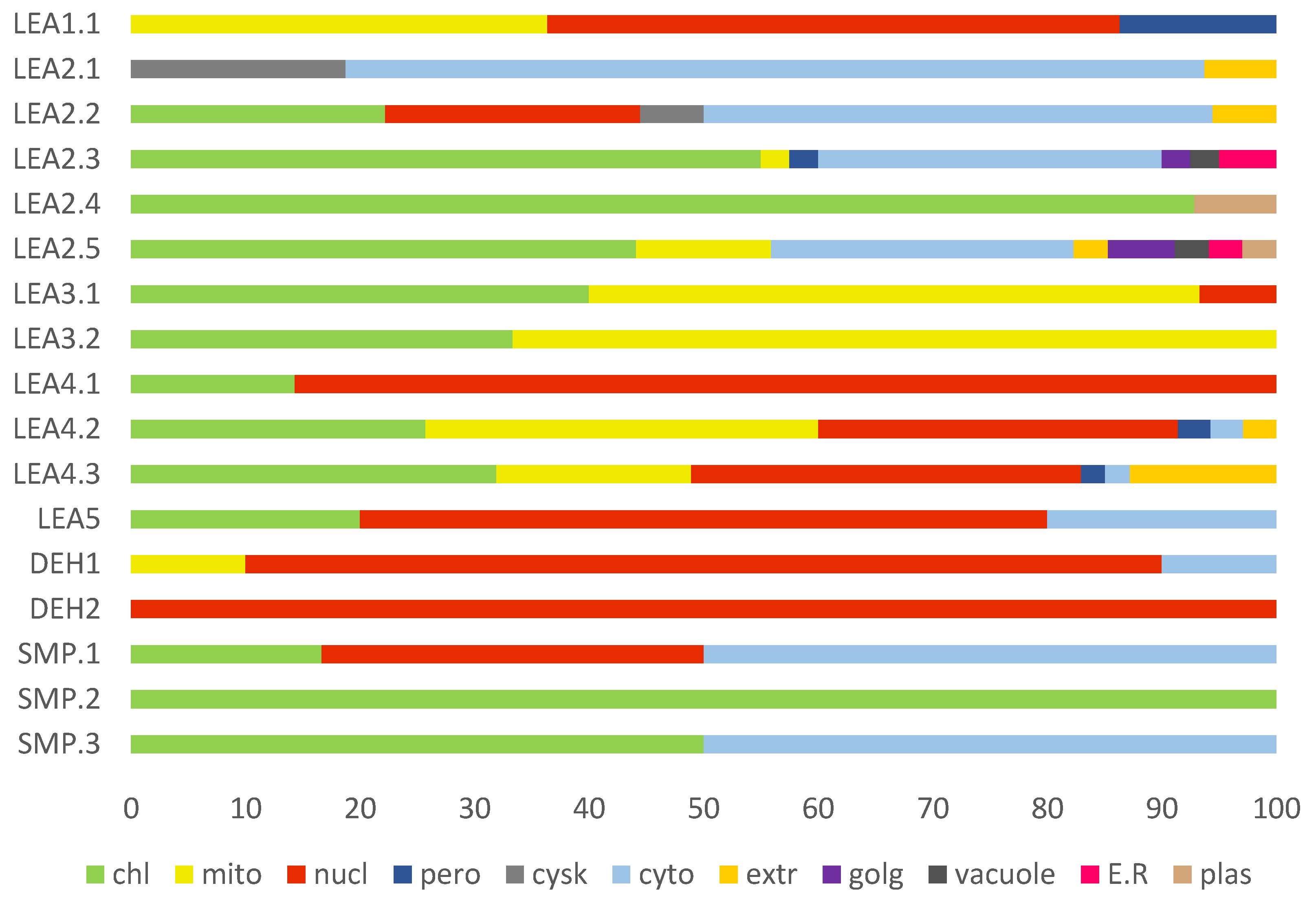
2.6. Cellular Compartmentalisation of R. serbica LEAPs
2.7. Analysis of Differentially Expressed R. serbica LEA Genes
3. Discussion
3.1. Identification and Classification of R. serbica LEAPs
3.2. Analysis of Amino Acid Composition and Physicochemical Properties of R. serbica LEAPs
3.3. Protein Structure and Disorder Prediction of R. serbica LEA Proteins
3.4. Subcellular Localisation of R. serbica LEA Proteins
3.5. Characterisation of the Individual R. serbica LEA Protein Family Groups and Estimation of Their Physiological Function under Desiccation
3.5.1. R. serbica Dehydrins
3.5.2. R. serbica LEA1 Protein Family Group
3.5.3. R. serbica LEA2 Protein Family Group
3.5.4. R. serbica LEA3 Protein Family Group
3.5.5. R. serbica LEA4 Protein Family Group
3.5.6. R. serbica LEA5 Protein Family Group
3.5.7. R. serbica SMPs
4. Materials and Methods
4.1. Plant Material and Treatment
4.2. De Novo Transcriptome Analysis of R. serbica HL and DL
4.2.1. RNA Extraction, cDNA Library Construction, and Illumina High-Throughput Sequencing
4.2.2. Transcriptome De Novo Assembly and Sequence Annotation
4.2.3. Differential Expression Analysis and Functional Enrichment
4.3. Identification and Classification of R. serbica LEAPs
4.4. Physiochemical Characterisation of R. serbica LEAPs
4.5. Phylogenetic Identification of R. serbica LEAPs
4.6. Conserved Motif Composition in R. serbica LEAPs
4.7. Secondary Structure and Disorder Predictions of R. serbica LEAPs
4.8. Modelling 3D Protein Structure
4.9. Annotation of the Subcellular Localisation of R. serbica LEAPs
4.10. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Farrant, J.M.; Hilhorst, H.W.M. What is dry? Exploring metabolism and molecular mobility at extremely low water contents. J. Exp. Bot. 2021, 72, 1507–1510. [Google Scholar] [CrossRef] [PubMed]
- Artur, M.A.S.; Zhao, T.; Ligterink, W.; Schranz, E.; Hilhorst, H.W.M. Dissecting the genomic diversification of late embryogenesis abundant (LEA) protein gene families in plants. Genome. Biol. Evol. 2019, 11, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Dirk, L.M.A.; Abdel, C.G.; Ahmad, I.; Neta, I.C.S.; Pereira, C.C.; Pereira, F.E.C.B.; Unêda-Trevisoli, S.H.; Pinheiro, D.G.; Downie, A.B. Late embryogenesis abundant protein-client protein interactions. Plants 2020, 9, 814. [Google Scholar] [CrossRef] [PubMed]
- Veljović-Jovanović, S.; Kukavica, B.; Stevanović, B.; Navari-Izzo, F. Senescence-and drought-related changes in peroxidase and superoxide dismutase isoforms in leaves of Ramonda serbica. J. Exp. Bot. 2006, 57, 1759–1768. [Google Scholar] [CrossRef]
- Vidović, M.; Ćuković, K. Isolation of high-quality RNA from recalcitrant leaves of variegated and resurrection plants. 3 Biotech 2020, 10, 286–294. [Google Scholar] [CrossRef]
- Liu, J.; Moyankova, D.; Lin, C.T.; Mladenov, P.; Sun, R.Z.; Djilianov, D.; Deng, X. Transcriptome reprogramming during severe dehydration contributes to physiological and metabolic changes in the resurrection plant Haberlea rhodopensis. BMC Plant Biol. 2018, 18, 351–367. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, B.; Phillips, J.; Zhang, Z.N.; Du, H.; Xu, T.; Huang, L.C.; Zhang, X.F.; Xu, G.H.; Li, W.L.; et al. Global transcriptome analysis reveals acclimation–primed processes involved in the acquisition of desiccation tolerance in Boea hygrometrica. Plant Cell Physiol. 2015, 56, 1429–1441. [Google Scholar] [CrossRef]
- Xiao, L.; Yang, G.; Zhang, L.; Yang, X.; Zhao, S.; Ji, Z.; Zhou, Q.; Hu, M.; Wang, Y.; Chen, M.; et al. The resurrection genome of Boea hygrometrica: A blueprint for survival of dehydration. Proc. Natl. Acad. Sci. USA 2015, 112, 5833–5837. [Google Scholar] [CrossRef]
- Rakić, T.; Lazarević, M.; Jovanović, Z.S.; Radović, S.; Siljak–Yakovlev, S.; Stevanović, B.; Stevanović, V. Resurrection plants of the genus Ramonda: Prospective survival strategies—Unlock further capacity of adaptation, or embark on the path of evolution? Front. Plant Sci. 2014, 4, 550–560. [Google Scholar] [CrossRef]
- Chandra, J.; Keshavkant, S. Desiccation-induced ROS accumulation and lipid catabolism in recalcitrant. Physiol. Mol. Biol. Plants 2018, 24, 75–87. [Google Scholar] [CrossRef]
- Farrant, J.M. A comparison of mechanisms of desiccation tolerance among three angiosperm resurrection plant species. Plant Ecol. 2000, 151, 29–39. [Google Scholar] [CrossRef]
- Ingram, J.; Bartels, D. The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 377–403. [Google Scholar] [CrossRef] [PubMed]
- Olvera-Carrillo, Y.; Luis Reyes, J.; Covarrubias, A.A. Late embryogenesis abundant proteins: Versatile players in the plant adaptation to water limiting environments. Plant Signal. Behav. 2011, 6, 586–589. [Google Scholar] [CrossRef] [PubMed]
- Hundertmark, M.; Hincha, D.K. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genom. 2008, 9, 118–140. [Google Scholar] [CrossRef]
- Magwanga, R.O.; Lu, P.; Kirungu, J.N.; Lu, H.; Wang, X.; Cai, X.; Zhou, Z.; Zhang, Z.; Salih, H.; Wang, K.; et al. Characterization of the late embryogenesis abundant (LEA) proteins family and their role in drought stress tolerance in upland cotton. BMC Genet. 2018, 19, 6–37. [Google Scholar] [CrossRef]
- Chen, Y.; Li, C.; Zhang, B.; Yi, J.; Yang, Y.; Kong, C.; Lei, C.; Gong, M. The role of the late embryogenesis-abundant (LEA) protein family in development and the abiotic stress response: A comprehensive expression analysis of potato. Genes 2019, 10, 148. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Dhar, S.; Banerjee, A.; Ray, S. Structural, functional, and evolutionary analysis of late embryogenesis abundant proteins (LEA) in Triticum aestivum: A detailed molecular level biochemistry using in silico approach. Comput. Biol. Chem. 2019, 82, 9–24. [Google Scholar] [CrossRef]
- Wang, W.; Gao, T.; Chen, J.; Yang, J.; Huang, H.; Yu, Y. The late embryogenesis abundant gene family in tea plant (Camellia sinensis): Genome-wide characterization and expression analysis in response to cold and dehydration stress. Plant Physiol. Biochem. 2019, 135, 277–286. [Google Scholar] [CrossRef]
- Wang, X.-S.; Zhu, H.-B.; Jin, G.-L.; Liu, H.-L.; Wu, W.-R.; Zhu, J. Genome-scale identification and analysis of LEA genes in rice (Oryza sativa L.). Plant Sci. 2007, 172, 414–420. [Google Scholar] [CrossRef]
- Divya, K.; Palakolanu, S.R.; Kavi Kishor, P.; Rajesh, A.S.; Vadez, V.; Sharma, K.K.; Mathur, P.B. Functional characterization of late embryogenesis abundant genes and promoters in pearl millet (Pennisetum glaucum L.) for abiotic stress tolerance. Physiol. Plant. 2021, 173, 1616–1628. [Google Scholar] [CrossRef]
- Nagaraju, M.; Kumar, S.A.; Reddy, P.S.; Kumar, A.; Rao, D.M.; Kavi Kishor, P.B. Genome-scale identification, classification, and tissue specific expression analysis of late embryogenesis abundant (LEA) genes under abiotic stress conditions in Sorghum bicolor L. PLoS ONE 2019, 14, e0209980. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, M.; Covarrubias, A.A. Late embryogenesis abundant (LEA) proteins in legumes. Front. Plant Sci. 2013, 4, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Muvunyi, B.P.; Yan, Q.; Wu, F.; Min, X.; Yan, Z.Z.; Kanzana, G.; Wang, Y.; Zhang, J. Mining late embryogenesis abundant (LEA) family genes in Cleistogenes songorica, a Xerophyte perennial desert plant. Int. J. Mol. Sci. 2018, 19, 3430. [Google Scholar] [CrossRef] [PubMed]
- Dure, L.; Greenway, S.C.; Galau, G.A. Developmental biochemistry of cottonseed embryogenesis and germination: Changing messenger ribonucleic acid populations as shown by in vitro and in vivo protein synthesis. Biochemistry 1981, 20, 4162–4168. [Google Scholar] [CrossRef]
- Salleh, F.M.; Evans, K.; Goodall, B.; Machin, H.; Mowla, S.B.; Mur, L.A.; Runions, J.; Theodoulou, F.L.; Foyer, C.H.; Rogers, H.J. A novel function for a redox-related LEA protein (SAG21/AtLEA5) in root development and biotic stress responses. Plant Cell Environ. 2012, 35, 418–429. [Google Scholar] [CrossRef]
- Liu, Y.; Liang, J.; Sun, L.; Yang, X.; Li, D. Group 3 LEA protein, zmlea3, is involved in protection from low temperature stress. Front. Plant Sci. 2016, 7, 1011–1021. [Google Scholar] [CrossRef][Green Version]
- Lin, R.; Zou, T.; Mei, Q.; Wang, Z.; Zhang, M.; Jian, S. Genome-wide analysis of the late embryogenesis abundant (LEA) and abscisic acid-, stress-, and ripening-induced (ASR) gene superfamily from Canavalia rosea and their roles in salinity/alkaline and drought tolerance. Int. J. Mol. Sci. 2021, 22, 4554. [Google Scholar] [CrossRef]
- Belott, C.; Janis, B.; Menze, M.A. Liquid-liquid phase separation promotes animal desiccation tolerance. Proc. Natl. Acad. Sci. USA 2020, 117, 27676–27684. [Google Scholar] [CrossRef]
- Darling, A.L.; Zaslavsky, B.Y.; Uversky, V.N. Intrinsic disorder-based emergence in cellular biology: Physiological and pathological liquid-liquid phase transitions in cells. Polymers 2019, 11, 990. [Google Scholar] [CrossRef]
- Vidović, M.; Milić-Komić, S. Regulation of proteolysis of intrinsically disordered proteins: Physiological consequences. In A Closer Look at Proteolysis, 1st ed.; Radosavljević, J., Ed.; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2021; Volume 1, pp. 111–157. [Google Scholar]
- Cuevas-Velazquez, C.L.; Reyes, J.L.; Covarrubias, A.A. Group 4 late embryogenesis abundant proteins as a model to study intrinsically disordered proteins in plants. Plant Signal. Behav. 2017, 12, 10893–10903. [Google Scholar] [CrossRef]
- Hundertmark, M.; Dimova, R.; Lengefeld, J.; Seckler, R.; Hincha, D.K. The intrinsically disordered late embryogenesis abundant protein LEA18 from Arabidopsis thaliana modulates membrane stability through binding and folding. Biochim. Biophys. Acta 2011, 1808, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Tolleter, D.; Hincha, D.K.; Macherel, D. A mitochondrial late embryogenesis abundant protein stabilizes model membranes in the dry state. Biochim. Biophys. Acta 2010, 1798, 1926–1933. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.E. The Investigation of Group 1 Late Embryogenesis Abundant Protein 6 and Its Role in Arabidopsis thaliana Desiccation Tolerance. Master’s Thesis, Grand Valley State University, Allendale, MI, USA, 2020. [Google Scholar]
- Ginsawaeng, O.; Heise, C.; Sangwan, R.; Karcher, D.; Hernández-Sánchez, I.E.; Sampathkumar, A.; Zuther, E. Subcellular localization of seed-expressed LEA_4 proteins reveals liquid-liquid phase separation for LEA9 and for LEA48 homo- and LEA42-LEA48 heterodimers. Biomolecules 2021, 11, 1770. [Google Scholar] [CrossRef] [PubMed]
- Thalhammer, A.; Hundertmark, M.; Popova, A.V.; Seckler, R.; Hincha, D.K. Interaction of two intrinsically disordered plant stress proteins (COR15A and COR15B) with lipid membranes in the dry state. Biochim Biophys Acta 2010, 1798, 1812–1820. [Google Scholar] [CrossRef]
- Graether, S.P.; Boddington, K.F. Disorder and function: A review of the dehydrin protein family. Front. Plant Sci. 2014, 5, 576–588. [Google Scholar] [CrossRef]
- Bremer, A.; Wolff, M.; Thalhammer, A.; Hincha, D.K. Folding of intrinsically disordered plant LEA proteins is driven by glycerol-induced crowding and the presence of membranes. FEBS J. 2017, 284, 919–936. [Google Scholar] [CrossRef]
- Furuki, T.; Niwa, T.; Taguchi, H.; Hatanaka, R.; Kikawada, T.; Sakurai, M. A LEA model peptide protects the function of a red fluorescent protein in the dry state. Biochem. Biophys. Rep. 2019, 17, 27–31. [Google Scholar] [CrossRef]
- Olvera-Carrillo, Y.; Campos, F.; Reyes, J.L.; Garciarrubio, A.; Covarrubias, A.A. Functional analysis of the group 4 late embryogenesis abundant proteins reveals their relevance in the adaptive response during water deficit in Arabidopsis. Plant Physiol. 2010, 154, 373–390. [Google Scholar] [CrossRef]
- Chakrabortee, S.; Tripathi, R.; Watson, M.; Schierle, G.S.; Kurniawan, D.P.; Kaminski, C.F.; Wise, M.J.; Tunnacliffe, A. Intrinsically disordered proteins as molecular shields. Mol. Biosyst. 2012, 8, 210–219. [Google Scholar] [CrossRef]
- Yuen, F.; Watson, M.; Barker, R.; Grillo, I.; Heenan, R.K.; Tunnacliffe, A.; Routh, A.F. Preferential adsorption to air-water interfaces: A novel cryoprotective mechanism for LEA proteins. Biochem. J. 2019, 476, 1121–1135. [Google Scholar] [CrossRef]
- Goyal, K.; Walton, L.J.; Tunnacliffe, A. LEA proteins prevent protein aggregation due to water stress. Biochem. J. 2005, 388, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Cuevas-Velazquez, C.L.; Saab-Rincón, G.; Reyes, J.L.; Covarrubias, A.A. The unstructured N-terminal region of Arabidopsis group 4 late embryogenesis abundant (LEA) proteins is required for folding and for chaperone-like activity under water deficit. J. Biol. Chem. 2016, 291, 10893–10903. [Google Scholar] [CrossRef] [PubMed]
- Cuevas-Velazquez, C.L.; Dinneny, J.R. Organization out of disorder: Liquid-liquid phase separation in plants. Curr. Opin. Plant Biol. 2018, 45, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Vidović, M.; Franchin, C.; Morina, F.; Veljović-Jovanović, S.; Masi, A.; Arrigoni, G. Efficient protein extraction for shotgun proteomics from hydrated and desiccated leaves of resurrection Ramonda serbica plants. Anal. Bioanal. Chem. 2020, 412, 8299–8312. [Google Scholar] [CrossRef]
- Gautier, R.; Douguet, D.; Antonny, B.; Drin, G. HELIQUEST: A web server to screen sequences with specific alpha-helical properties. Bioinformatics 2008, 24, 2101–2102. [Google Scholar] [CrossRef]
- Milić, S.; Bogdanović Pristov, J.; Mutavdžić, D.; Savić, A.; Spasić, M.; Spasojević, I. The relationship of physicochemical properties to the antioxidative activity of free amino acids in Fenton system. Environ. Sci. Technol. 2015, 49, 4245–4254. [Google Scholar] [CrossRef]
- Lazarević, M.; Siljak-Yakovlev, S.; Lazarević, P.; Stevanović, B.; Stevanović, V. Pollen and seed morphology of resurrection plants from the genus Ramonda (Gesneriaceae): Relationship with ploidy level and relevance to their ecology and identification. Turk. J. Bot. 2013, 37, 872–885. [Google Scholar] [CrossRef]
- Rodriguez, M.C.; Edsgärd, D.; Hussain, S.S.; Alquezar, D.; Rasmussen, M.; Gilbert, T.; Nielsen, B.H.; Bartels, D.; Mundy, J. Transcriptomes of the desiccation-tolerant resurrection plant Craterostigma plantagineum. Plant J. 2010, 63, 212–228. [Google Scholar] [CrossRef]
- Fujioka, H.; Samejima, H.; Suzuki, H.; Mizutani, M.; Okamoto, M.; Sugimoto, Y. Aberrant protein phosphatase 2C leads to abscisic acid insensitivity and high transpiration in parasitic Striga. Nat. Plants 2019, 5, 258–262. [Google Scholar] [CrossRef]
- Bray, E.A. Molecular responses to water deficit. Plant Physiol. 1993, 103, 1035–1040. [Google Scholar] [CrossRef]
- Dure, L. A repeating 11-mer amino acid motif and plant desiccation. Plant J. 1993, 3, 363–369. [Google Scholar] [CrossRef]
- Wise, M.J. LEAping to conclusions: A computational reanalysis of late embryogenesis abundant proteins and their possible roles. BMC Bioinform. 2003, 4, 52–71. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.K.; Graether, S.P. Conserved sequence motifs in the abiotic stress response protein late embryogenesis abundant 3. PLoS ONE 2020, 15, e0237177. [Google Scholar] [CrossRef] [PubMed]
- Candat, A.; Paszkiewicz, G.; Neveu, M.; Gautier, R.; Logan, D.C.; Avelange-Macherel, M.H.; Macherel, D. The ubiquitous distribution of late embryogenesis abundant proteins across cell compartments in Arabidopsis offers tailored protection against abiotic stress. Plant Cell 2014, 26, 3148–3166. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, M.; Olvera-Carrillo, Y.; Garciarrubio, A.; Campos, F.; Covarrubias, A.A. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 2008, 148, 6–24. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. RCSB protein data bank: Structural biology views for basic and applied research. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Piovesan, D.; Walsh, I.; Minervini, G.; Tosatto, S.C.E. FELLS: Fast estimator of latent local structure. Bioinformatics 2017, 33, 1889–1891. [Google Scholar] [CrossRef] [PubMed]
- NDong, C.; Danyluk, J.; Wilson, K.E.; Pocock, T.; Huner, N.P.; Sarhan, F. Cold-regulated cereal chloroplast late embryogenesis abundant-like proteins. Molecular characterization and functional analyses. Plant Physiol. 2002, 129, 1368–1381. [Google Scholar] [CrossRef]
- Wise, M.J.; Tunnacliffe, A. POPP the question: What do LEA proteins do? Trends Plant Sci. 2004, 9, 13–17. [Google Scholar] [CrossRef]
- Hara, M.; Shinoda, Y.; Tanaka, Y.; Kuboi, T. DNA binding of citrus dehydrin promoted by zinc ion. Plant Cell Environ. 2009, 32, 532–541. [Google Scholar] [CrossRef]
- Zhang, Q.; Bartels, D. Molecular responses to dehydration and desiccation in desiccation-tolerant angiosperm plants. J. Exp. Bot. 2018, 69, 3211–3222. [Google Scholar] [CrossRef] [PubMed]
- Mouillon, J.M.; Gustafsson, P.; Harryson, P. Structural investigation of disordered stress proteins. Comparison of full-length dehydrins with isolated peptides of their conserved segments. Plant Physiol. 2006, 141, 638–650. [Google Scholar] [CrossRef] [PubMed]
- Koag, M.C.; Wilkens, S.; Fenton, R.D.; Resnik, J.; Vo, E.; Close, T.J. The K-segment of maize DHN1 mediates binding to anionic phospholipid vesicles and concomitant structural changes. Plant Physiol. 2009, 150, 1503–1514. [Google Scholar] [CrossRef] [PubMed]
- Hara, M.; Fujinaga, M.; Kuboi, T. Metal binding by citrus dehydrin with histidine-rich domains. J. Exp. Bot. 2005, 56, 2695–2703. [Google Scholar] [CrossRef]
- Ueda, E.K.; Gout, P.W.; Morganti, L. Current and prospective applications of metal ion-protein binding. J. Chromatogr. A 2003, 988, 1–23. [Google Scholar] [CrossRef]
- Hanin, M.; Brini, F.; Ebel, C.; Toda, Y.; Takeda, S.; Masmoudi, K. Plant dehydrins and stress tolerance: Versatile proteins for complex mechanisms. Plant Signal. Behav. 2011, 6, 1503–1509. [Google Scholar] [CrossRef]
- Malik, A.A.; Veltri, M.; Boddington, K.F.; Singh, K.K.; Graether, S.P. Genome analysis of conserved dehydrin motifs in vascular plants. Front. Plant Sci. 2017, 8, 709–727. [Google Scholar] [CrossRef]
- Goday, A.; Jensen, A.B.; Culiáñez-Macià, F.A.; Mar Albà, M.; Figueras, M.; Serratosa, J.; Torrent, M.; Pagès, M. The maize abscisic acid-responsive protein Rab17 is located in the nucleus and interacts with nuclear localization signals. Plant Cell 1994, 6, 351–360. [Google Scholar]
- Alsheikh, M.K.; Heyen, B.J.; Randall, S.K. Ion binding properties of the dehydrin ERD14 are dependent upon phosphorylation. J. Biol. Chem. 2003, 278, 40882–40889. [Google Scholar] [CrossRef]
- Clarke, M.W.; Boddington, K.F.; Warnica, J.M.; Atkinson, J.; McKenna, S.; Madge, J.; Barker, C.H.; Graether, S.P. Structural and functional insights into the cryoprotection of membranes by the intrinsically disordered dehydrins. J. Biol. Chem. 2015, 290, 26900–26913. [Google Scholar] [CrossRef]
- Peng, A.; Weber, S.C. Evidence for and against liquid-liquid phase separation in the nucleus. Noncoding RNA 2019, 5, 50–64. [Google Scholar]
- Jaspard, E.; Hunault, G. Comparison of amino acids physico-chemical properties and usage of late embryogenesis abundant proteins, hydrophilins and WHy domain. PLoS ONE 2014, 9, e109570. [Google Scholar]
- Liu, Y.; Song, Q.; Li, D.; Yang, X. Multifunctional roles of plant dehydrins in response to environmental stresses. Front. Plant Sci. 2017, 8, 1018–1022. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, Z.; Wang, L.; Wu, R.; Phillips, J.; Deng, X. LEA 4 group genes from the resurrection plant Boea hygrometrica confer dehydration tolerance in transgenic tobbaco. Plant Sci. 2009, 176, 90–98. [Google Scholar] [CrossRef]
- Jiang, Q.; Li, X.; Niu, F.; Sun, X.; Hu, Z.; Zhang, H. iTRAQ-based quantitative proteomic analysis of wheat roots in response to salt stress. Proteomics 2017, 17, 1600265. [Google Scholar] [CrossRef]
- Scholtz, J.M.; Baldwin, R.L. The mechanism of alpha-helix formation by peptides. Annu. Rev. Biophys. Biomol. Struct. 1992, 21, 95–118. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Davidson, N.M.; Oshlack, A. Corset: Enabling differential gene expression analysis for de novo assembled transcriptomes. Genome Biol. 2014, 15, 410–424. [Google Scholar]
- Bateman, A.; Birney, E.; Cerruti, L.; Durbin, R.; Etwiller, L.; Eddy, S.R.; Griffiths-Jones, S.; Howe, K.L.; Marshall, M.; Sonnhammer, E.L. The Pfam protein families database. Nucleic Acids Res. 2002, 30, 276–280. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323–339. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39 (Suppl. S2), W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Mulder, N.J.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Barrell, D.; Bateman, A.; Binns, D.; Biswas, M.; Bradley, P.; Bork, P.; et al. The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res. 2003, 31, 315–318. [Google Scholar] [CrossRef]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef]
- Cock, P.J.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, 293–296. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37 (Suppl. S2), W202–W208. [Google Scholar] [CrossRef]
- Geourjon, C.; Deléage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 1995, 11, 681–684. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999, 292, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 2015, 43, W389–W394. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Erdős, G.; Pajkos, M.; Dosztányi, Z. IUPred3: Prediction of protein disorder enhanced with unambiguous experimental annotation and visualization of evolutionary conservation. Nucleic Acids Res. 2021, 49, W297–W303. [Google Scholar] [CrossRef]
- Pollastri, G.; Przybylski, D.; Rost, B.; Baldi, P. Improving the prediction of protein secondary structure in three and eight classes using recurrent neural networks and profiles. Proteins 2002, 47, 228–235. [Google Scholar] [CrossRef]
- Sickmeier, M.; Hamilton, J.A.; LeGall, T.; Vacic, V.; Cortese, M.S.; Tantos, A.; Szabo, B.; Tompa, P.; Chen, J.; Uversky, V.N.; et al. DisProt: The database of disordered proteins. Nucleic Acids Res. 2007, 35 (Suppl. S1), D786–D793. [Google Scholar] [CrossRef]
- Mika, S.; Rost, B. NMPdb: Database of nuclear matrix proteins. Nucleic Acids Res. 2005, 33 (Suppl. S1), D160–D163. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold-Making protein folding accessible to all. bioRxiv 2021. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Emanuelsson, O.; Brunak, S.; von Heijne, G.; Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef] [PubMed]
- Bodén, M.; Hawkins, J. Prediction of subcellular localization using sequence-biased recurrent networks. Bioinformatics 2005, 21, 2279–2286. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35 (Suppl. S2), W585–W587. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
| LEA Protein Family Group | Pfam ID | Protein Number | A. thaliana Similarity, % | G. hirsutum Similarity, % |
|---|---|---|---|---|
| LEA1 | PF03760 | 24 | 41.5 ± 0.9 | 28.8 ± 0.4 |
| LEA2 | PF03168 | 127 | 32.8 ± 0.9 | 29.8 ± 0.3 |
| LEA3 | PF03242 | 18 | 34.7 ± 1.6 | 27.7 ± 0.5 |
| LEA4 | PF02987 | 96 | 29.8 ± 0.3 | 28.2 ± 0.3 |
| LEA5 | PF00477 | 11 | 58.6 ± 4.4 | 27.8 ± 0.5 |
| Dehydrin | PF04927 | 25 | 41.6 ± 1.6 | 34.4 ± 0.9 |
| SMP | PF00257 | 17 | 37.9 ± 0.9 | 24.7 ± 0.3 |
| LEA Protein Group | aa # | Calculated pI | Mw (kDa) | GRAVY Index | Amino Acid (aa) Composition | ||||
|---|---|---|---|---|---|---|---|---|---|
| % | % | % | % | % | |||||
| Charged | Polar | Nonpolar | Aromatic | Cys | |||||
| LEA1 | 139 ± 5 | 8.2 ± 0.4 d | 14.4 ± 0.5 a | −0.93 ± 0.05 b | 23.2 ± 1.3 a | 33.2 ± 0.9 c,d | 40.5 ± 0.8 c | 5.1 ± 0.5 a | 0.10 ± 0.06 a |
| LEA2 | 226 ± 5 | 8.4 ± 0.3 d | 25.2 ± 0.7 b | −0.09 ± 0.03 e | 21.4 ± 0.5 a | 29.3 ± 0.5 b,c | 38.8 ± 0.4 b,c | 9.7 ± 0.2 c | 1.66 ± 0.10 c |
| LEA3 | 126 ± 5 | 7.0 ± 0.4 b,c,d | 14.0 ± 0.6 a | −0.59 ± 0.03 c | 23.6 ± 0.5 a | 35.6 ± 1.0 d | 32.3 ± 0.8 a | 10.5 ± 0.4 c | 0.56 ± 0.09 a,b |
| LEA4 | 187 ± 7 | 6.1 ± 0.2 a,b | 17.9 ± 0.9 a | −1.01 ± 0.03 b | 35.2 ± 0.6 b,c | 23.6 ± 0.5 a | 36.7 ± 0.5 b | 3.7 ± 0.2 a | 0.33 ± 0.06 a,b |
| LEA5 | 118 ± 7 | 8.1 ± 0.5 c,d | 12.7 ± 0.8 a | −1.02 ± 0.14 b | 30.0 ± 1.4 b | 29.2 ± 1.7 b,c | 36.9 ± 1.7 b,c | 4.7 ± 0.9 a | 0.56 ± 0.28 a,b |
| Dehydrin | 143 ± 9 | 6.7 ± 0.5 b,c | 15.6 ± 1.0 a | −1.40 ± 0.05 a | 37.2 ± 2.7 c | 28.7 ± 1.5 b,c | 29.3 ± 1.5 a | 9.5 ± 0.5 c | 1.00 ± 0.19 b,c |
| SMP | 157 ± 15 | 4.9 ± 0.3 a | 16.4 ± 1.6 a | −0.27 ± 0.04 d | 22.9 ± 0.8 a | 26.9 ± 1.3 a,b | 46.6 ± 1.0 d | 6.8 ± 0.4 b | 0.88 ± 0.21 b |
| Protein Family | Motif | aa no. | Motif e-Value | Consensus Sequence | Gravy Index | Consensus Logo * |
|---|---|---|---|---|---|---|
| LEA1 | M1.1 | 50 | 1.25 × 10−52 | TKATVQEKAEQMKTRDPLQKEMATQKKEAKIQEAERQKQEARQQNSAAKH | −1.786 | 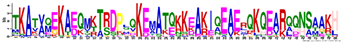 |
| M1.2 | 21 | 3.8 × 10−21 | MQAAKETAANVAASANSGMEK | −0.352 |  | |
| LEA2 | M2.1 | 50 | 5.4 × 10−55 | IEETIGFGKPTADVTDVDLKDINLEKADYVVDVLVKNPYPIPIPLIDINY | 0.048 | 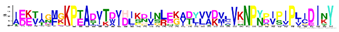 |
| M2.2 | 50 | 2.7 × 10−55 | KSTYADIGPGWIIPYRLKVDLIVDVPVFGRLTLPLEKKGEIPIPYKPDID | −0.018 |  | |
| M2.3 | 50 | 5.1 × 10−63 | IRFDKFSFEETVATLHLKLENKNDFDLGLKDLDYEVWLCNVSIGGAYMKK | −0.268 | 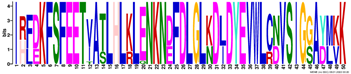 | |
| M2.4 | 50 | 1.1 × 10−44 | TLNLTVTVRNPNFYSIKYDSSTVSIGYRGNKLGRVTIPAGRIGARSSQRV | −0.328 |  | |
| M2.5 | 50 | 1.9 × 10−64 | ITFRPKDFGSALWDMIRGKGTGYTIKGNINVDTPFGFMKLPISKEGGTTC | −0.238 |  | |
| M2.6 | 29 | 1.9 × 10−34 | SGLIPDAGSLKAHGSTTVKVPICLIYDDI | 0.444 | 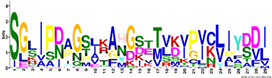 | |
| M2.7 | 50 | 3.1 × 10−57 | NATLQLERVEIMSDVILLLEDLAKGEIMFDTEVDISGKLRVFFFDLPLKT | 0.376 |  | |
| M2.8 | 21 | 1.6 × 10−22 | RNPNKRIGIYYDQIDAYASYK | −1.200 | 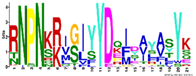 | |
| M2.9 | 24 | 1.2e−28 | GGGKRINDKGWPECNVIMEEGKYD | −1.204 |  | |
| LEA3 | M3.1 | 41 | 1.8 × 10−53 | TYDKNPDEEHAFSAVVPDNVIPPQTQQYWAPHPKTGVFGPA | −0.817 |  |
| M3.2 | 29 | 1.6 × 10−36 | SVSNGGADSVLEQKAFFRPLEDLDKPHHP | −0.766 | 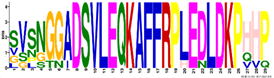 | |
| M3.3 | 29 | 2.4 × 10−35 | MAANLQSRGLASFSKQFVIRVRSRDSTII | 0.048 |  | |
| M3.4 | 50 | 1.9 × 10−48 | IRMLNKESEEPTKISWVPDPVTGYYRPENKATEIDAAELRRILIKDNTRR | −0.994 |  | |
| M3.5 | 6 | 1 × 10−8 | RRGVHV | −0.700 | 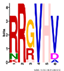 | |
| LEA4 | M4.1 | 29 | 8.5 × 10−29 | AKDYVADKAKEAKDSAAEKAKETKDKAGE | −1.617 | 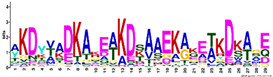 |
| M4.2 | 29 | IIGSLIGTVQGTVEHAKEAVLGKSQEASE | 0.059 |  | ||
| M4.3 | 36 | AKMKAEDTTEAAKETYEETKENARKKMEEMKIVGEG | −1.962 |  | ||
| M4.4 | 21 | AKEKAKEAKDSAKDKAGETKD | −1.438 | 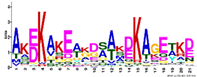 | ||
| LEA5 | M5.1 | 50 | 1.8 × 10−60 | QDKRAELDAKASQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQMGT | −1.228 | 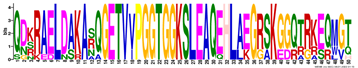 |
| M5.2 | 21 | 3.4 × 10−36 | YQEMGRKGGLSSNDKSGAERAEEEGITID | −1.256 |  | |
| Dehydrins | M6.1 | 29 | 2.3 × 10−30 | GGGGVAGQEEPEKKGMMDKIKEKLPGGHH | −1.214 | 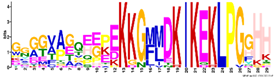 |
| M6.2 | 29 | 8.4 × 10−33 | GPTTGPPKHRRSGSSSSSSSEDDGMGGRR | −1.679 | 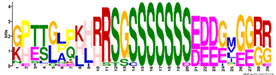 | |
| M6.3 | 29 | 7.2 × 10−37 | MAEYGGNYGNETKQTDEYGNPVHHPQGGG | −1.559 |  | |
| M6.4 | 21 | 5.4 × 10−23 | KGLKDKIKEKLPGGKKETEPP | −1.710 | 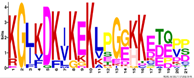 | |
| SMP | M7.1 | 50 | 3.5 × 10−60 | PQDAATMQAAENSVLGQTQKGGVAATMQSAANRNERAGVVGHNDVTDIIS | −0.402 | 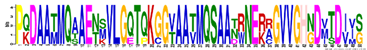 |
| M7.2 | 41 | 1.3 × 10−48 | SAAGDKPVDESDAAAIQAAEARATGLGRVVPGGLGAEAKSA | −0.090 |  |
| Subgroup | Rs_id | LEAP_id | log2(DL/HL) | Subgroup | Rs_id | LEA_id | log2(DL/HL) |
|---|---|---|---|---|---|---|---|
| LEA1.1 | Rs_164046 | RsLEA86 | 6.97 | LEA3.1 | Rs_161911 | RsLEA85 | −3.48 |
| LEA1.1 | Rs_152347 | RsLEA78 | 6.39 | LEA3.2 | Rs_114021 | RsLEA128 | −9.07 |
| LEA1.1 | Rs_185287 | RsLEA104 | 5.24 | LEA4.2 | Rs_146887 | RsLEA75 | 6.42 |
| LEA1.1 | Rs_186228 | RsLEA277 | 3.91 | LEA4.2 | Rs_131921 | RsLEA312 | 5.87 |
| LEA1.1 | Rs_116928 | RsLEA44 | 3.74 | LEA4.2 | Rs_194183 | RsLEA188 | 4.99 |
| LEA1.1 | Rs_105968 | RsLEA146 | 2.52 | LEA4.2 | Rs_186681 | RsLEA310 | 4.94 |
| LEA1.1 | Rs_125102 | RsLEA52 | 2.45 | LEA4.2 | Rs_148951 | RsLEA76 | 3.10 |
| LEA1.1 | Rs_172584 | RsLEA267 | 2.43 | LEA4.2 | Rs_146172 | RsLEA316 | −4.35 |
| LEA1.1 | Rs_183967 | RsLEA101 | 2.35 | LEA4.2 | Rs_182435 | RsLEA51 | −4.91 |
| LEA1.1 | Rs_156613 | RsLEA266 | 2.21 | LEA4.3 | Rs_190897 | RsLEA110 | 6.04 |
| LEA1.3 | Rs_170082 | RsLEA129 | 3.00 | LEA4.3 | Rs_189187 | RsLEA109 | 5.64 |
| LEA1.3 | Rs_108065 | RsLEA26 | 2.55 | LEA4.3 | Rs_131918 | RsLEA311 | 5.12 |
| LEA2.0 | Rs_130914 | RsLEA122 | −2.24 | LEA4.3 | Rs_109487 | RsLEA175 | 4.90 |
| LEA2.1 | Rs_169359 | RsLEA232 | 2.93 | LEA4.3 | Rs_109602 | RsLEA301 | 4.29 |
| LEA2.1 | Rs_127322 | RsLEA55 | 2.33 | LEA4.3 | Rs_184475 | RsLEA309 | 3.77 |
| LEA2.2 | Rs_151841 | RsLEA154 | 4.64 | LEA4.3 | Rs_149505 | RsLEA130 | 2.95 |
| LEA2.2 | Rs_104785 | RsLEA275 | 4.51 | LEA4.3 | Rs_181059 | RsLEA302 | 2.74 |
| LEA2.2 | Rs_125141 | RsLEA276 | 3.66 | LEA4.3 | Rs_136891 | RsLEA314 | 2.73 |
| LEA2.2 | Rs_164865 | RsLEA272 | 2.50 | LEA4.3 | Rs_190898 | RsLEA111 | 2.31 |
| LEA2.2 | Rs_187807 | RsLEA125 | 2.07 | LEA4.3 | Rs_108999 | RsLEA36 | 2.18 |
| LEA2.2 | Rs_173883 | RsLEA265 | −2.98 | LEA4.3 | Rs_166537 | RsLEA49 | 1.99 |
| LEA2.2 | Rs_166384 | RsLEA269 | −4.06 | LEA4.3 | Rs_172003 | RsLEA95 | −3.63 |
| LEA2.3 | Rs_194495 | RsLEA270 | 3.77 | LEA5 | Rs_188268 | RsLEA202 | 11.80 |
| LEA2.3 | Rs_110370 | RsLEA262 | 3.19 | LEA5 | Rs_159833 | RsLEA196 | 8.53 |
| LEA2.3 | Rs_121097 | RsLEA261 | 3.09 | LEA5 | Rs_128109 | RsLEA200 | 8.13 |
| LEA2.3 | Rs_118201 | RsLEA230 | 2.44 | LEA5 | Rs_193475 | RsLEA204 | 8.04 |
| LEA2.3 | Rs_183071 | RsLEA264 | 2.39 | LEA5 | Rs_124807 | RsLEA201 | 5.55 |
| LEA2.3 | Rs_193485 | RsLEA291 | −2.14 | LEA5 | Rs_125649 | RsLEA199 | 4.35 |
| LEA2.3 | Rs_171129 | RsLEA256 | −2.59 | LEA5 | Rs_176248 | RsLEA203 | 2.86 |
| LEA2.3 | Rs_138912 | RsLEA296 | −2.71 | DEH1 | Rs_131408 | RsLEA166 | 3.78 |
| LEA2.3 | Rs_145248 | RsLEA285 | −2.89 | DEH1 | Rs_172145 | RsLEA139 | 3.38 |
| LEA2.3 | Rs_180651 | RsLEA98 | −3.46 | DEH1 | Rs_134636 | RsLEA298 | 3.09 |
| LEA2.3 | Rs_138298 | RsLEA68 | −5.55 | DEH1 | Rs_107019 | RsLEA152 | 2.78 |
| LEA2.4 | Rs_110833 | RsLEA254 | −2.55 | DEH1 | Rs_181340 | RsLEA151 | 2.50 |
| LEA2.4 | Rs_181906 | RsLEA257 | −3.77 | DEH1 | Rs_113392 | RsLEA163 | 2.30 |
| LEA2.5 | Rs_160078 | RsLEA228 | 8.12 | DEH2 | Rs_156753 | RsLEA172 | 4.35 |
| LEA2.5 | Rs_159852 | RsLEA239 | 3.66 | SMP1 | Rs_140935 | RsLEA70 | 7.78 |
| LEA2.5 | Rs_162712 | RsLEA211 | 3.34 | SMP1 | Rs_106521 | RsLEA33 | 3.67 |
| LEA2.5 | Rs_139255 | RsLEA244 | −2.32 | SMP2 | Rs_135719 | RsLEA66 | 8.03 |
| LEA2.5 | Rs_186090 | RsLEA121 | −3.49 | SMP2 | Rs_134737 | RsLEA65 | 3.22 |
| LEA2.5 | Rs_140027 | RsLEA212 | −3.75 | SMP2 | Rs_134736 | RsLEA64 | 2.98 |
| LEA2.5 | Rs_149607 | RsLEA103 | −4.03 | SMP2 | Rs_156298 | RsLEA83 | 2.45 |
| LEA3.1 | Rs_153025 | RsLEA80 | 2.23 | SMP3 | Rs_140941 | RsLEA71 | 9.22 |
| LEA3.1 | Rs_125374 | RsLEA53 | −2.00 | SMP3 | Rs_106559 | RsLEA34 | 5.99 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pantelić, A.; Stevanović, S.; Komić, S.M.; Kilibarda, N.; Vidović, M. In Silico Characterisation of the Late Embryogenesis Abundant (LEA) Protein Families and Their Role in Desiccation Tolerance in Ramonda serbica Panc. Int. J. Mol. Sci. 2022, 23, 3547. https://doi.org/10.3390/ijms23073547
Pantelić A, Stevanović S, Komić SM, Kilibarda N, Vidović M. In Silico Characterisation of the Late Embryogenesis Abundant (LEA) Protein Families and Their Role in Desiccation Tolerance in Ramonda serbica Panc. International Journal of Molecular Sciences. 2022; 23(7):3547. https://doi.org/10.3390/ijms23073547
Chicago/Turabian StylePantelić, Ana, Strahinja Stevanović, Sonja Milić Komić, Nataša Kilibarda, and Marija Vidović. 2022. "In Silico Characterisation of the Late Embryogenesis Abundant (LEA) Protein Families and Their Role in Desiccation Tolerance in Ramonda serbica Panc" International Journal of Molecular Sciences 23, no. 7: 3547. https://doi.org/10.3390/ijms23073547
APA StylePantelić, A., Stevanović, S., Komić, S. M., Kilibarda, N., & Vidović, M. (2022). In Silico Characterisation of the Late Embryogenesis Abundant (LEA) Protein Families and Their Role in Desiccation Tolerance in Ramonda serbica Panc. International Journal of Molecular Sciences, 23(7), 3547. https://doi.org/10.3390/ijms23073547








