Overexpression of SgDREB2C from Stylosanthes guianensis Leads to Increased Drought Tolerance in Transgenic Arabidopsis
Abstract
:1. Introduction
2. Results
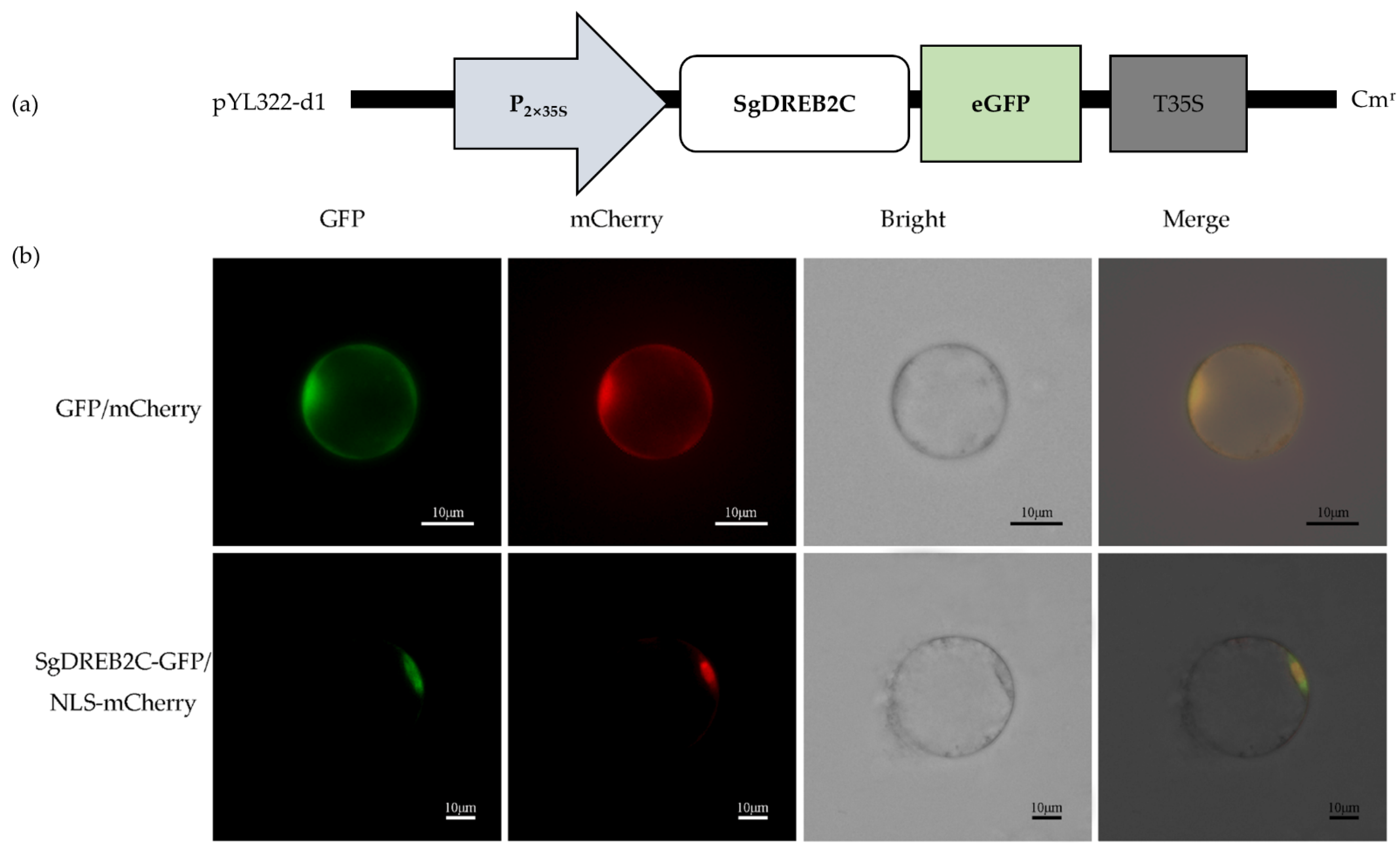
2.1. Characterization of SgDREB2C
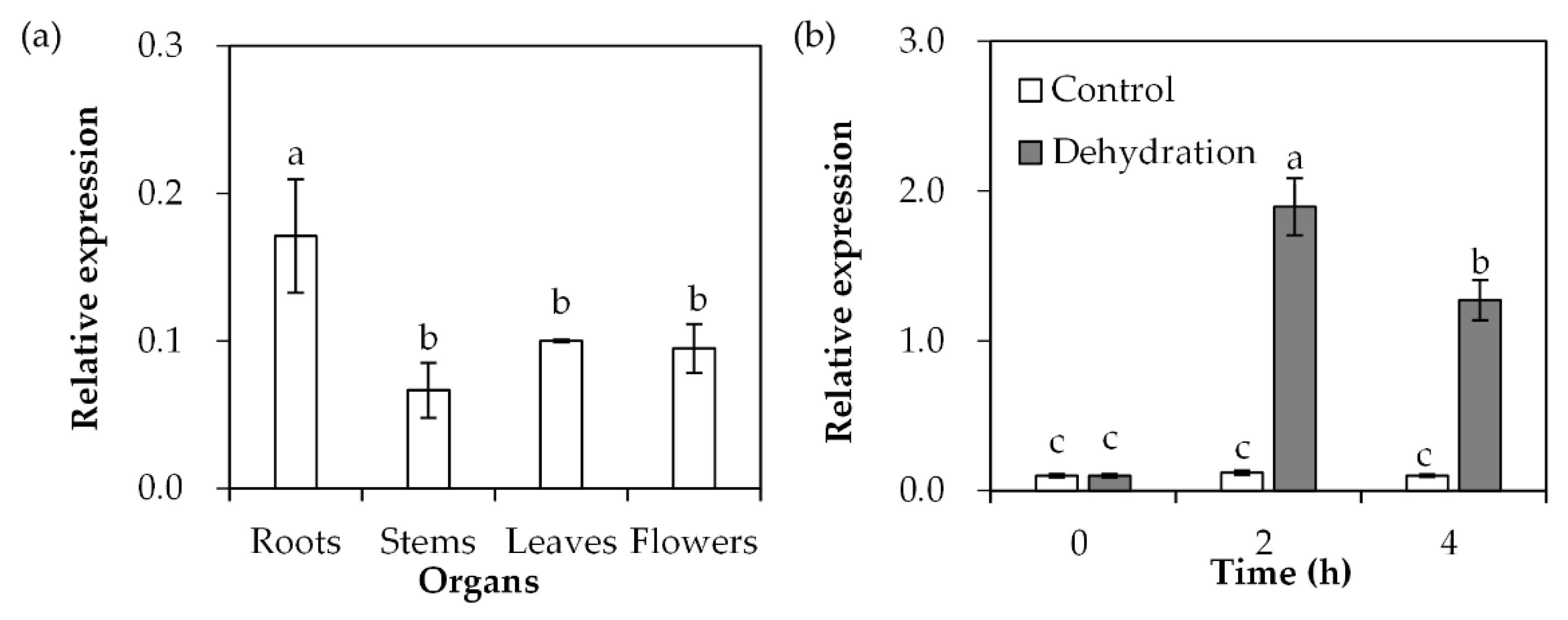
2.2. Analysis of Spatial Expression of SgDREB2C and Response to Dehydration
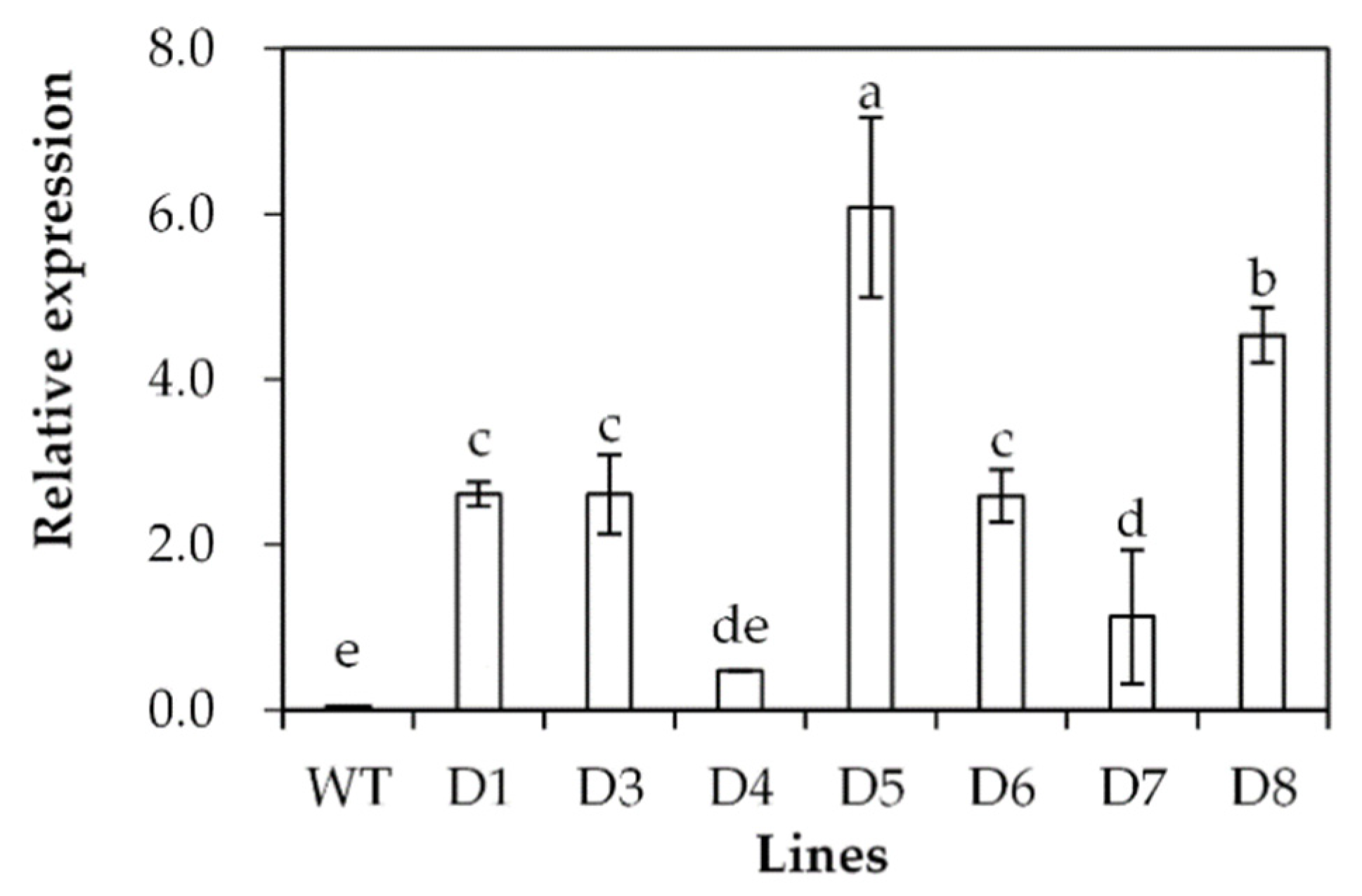
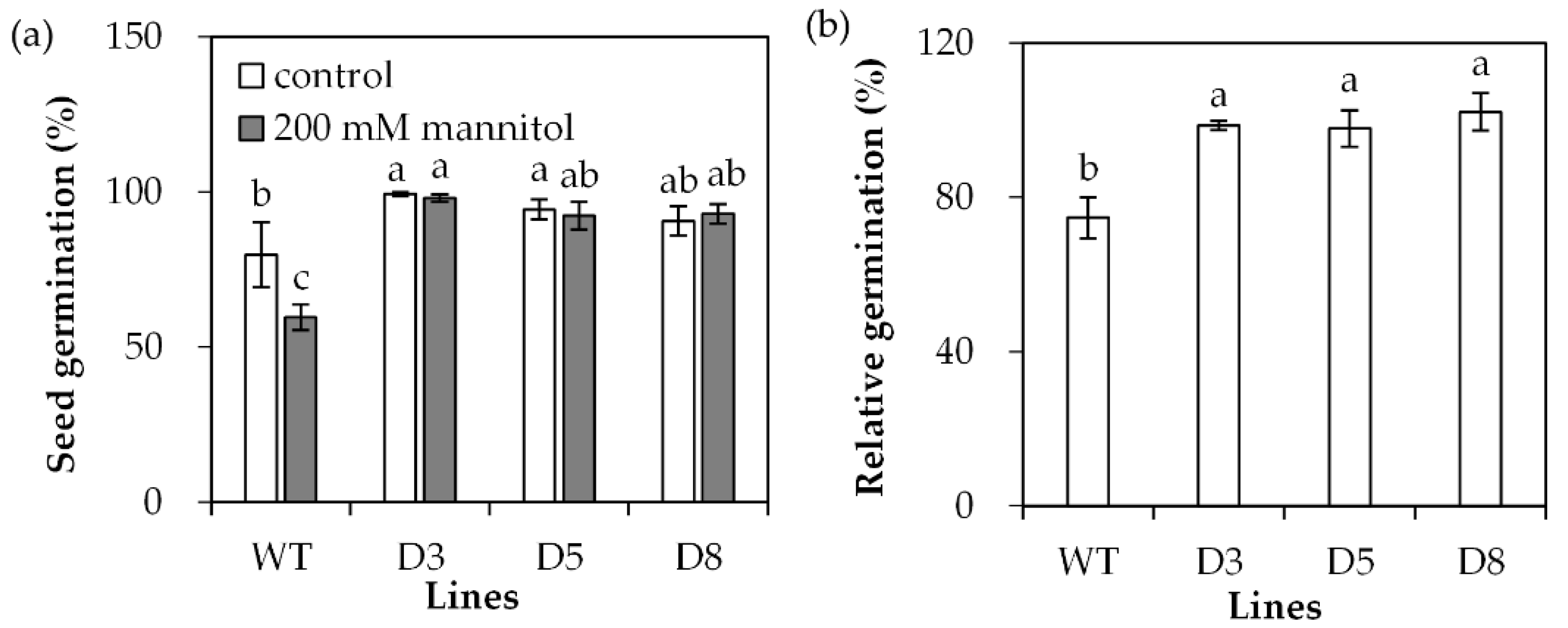
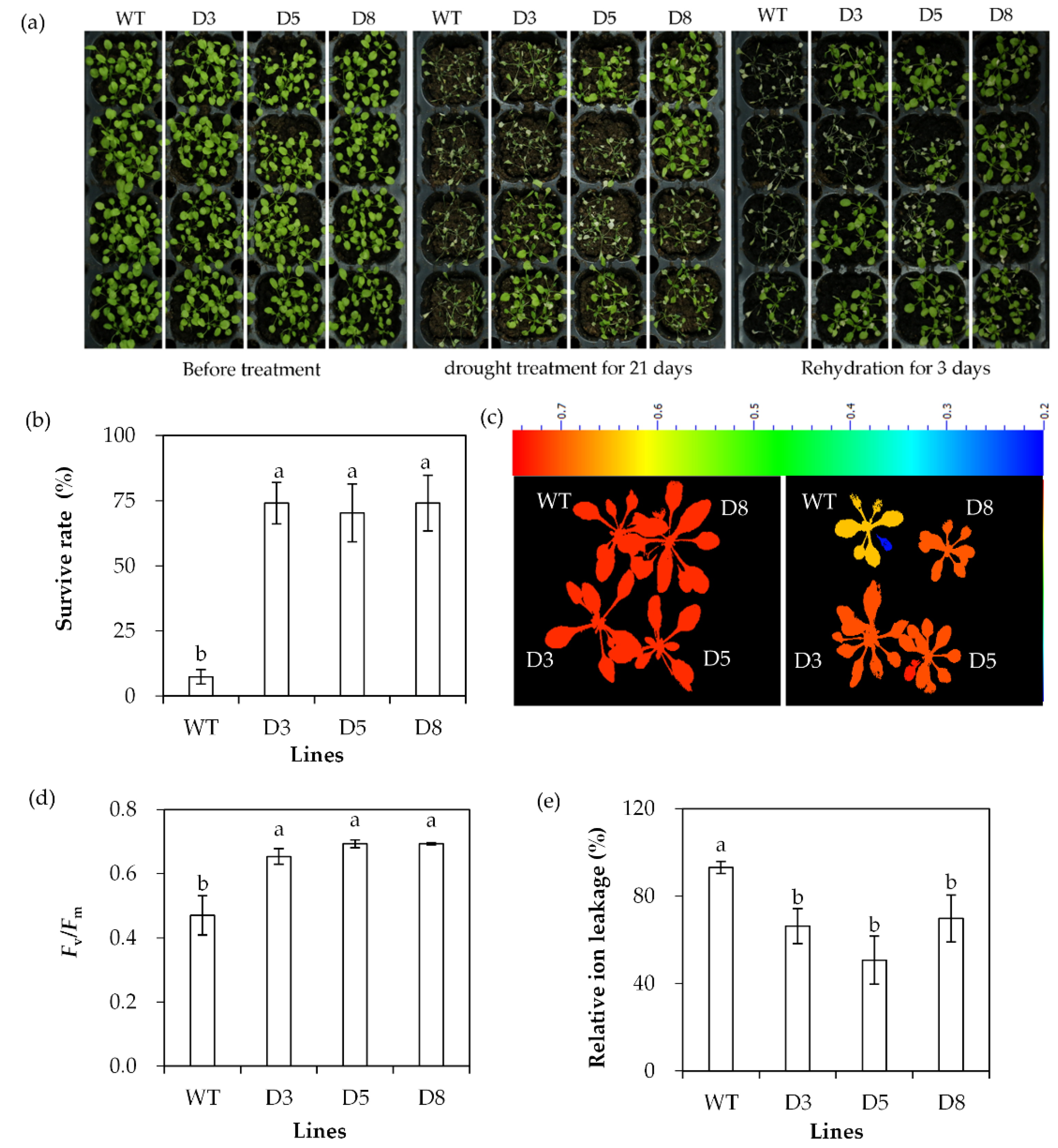
2.3. Analysis of Drought Tolerance in Transgenic Plants Overexpressing SgDREB2C
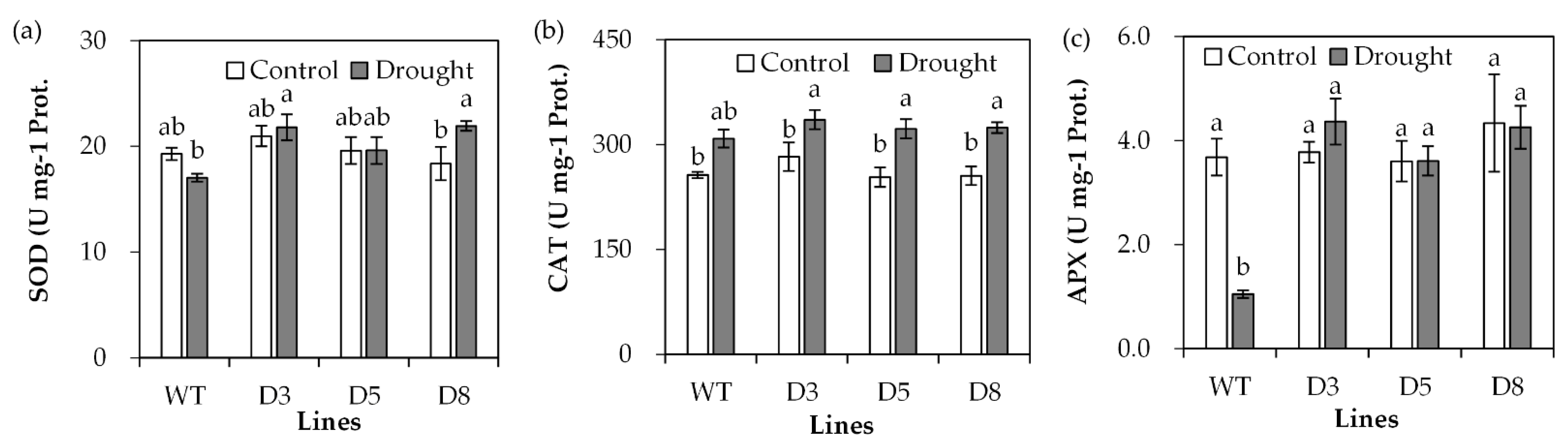
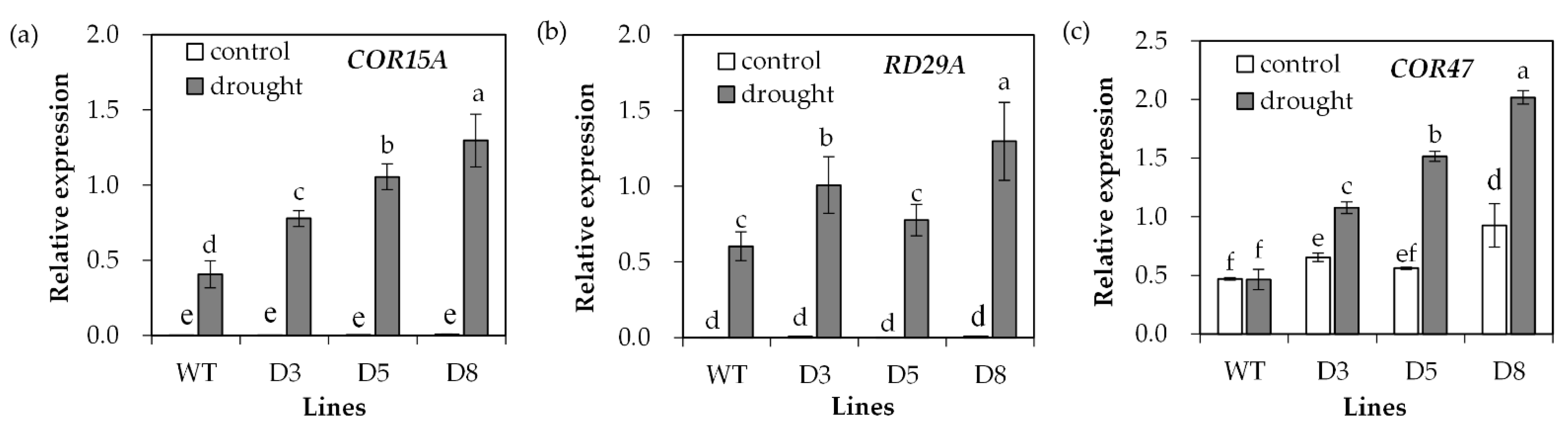
2.4. Analysis of Antioxidant Defense System and Stress-Responsive Genes
3. Discussion
4. Materials and Methods
4.1. Plant Growth and Dehydration Treatment
4.2. Cloning and Sequence Analysis of SgDREB2C
4.3. Subcellular Localization Assay
4.4. Generation of Transgenic Arabidopsis
4.5. Evaluation of Drought Tolerance
4.6. Measurements of Antioxidant Enzyme Activities
4.7. Real-Time Quantitative PCR (qPCR)
4.8. Statistic Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gong, Z.Z.; Xiong, L.M.; Shi, H.Z.; Yang, S.H.; Herrera-Estrella, L.R.; Xu, G.H.; Chao, D.Y.; Li, J.R.; Wang, P.Y.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci.China-Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, T.; Khalid, S.; Abdullah, M.; Ahmed, Z.; Shah, M.K.; Ghafoor, A.; Du, X. Insights into drought stress signaling in plants and the molecular genetic basis of cotton drought tolerance. Cells 2020, 9, 105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhang, Z.; Xie, S.; Si, T.; Li, Y.; Zhu, J.-K. Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 2016, 171, 2744–2759. [Google Scholar] [CrossRef] [Green Version]
- Jia, Y.X.; Ding, Y.L.; Shi, Y.T.; Zhang, X.Y.; Gong, Z.Z.; Yang, S.H. The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 2016, 212, 345–353. [Google Scholar] [CrossRef] [Green Version]
- Kidokoro, S.; Watanabe, K.; Ohori, T.; Moriwaki, T.; Maruyama, K.; Mizoi, J.; Htwe, N.M.P.S.; Fujita, Y.; Sekita, S.; Shinozaki, K.; et al. Soybean DREB1/CBF-type transcription factors function in heat and drought as well as cold stress-responsive gene expression. Plant J. 2015, 81, 505–518. [Google Scholar] [CrossRef]
- Zhou, Y.B.; Chen, M.; Guo, J.K.; Wang, Y.X.; Min, D.H.; Jiang, Q.Y.; Ji, H.T.; Huang, C.Y.; Wei, W.; Xu, H.J.; et al. Overexpression of soybean DREB1 enhances drought stress tolerance of transgenic wheat in the field. J. Exp. Bot. 2020, 71, 1842–1857. [Google Scholar] [CrossRef]
- Qin, F.; Kakimoto, M.; Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Tran, L.S.P.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J. 2007, 50, 54–69. [Google Scholar] [CrossRef]
- Song, C.; Lee, J.; Kim, T.; Hong, J.C.; Lim, C.O. VOZ1, a transcriptional repressor of DREB2C, mediates heat stress responses in Arabidopsis. Planta 2018, 247, 1439–1448. [Google Scholar] [CrossRef]
- Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Seki, M.; Miura, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration- and high-salinity-responsive gene expression. Plant Mol. Biol. 2000, 42, 657–665. [Google Scholar] [CrossRef]
- Larkindale, J.; Vierling, E. Core genome responses involved in acclimation to high temperature. Plant Physiol. 2008, 146, 748. [Google Scholar] [CrossRef] [Green Version]
- Sakuma, Y.; Maruyama, K.; Qin, F.; Osakabe, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. USA 2006, 103, 18822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell. 2006, 18, 1292–1309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.-X.; Tang, Y.-J.; Ma, Q.-B.; Yang, C.-Y.; Mu, Y.-H.; Suo, H.-C.; Luo, L.-H.; Nian, H. OsDREB2A, a rice transcription factor, significantly affects salt tolerance in transgenic soybean. PLoS ONE 2013, 8, e83011. [Google Scholar] [CrossRef] [PubMed]
- Dubouzet, J.G.; Sakuma, Y.; Ito, Y.; Kasuga, M.; Dubouzet, E.G.; Miura, S.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003, 33, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-Q.; Meng, X.-P.; Zhang, Y.; Xia, M.; Wang, X.-P. Over-expression of OsDREB genes lead to enhanced drought tolerance in rice. Biotechnol. Lett. 2008, 30, 2191–2198. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.Y.; Wang, X.W.; Wu, Z.J.; Chen, M.; Chai, T.Y.; Wang, H. Overexpression of the Polygonum cuspidatum PcDREB2A gene encoding a DRE-Binding transcription factor enhances the drought tolerance of transgenic Arabidopsis thaliana. J. Plant. Biol. 2021, in press. [CrossRef]
- Li, X.S.; Zhang, D.Y.; Li, H.Y.; Wang, Y.C.; Zhang, Y.M.; Wood, A.J. EsDREB2B, a novel truncated DREB2-type transcription factor in the desert legume Eremosparton songoricum, enhances tolerance to multiple abiotic stresses in yeast and transgenic tobacco. BMC Plant Biol. 2014, 14, 44. [Google Scholar] [CrossRef] [Green Version]
- Hwang, J.E.; Lim, C.J.; Chen, H.; Je, J.; Song, C.; Lim, C.O. Overexpression of Arabidopsis dehydration-responsive element-binding protein 2C confers tolerance to oxidative stress. Mol. Cells 2012, 33, 135–140. [Google Scholar] [CrossRef] [Green Version]
- Lim, C.J.; Hwang, J.E.; Chen, H.; Hong, J.K.; Yang, K.A.; Choi, M.S.; Lee, K.O.; Chung, W.S.; Lee, S.Y.; Lim, C.O. Over-expression of the Arabidopsis DRE/CRT-binding transcription factor DREB2C enhances thermotolerance. Biochem. Biophys. Res. Commun. 2007, 362, 431–436. [Google Scholar] [CrossRef]
- Lee, S.-J.; Kang, J.-Y.; Park, H.-J.; Kim, M.D.; Bae, M.S.; Choi, H.-I.; Kim, S.Y. DREB2C interacts with ABF2, a bZIP protein regulating abscisic acid-responsive gene expression, and its overexpression affects abscisic acid sensitivity. Plant Physiol. 2010, 153, 716–727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Hwang, J.E.; Lim, C.J.; Kim, D.Y.; Lee, S.Y.; Lim, C.O. Arabidopsis DREB2C functions as a transcriptional activator of HsfA3 during the heat stress response. Biochem. Biophys. Res. Commun. 2010, 401, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Je, J.; Song, C.; Hwang, J.E.; Chung, W.S.; Lim, C.O. DREB2C acts as a transcriptional activator of the thermo tolerance-related phytocystatin 4 (AtCYS4) gene. Transgenic Res. 2014, 23, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Nguyen, T.T.; Baek, J.; Song, Y.H.; Hong, J.C.; Lim, C.O. DREB2 family transcription factors promote the expression of Phytocystatin1 in Arabidopsis thaliana during seed germination. J. Plant Biol. 2020, 64, 45–53. [Google Scholar] [CrossRef]
- Je, J.; Chen, H.; Song, C.; Lim, C.O. Arabidopsis DREB2C modulates ABA biosynthesis during germination. Biochem. Biophys. Res. Commun. 2014, 452, 91–98. [Google Scholar] [CrossRef]
- Herath, V. Small family, big impact: In silico analysis of DREB2 transcription factor family in rice. Comput. Biol. Chem. 2016, 65, 128–139. [Google Scholar] [CrossRef]
- Zhao, K.; Shen, X.J.; Yuan, H.Z.; Liu, Y.; Liao, X.; Wang, Q.; Liu, L.L.; Li, F.; Li, T.H. Isolation and characterization of dehydration-responsive element-binding factor 2C (MsDREB2C) from Malus sieversii Roem. Plant Cell Physiol. 2013, 54, 1415–1430. [Google Scholar] [CrossRef] [Green Version]
- Zhao, K.; Guo, X.W.; Liao, X.; Wang, Q.; Liu, D.; Li, T.H. Arabidopsis plants overexpressing the MsDREB2C exhibit increased susceptibility to alternaria mali infection. J. Plant Growth Regul. 2015, 34, 78–87. [Google Scholar] [CrossRef]
- Zhang, C.; Shi, S.; Liu, Z.; Yang, F.; Yin, G. Drought tolerance in alfalfa (Medicago sativa L.) varieties is associated with enhanced antioxidative protection and declined lipid peroxidation. J. Plant Physiol. 2019, 232, 226–240. [Google Scholar] [CrossRef]
- Sharma, P.; Sharma, P.; Arora, P.; Verma, V.; Khanna, K.; Saini, P.; Bhardwaj, R. Role and Regulation of ROS and Antioxidants as Signaling Molecules in Response to Abiotic Stresses. In Plant Signaling Molecules, 1st ed.; Khan, M.I., Reddy, P.S., Ferrante, A., Khan, N., Eds.; Woodhead Publishing: Sawston, Cambridge, UK, 2019; pp. 141–156. [Google Scholar]
- Da Costa, M.; Huang, B.R. Changes in antioxidant enzyme activities and lipid peroxidation for bentgrass species in response to drought stress. J. Am. Soc. Hort. Sci. 2007, 132, 319–326. [Google Scholar] [CrossRef] [Green Version]
- Sheoran, S.; Thakur, V.; Narwal, S.; Turan, R.; Mamrutha, H.M.; Singh, V.; Tiwari, V.; Sharma, I. Differential activity and expression profile of antioxidant enzymes and physiological changes in wheat (Triticum aestivum L.) under drought. Appl. Biochem. Biotechnol. 2015, 177, 1282–1298. [Google Scholar] [CrossRef]
- Bao, G.; Zhuo, C.; Qian, C.; Xiao, T.; Guo, Z.; Lu, S. Co-expression of NCED and ALO improves vitamin C level and tolerance to drought and chilling in transgenic tobacco and stylo plants. Plant Biotechnol. J. 2016, 14, 206–214. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, J.; Lu, S.; Cai, J.; Guo, Z. Overexpressing SgNCED1 in tobacco increases ABA level, antioxidant enzyme activities, and stress tolerance. J. Plant Growth Regul. 2008, 27, 151–158. [Google Scholar] [CrossRef]
- Zhou, B.; Guo, Z.; Liu, Z. Effects of abscisic acid on antioxidant systems of Stylosanthes guianensis (Aublet) Sw. under chilling stress. Crop Sci. 2005, 45, 599–605. [Google Scholar] [CrossRef]
- Lu, S.; Wang, X.; Guo, Z. Differential responses to chilling in Stylosanthes guianensis (Aublet) Sw. and its mutants. Agron. J. 2013, 105, 377–382. [Google Scholar] [CrossRef]
- Yang, J.; Guo, Z. Cloning of a 9-cis-epoxycarotenoid dioxygenase gene (SgNCED1) from Stylosanthes guianensis and its expression in response to abiotic stresses. Plant Cell Rep. 2007, 26, 1383–1390. [Google Scholar] [CrossRef]
- Chen, H.; Je, J.; Song, C.; Hwang, J.E.; Lim, C.O. A proximal promoter region of Arabidopsis DREB2C confers tissue—Specific expression under heat stress. J. Integr. Plant Biol. 2012, 54, 640–651. [Google Scholar] [CrossRef]
- Ghotbi-Ravandi, A.A.; Shahbazi, M.; Shariati, M.; Mulo, P. Effects of mild and severe drought stress on photosynthetic efficiency in tolerant and susceptible barley (Hordeum vulgare L.) genotypes. J. Agron. Crop Sci. 2014, 200, 403–415. [Google Scholar] [CrossRef]
- Daszkowska-Golec, A.; Collin, A.; Sitko, K.; Janiak, A.; Kalaji, H.M.; Szarejko, I. Genetic and physiological dissection of photosynthesis in barley exposed to drought stress. Int. J. Mol. Sci. 2019, 20, 6341. [Google Scholar] [CrossRef] [Green Version]
- Saglam, A.; Terzi, R.; Demiralay, M. Effect of polyethylene glycol induced drought stress on photosynthesis in two chickpea genotypes with different drought tolerance. Acta Biol. Hung. 2014, 65, 178–188. [Google Scholar] [CrossRef]
- Laxa, M.; Liebthal, M.; Telman, W.; Chibani, K.; Dietz, K.-J. The role of the plant antioxidant system in drought tolerance. Antioxidants 2019, 8, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Huang, Y.; Khadr, A.; Wang, Y.-H.; Xu, Z.-S.; Xiong, A.-S. DcDREB1A, a DREB-binding transcription factor from Daucus carota, enhances drought tolerance in transgenic Arabidopsis thaliana and modulates lignin levels by regulating lignin-biosynthesis-related genes. Environ. Exp. Bot. 2020, 169, 103896. [Google Scholar] [CrossRef]
- Seki, M.; Narusaka, M.; Abe, H.; Kasuga, M.; Yamaguchi-Shinozaki, K.; Carninci, P.; Hayashizaki, Y.; Shinozaki, K. Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell. 2001, 13, 61–72. [Google Scholar] [CrossRef] [Green Version]
- Ju, Y.L.; Min, Z.; Yue, X.F.; Zhang, Y.L.; Zhang, J.X.; Zhang, Z.Q.; Fang, Y.L. Overexpression of grapevine VvNAC08 enhances drought tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2020, 151, 214–222. [Google Scholar] [CrossRef]
- Chen, C.C.; Cui, X.Y.; Zhang, P.Y.; Wang, Z.; Zhang, J.X. Expression of the pyrroline-5-carboxylate reductase (P5CR) gene from the wild grapevine Vitis yeshanensis promotes drought resistance in transgenic Arabidopsis. Plant Physiol. Biochem. 2021, 168, 188–201. [Google Scholar] [CrossRef]
- Lu, P.L.; Chen, N.Z.; An, R.; Su, Z.; Qi, B.S.; Ren, F.; Chen, J.; Wang, X.C. A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol. Biol. 2007, 63, 289–305. [Google Scholar] [CrossRef]
- Woo, J.W.; Kim, J.; Il Kwon, S.; Corvalan, C.; Cho, S.W.; Kim, H.; Kim, S.-G.; Kim, S.-T.; Choe, S.; Kim, J.-S. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 2015, 33, 1162–1164. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Veerappa, R.; Robert, D.; Slocum, R.D.; Siegenthaler, A.; Wang, J.; Clark, G.; Roux, S.J. Ectopic expression of a pea apyrase enhances root system architecture and drought survival in Arabidopsis and soybean. Plant Cell Environ. 2019, 42, 337–353. [Google Scholar] [CrossRef]
- Huang, S.; Chen, M.; Zhao, Y.; Wen, X.; Guo, Z.; Lu, S. CBL4-CIPK5 pathway confers salt but not drought and chilling tolerance by regulating ion homeostasis. Environ. Exp. Bot. 2020, 179, 104230. [Google Scholar] [CrossRef]
- Guo, Z.; Tan, J.; Zhuo, C.; Wang, C.; Xiang, B.; Wang, Z. Abscisic acid, H2O2 and nitric oxide interactions mediated cold-induced S-adenosylmethionine synthetase in Medicago sativa subsp. falcata that confers cold tolerance through up-regulating polyamine oxidation. Plant Biotechnol. J. 2014, 12, 601–612. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, Y.; Xiang, L.; Song, Z.; Lu, S. Overexpression of SgDREB2C from Stylosanthes guianensis Leads to Increased Drought Tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2022, 23, 3520. https://doi.org/10.3390/ijms23073520
Han Y, Xiang L, Song Z, Lu S. Overexpression of SgDREB2C from Stylosanthes guianensis Leads to Increased Drought Tolerance in Transgenic Arabidopsis. International Journal of Molecular Sciences. 2022; 23(7):3520. https://doi.org/10.3390/ijms23073520
Chicago/Turabian StyleHan, Yun, Leilei Xiang, Zhigang Song, and Shaoyun Lu. 2022. "Overexpression of SgDREB2C from Stylosanthes guianensis Leads to Increased Drought Tolerance in Transgenic Arabidopsis" International Journal of Molecular Sciences 23, no. 7: 3520. https://doi.org/10.3390/ijms23073520
APA StyleHan, Y., Xiang, L., Song, Z., & Lu, S. (2022). Overexpression of SgDREB2C from Stylosanthes guianensis Leads to Increased Drought Tolerance in Transgenic Arabidopsis. International Journal of Molecular Sciences, 23(7), 3520. https://doi.org/10.3390/ijms23073520




