Optimization of Short RNA Aptamers for TNBC Cell Targeting
Abstract
:1. Introduction
2. Results
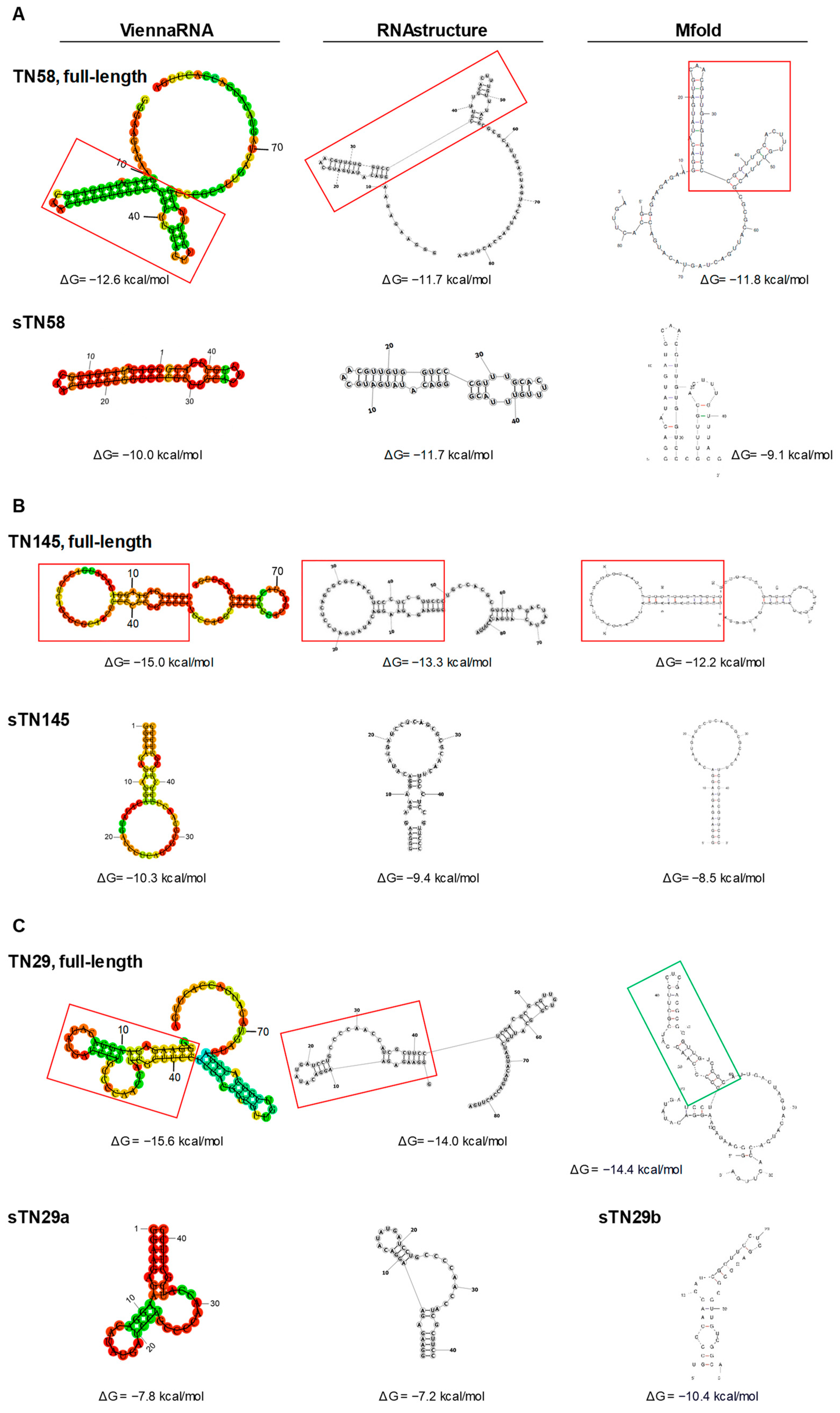
2.1. Identification of Potential Binding Sites
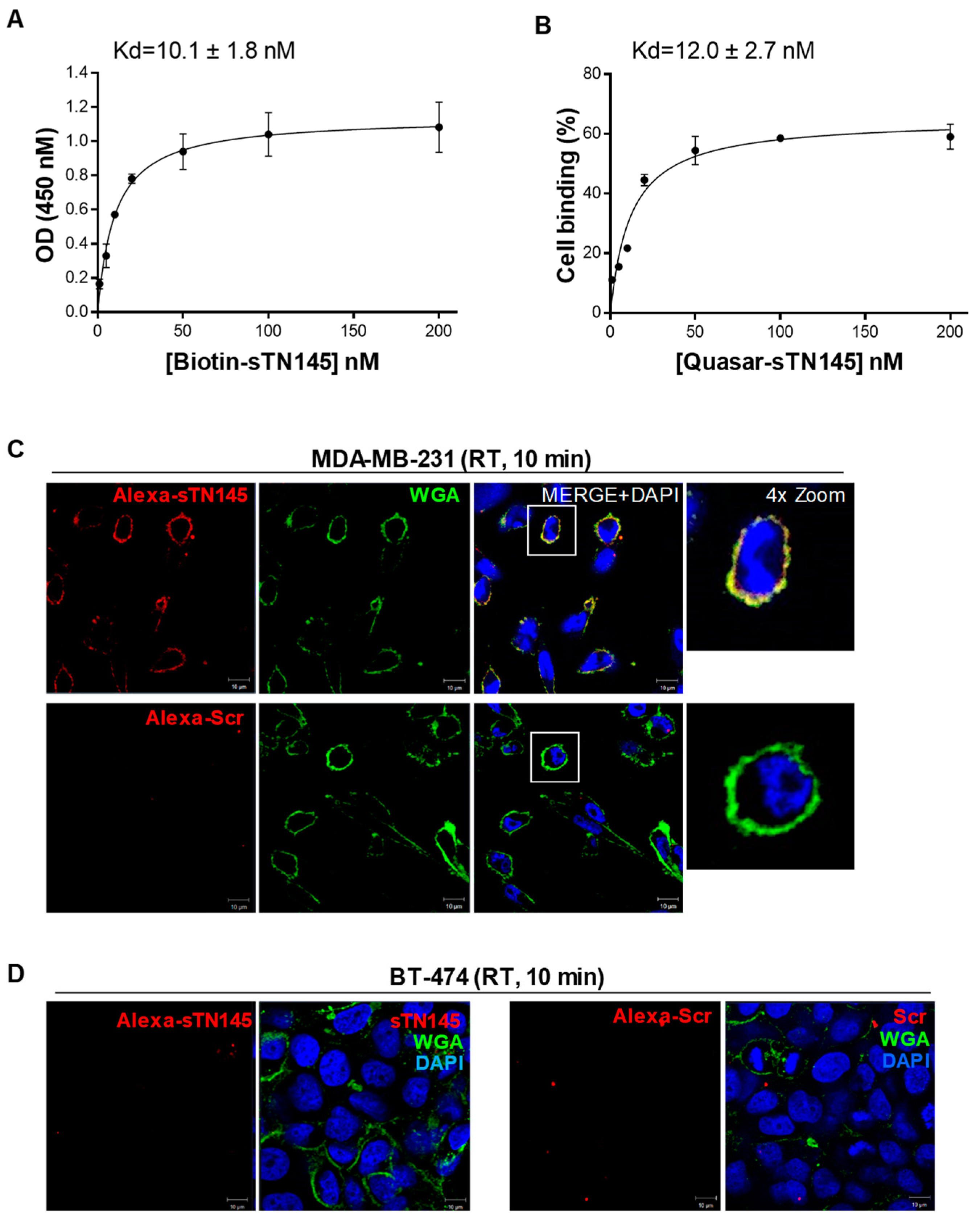
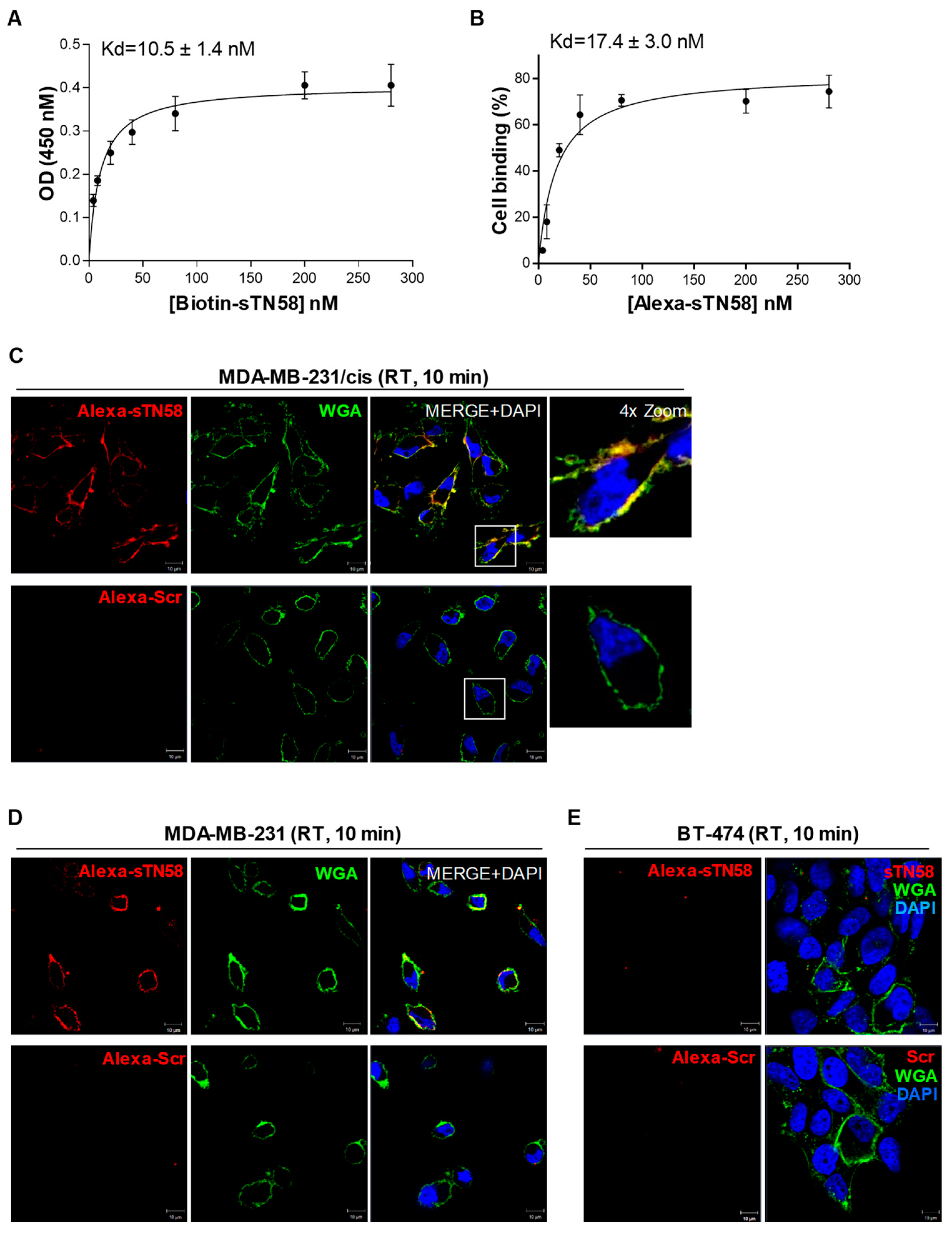
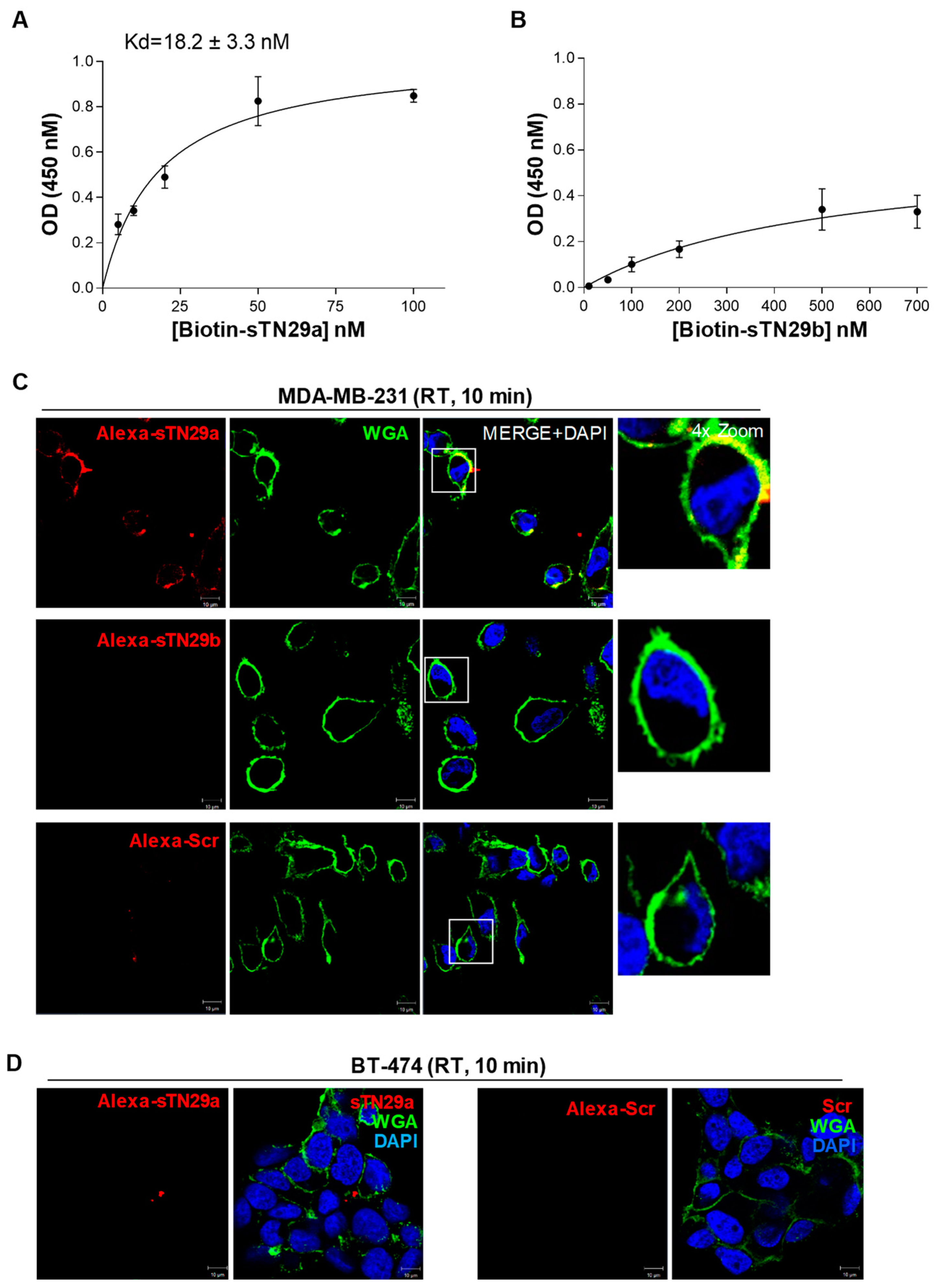
2.2. The Short Aptamers Specifically Target TNBC Cells
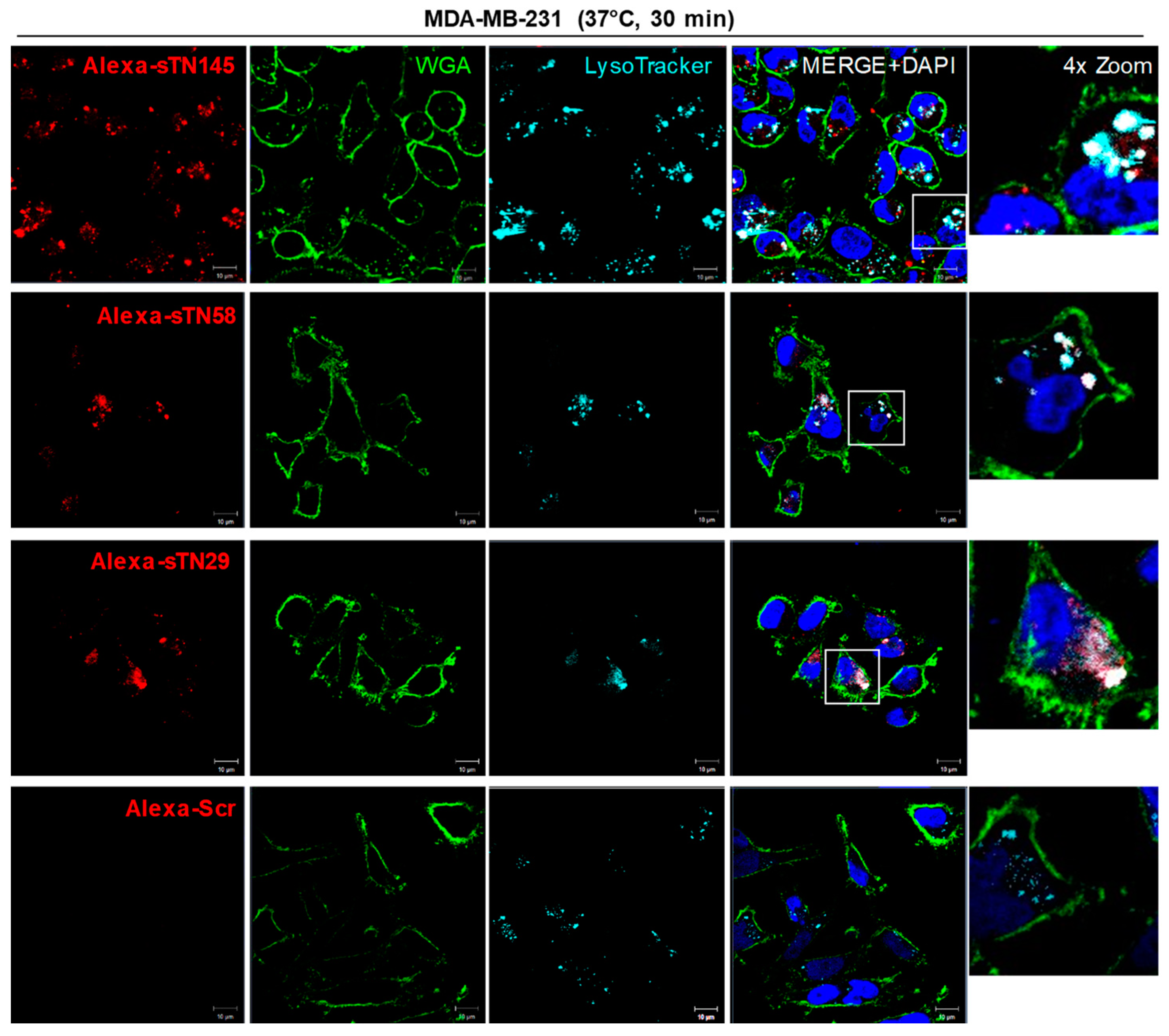
2.3. The Short Aptamers Actively Internalize into TNBC Cells
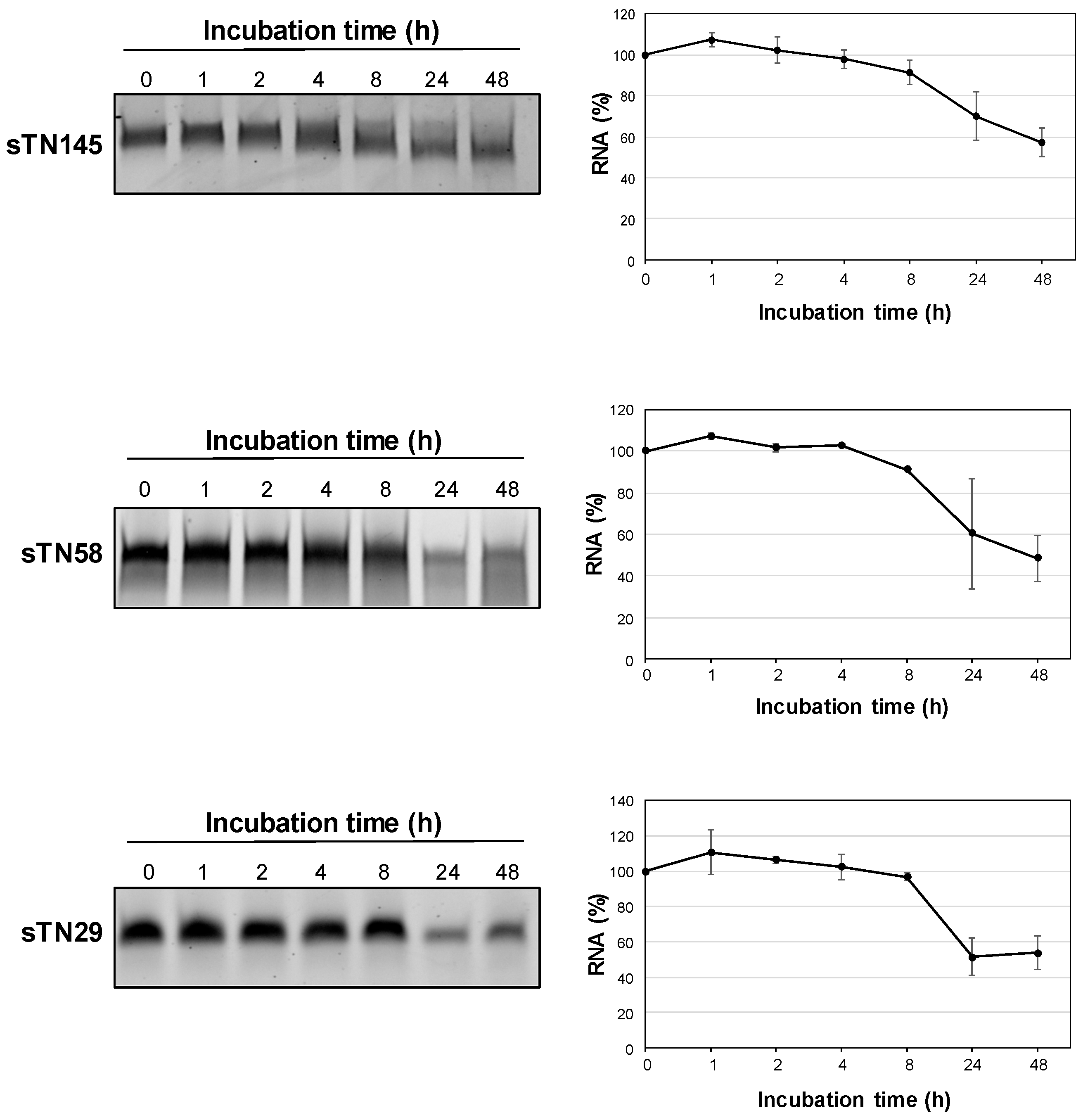
2.4. The Short Aptamers Are Highly Resistant to Nuclease Degradation
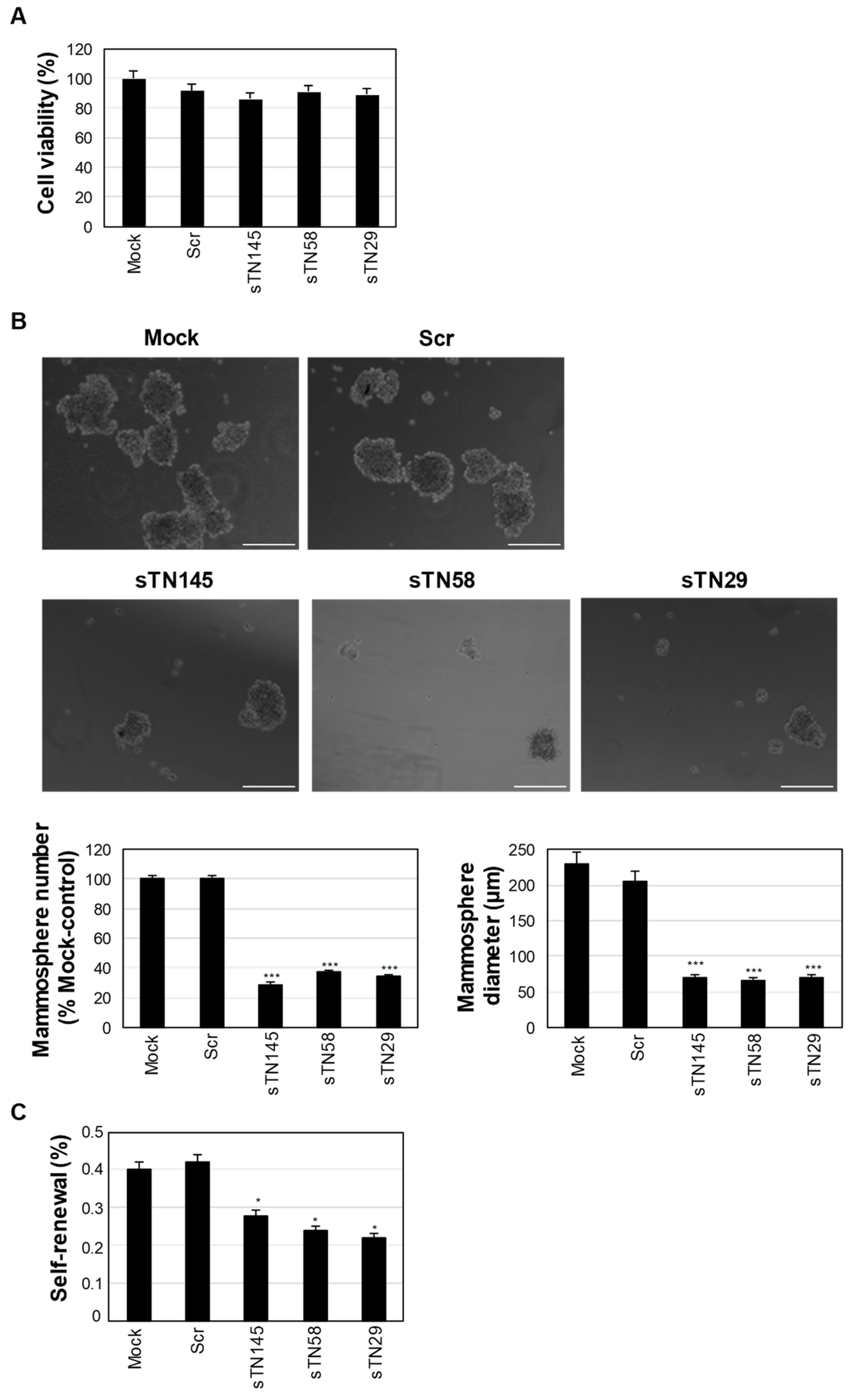
2.5. The Short Aptamers Efficiently Target TNBC CS.C.s
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Culture Conditions
4.2. Aptamer Synthesis, Use Conditions and Secondary Structure Prediction
4.3. Cell Binding Analyses
4.4. Confocal Microscopy
4.5. Cell Viability
4.6. Aptamer Stability in Human Serum
4.7. Mammosphere Formation Assay
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Newman, L.A.; Reis-Filho, J.S.; Morrow, M.; Carey, L.A.; King, T.A. The 2014 Society of Surgical Oncology Susan G. Komen for the Cure Symposium: Triple-negative breast cancer. Ann. Surg. Oncol. 2015, 22, 874–882. [Google Scholar] [PubMed]
- Camorani, S.; Fedele, M.; Zannetti, A.; Cerchia, L. TNBC Challenge: Oligonucleotide Aptamers for New Imaging and Therapy Modalities. Pharmaceuticals 2018, 11, 123. [Google Scholar]
- Gadi, V.K.; Davidson, N.E. Practical Approach to Triple-Negative Breast Cancer. J. Oncol. Pract. 2017, 13, 293–300. [Google Scholar] [PubMed]
- Echeverria, G.V.; Ge, Z.; Seth, S.; Zhang, X.; Jeter-Jones, S.; Zhou, X.; Cai, S.; Tu, Y.; McCoy, A.; Peoples, M.; et al. Resistance to neoadjuvant chemotherapy in triple-negative breast cancer mediated by a reversible drug-tolerant state. Sci. Transl. Med. 2019, 11, eaav0936. [Google Scholar]
- Garrido-Castro, A.C.; Lin, N.U.; Polyak, K. Insights into Molecular Classifications of Triple-Negative Breast Cancer: Improving Patient Selection for Treatment. Cancer Discov. 2019, 9, 176–198. [Google Scholar]
- Shigdar, S.; Agnello, L.; Fedele, M.; Camorani, S.; Cerchia, L. Profiling Cancer Cells by Cell-SELEX: Use of Aptamers for Discovery of Actionable Biomarkers and Therapeutic Applications Thereof. Pharmaceutics 2021, 14, 28. [Google Scholar]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar]
- Agnello, L.; Camorani, S.; Fedele, M.; Cerchia, L. Aptamers and antibodies: Rivals or allies in cancer targeted therapy? Explor. Target Antitumor Ther. 2021, 2, 107–121. [Google Scholar]
- Ravichandran, G.; Rengan, A.K. Aptamer-Mediated Nanotheranostics for Cancer Treatment: A Review. ACS Appl. Nano Mater. 2020, 3, 9542–9559. [Google Scholar]
- Agnello, L.; Tortorella, S.; d’Argenio, A.; Carbone, C.; Camorani, S.; Locatelli, E.; Auletta, L.; Sorrentino, D.; Fedele, M.; Zannetti, A.; et al. Optimizing cisplatin delivery to triple-negative breast cancer through novel EGFR aptamer-conjugated polymeric nanovectors. J. Exp. Clin. Cancer Res. 2021, 40, 239. [Google Scholar]
- Camorani, S.; Granata, I.; Collina, F.; Leonetti, F.; Cantile, M.; Botti, G.; Fedele, M.; Guarracino, M.R.; Cerchia, L. Novel Aptamers Selected on Living Cells for Specific Recognition of Triple-Negative Breast Cancer. iScience 2020, 23, 100979. [Google Scholar] [PubMed]
- Gao, S.; Zheng, X.; Jiao, B.; Wang, L. Post-SELEX optimization of aptamers. Anal. Bioanal. Chem. 2016, 408, 4567–4573. [Google Scholar] [PubMed]
- Ito, Y.; Fukusaki, E. DNA as a ‘Nanomaterial’. J. Mol. Catal. B Enzym. 2004, 28, 155–166. [Google Scholar]
- Sanità, G.; Carrese, B.; Lamberti, A. Nanoparticle Surface Functionalization: How to Improve Biocompatibility and Cellular Internalization. Front. Mol. Biosci. 2020, 7, 587012. [Google Scholar]
- Friedman, A.D.; Claypool, S.E.; Liu, R. The smart targeting of nanoparticles. Curr. Pharm. Des. 2013, 19, 6315–6329. [Google Scholar]
- Rockey, W.M.; Hernandez, F.J.; Huang, S.Y.; Cao, S.; Howell, C.A.; Thomas, G.S.; Liu, X.Y.; Lapteva, N.; Spencer, D.M.; McNamara, J.O.; et al. Rational truncation of an RNA aptamer to prostate-specific membrane antigen using computational structural modeling. Nucleic Acid Ther. 2011, 21, 299–314. [Google Scholar]
- Shangguan, D.; Tang, Z.; Mallikaratchy, P.; Xiao, Z.; Tan, W. Optimization and modifications of aptamers selected from live cancer cell lines. Chembiochem 2007, 8, 603–606. [Google Scholar]
- Afanasyeva, A.; Nagao, C.; Mizuguchi, K. Prediction of the secondary structure of short DNA aptamers. Biophys. Physicobiol. 2019, 16, 287–294. [Google Scholar]
- Gardner, P.P.; Giegerich, R. A comprehensive comparison of comparative RNA structure prediction approaches. BMC Bioinform. 2004, 5, 140. [Google Scholar]
- Mathews, D.H. RNA secondary structure analysis using RNAstructure. Curr. Protoc. Bioinform. 2006, 13, 12–16. [Google Scholar]
- Holliday, D.L.; Speirs, V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011, 13, 215. [Google Scholar] [PubMed] [Green Version]
- Camorani, S.; Hill, B.S.; Collina, F.; Gargiulo, S.; Napolitano, M.; Cantile, M.; Di Bonito, M.; Botti, G.; Fedele, M.; Zannetti, A.; et al. Targeted imaging and inhibition of triple-negative breast cancer metastases by a PDGFRβ aptamer. Theranostics 2018, 8, 5178–5199. [Google Scholar] [PubMed]
- Camorani, S.; Crescenzi, E.; Gramanzini, M.; Fedele, M.; Zannetti, A.; Cerchia, L. Aptamer-mediated impairment of EGFR-integrin αvβ3 complex inhibits vasculogenic mimicry and growth of triple-negative breast cancers. Sci. Rep. 2017, 7, 46659. [Google Scholar] [PubMed] [Green Version]
- Yoon, S.; Rossi, J.J. Aptamers: Uptake mechanisms and intracellular applications. Adv. Drug Deliv. Rev. 2018, 134, 22–35. [Google Scholar] [PubMed]
- Friedman, A.D.; Kim, D.; Liu, R. Highly stable aptamers selected from a 2’-fully modified fGmH RNA library for targeting biomaterials. Biomaterials 2015, 36, 110–123. [Google Scholar]
- White, R.R.; Sullenger, B.A.; Rusconi, C.P. Developing aptamers into therapeutics. J. Clin. Investig. 2000, 106, 929–934. [Google Scholar]
- Ponti, D.; Costa, A.; Zaffaroni, N.; Pratesi, G.; Petrangolini, G.; Coradini, D.; Pilotti, S.; Pierotti, M.A.; Daidone, M.G. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005, 65, 5506–5511. [Google Scholar]
- Cowperthwaite, M.C.; Ellington, A.D. Bioinformatic analysis of the contribution of primer sequences to aptamer structures. J. Mol. Evol. 2008, 67, 95–102. [Google Scholar]
- Shui, B.; Ozer, A.; Zipfel, W.; Sahu, N.; Singh, A.; Lis, J.T.; Shi, H.; Kotlikoff, M.I. RNA aptamers that functionally interact with green fluorescent protein and its derivatives. Nucleic Acids Res. 2012, 40, e39. [Google Scholar]
- Le, T.T.; Chumphukam, O.; Cass, A.E.G.; Anthony, E.G. Determination of minimal sequence for binding of an aptamer. A comparison of truncation and hybridization inhibition methods. RSC Adv. 2014, 4, 47227. [Google Scholar]
- Earnest, K.G.; McConnell, E.M.; Hassan, E.M.; Wunderlich, M.; Hosseinpour, B.; Bono, B.S.; Chee, M.J.; Mulloy, J.C.; Willmore, W.G.; DeRosa, M.C.; et al. Development and characterization of a DNA aptamer for MLL-AF9 expressing acute myeloid leukemia cells using whole cell-SELEX. Sci. Rep. 2021, 11, 19174. [Google Scholar] [PubMed]
- Ibarra, L.E.; Camorani, S.; Agnello, L.; Pedone, E.; Pirone, L.; Chesta, C.A.; Palacios, R.E.; Fedele, M.; Cerchia, L. Selective photo-assisted eradication of Triple-Negative breast cancer cells through aptamer decoration of doped conjugated polymer nanoparticles. Pharmaceutics 2022, 14, 626. [Google Scholar]
- Passariello, M.; Camorani, S.; Vetrei, C.; Cerchia, L.; De Lorenzo, C. Novel Human Bispecific Aptamer-Antibody Conjugates for Efficient Cancer Cell Killing. Cancers 2019, 11, 1268. [Google Scholar]
- Passariello, M.; Camorani, S.; Vetrei, C.; Ricci, S.; Cerchia, L.; De Lorenzo, C. Ipilimumab and Its Derived EGFR Aptamer-Based Conjugate Induce Efficient NK Cell Activation against Cancer Cells. Cancers 2020, 12, 331. [Google Scholar]
- Camorani, S.; Passariello, M.; Agnello, L.; Esposito, S.; Collina, F.; Cantile, M.; Di Bonito, M.; Ulasov, I.V.; Fedele, M.; Zannetti, A.; et al. Aptamer targeted therapy potentiates immune checkpoint blockade in triple-negative breast cancer. J. Exp. Clin. Cancer Res. 2020, 39, 180. [Google Scholar] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Camorani, S.; d’Argenio, A.; Agnello, L.; Nilo, R.; Zannetti, A.; Ibarra, L.E.; Fedele, M.; Cerchia, L. Optimization of Short RNA Aptamers for TNBC Cell Targeting. Int. J. Mol. Sci. 2022, 23, 3511. https://doi.org/10.3390/ijms23073511
Camorani S, d’Argenio A, Agnello L, Nilo R, Zannetti A, Ibarra LE, Fedele M, Cerchia L. Optimization of Short RNA Aptamers for TNBC Cell Targeting. International Journal of Molecular Sciences. 2022; 23(7):3511. https://doi.org/10.3390/ijms23073511
Chicago/Turabian StyleCamorani, Simona, Annachiara d’Argenio, Lisa Agnello, Roberto Nilo, Antonella Zannetti, Luis Exequiel Ibarra, Monica Fedele, and Laura Cerchia. 2022. "Optimization of Short RNA Aptamers for TNBC Cell Targeting" International Journal of Molecular Sciences 23, no. 7: 3511. https://doi.org/10.3390/ijms23073511
APA StyleCamorani, S., d’Argenio, A., Agnello, L., Nilo, R., Zannetti, A., Ibarra, L. E., Fedele, M., & Cerchia, L. (2022). Optimization of Short RNA Aptamers for TNBC Cell Targeting. International Journal of Molecular Sciences, 23(7), 3511. https://doi.org/10.3390/ijms23073511









