Immunometabolism of Myeloid-Derived Suppressor Cells: Implications for Mycobacterium tuberculosis Infection and Insights from Tumor Biology
Abstract
1. Introduction
2. MDSC Classification
2.1. Origin and Identification
2.2. Recruitment and Expansion of MDSC
2.3. Immunosuppressive Mechanisms of MDSC
- (i)
- Sequestration of cardinal amino acids (AA) such as L-arginine, L-cysteine (Cys), and L-tryptophan by the activity of inducible enzymes such as arginase 1 (ARG1), inducible nitric oxide synthase (iNOS/NOS2) and indolamine dioxygenase (IDO) [27,39]. The reduction of these AA leads to the inhibition of T-cell activation and proliferation and reduced expression of T-cell receptor TCR-CD3 ζ chain, inducing the cell to undergo proliferative arrest [40,41,42];
- (ii)
- (iii)
- Indirect suppression of T and effector B cells through the induction of tolerogenic immune cells such as de novo generation of Fox-P3+ regulatory T cells (Tregs) [45], regulatory B cells, and tumor-associated macrophages (TAMs) [41,46]. Recently, a novel mechanism was elucidated through which MDSC-dependent metabolic and functional paralysis of CD8+ T cells occurs [47]. The mechanism involves methylglyoxal-derived glycation of L-arginine products such as argpyrimidine and hydroimidazolone, thereby depleting cytosolic amino acids such as L-arginine, resulting in T-cell paralysis [47,48]. The transfer of methylglyoxal from MDSC to T cells is dependent on cell–cell contact, resulting in more T-cell suppression at sites where MDSC may accumulate, such as tumor tissue [47];
- (iv)
- Secretion of suppressive cytokines such as TGF-β and IL-10 that exert direct suppressive effects on T-cell responses [45];
- (v)
- Induction of T-cell apoptosis through the induction of the B7 family of immune-regulatory ligands, a co-signaling network superfamily that plays an essential role in the modification of T-cell activation and tolerance [49], such as B7-H1 (programmed cell death ligand 1 (PD-L1)), B7-H3, and B7-H4 [50], and impairment of T-cell migration through the reduction of CD62L expression [29].
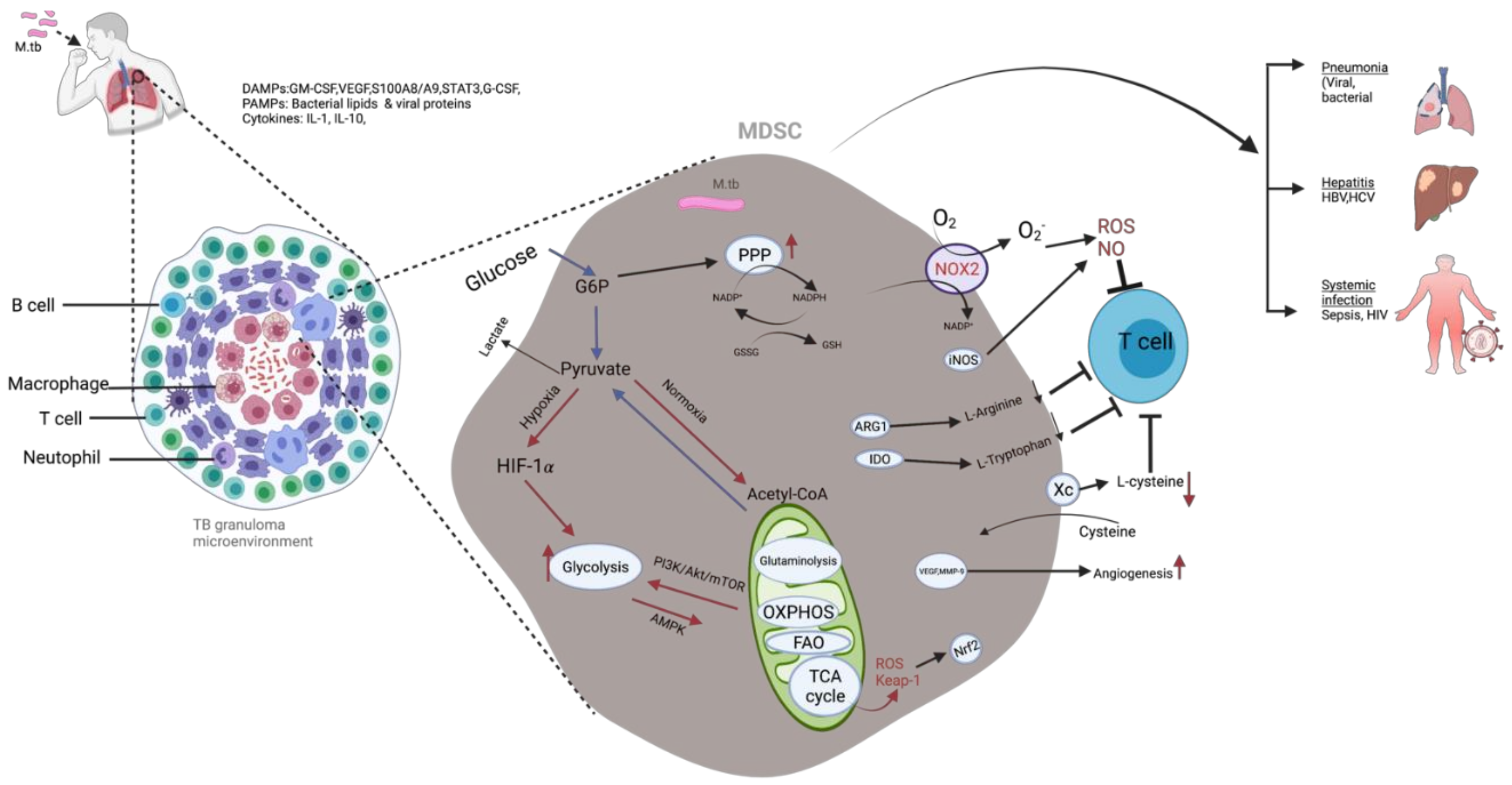
3. MDSC during M. tb Infection
M. tb Infection and Metabolic Reprogramming of Myeloid Cells
4. Immunometabolism of MDSC in Oncology
4.1. Metabolic Reprogramming of MDSC
4.2. Metabolism of Glucose, Lipids, and Amino Acids by Tumor-Derived MDSC
4.3. ROS regulates MDSC-Mediated Immune Suppression
5. An Intersection Point of MDSC in Tumor Biology and MDSC in Tuberculosis
5.1. Crosstalk between MDSC and B Cells
5.2. The Impact of MDSC in TB Infection and Prospects of Immune Metabolic Targeting Therapy of Drug-Resistant TB
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Emens, L.A.; Silverstein, S.C.; Khleif, S.; Marincola, F.M.; Galon, J. Toward integrative cancer immunotherapy: Targeting the tumor microenvironment. J. Transl. Med. 2012, 10, 70. [Google Scholar] [CrossRef] [PubMed]
- Donkor, M.K.; Lahue, E.; Hoke, T.A.; Shafer, L.R.; Coskun, U.; Solheim, J.C.; Gulen, D.; Bishay, J.; Talmadge, J.E. Mammary tumor heterogeneity in the expansion of myeloid-derived suppressor cells. Int. Immunopharmacol. 2009, 9, 937–948. [Google Scholar] [CrossRef]
- Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Bronte, V. Coordinated regulation of myeloid cells by Tumors. Nat. Rev. Immunol. 2012, 12, 253–268. [Google Scholar] [CrossRef] [PubMed]
- Umansky, V.; Blattner, C.; Gebhardt, C.; Utikal, J. The role of myeloid-derived suppressor cells (MDSC) in cancer progression. Vaccines 2016, 4, 36. [Google Scholar] [CrossRef] [PubMed]
- Hossain, F.; Al-Khami, A.A.; Wyczechowska, D.; Hernandez, C.; Zheng, L.; Reiss, K.; Del Valle, L.; Trillo-Tinoco, J.; Maj, T.; Zou, W.; et al. Inhibition of Fatty Acid Oxidation Modulates Immunosuppressive Functions of Myeloid-Derived Suppressor Cells and Enhances Cancer Therapies. Cancer Immunol. Res. 2015, 3, 1236–1247. [Google Scholar] [CrossRef] [PubMed]
- Park, M.-J.; Baek, J.-A.; Kim, S.-Y.; Park, S.-H.; Cho, M.-L.; Kwok, S.-K. 268 PD-L1 expressing myeloid-derived suppressor cells (MDSCs) have potent immuoregulatory activity and control lupus-like autoimmunity. Abstracts 2019, 6, A196. [Google Scholar] [CrossRef]
- Forghani, P.; Petersen, C.T.; Waller, E.K. Activation of VIP signaling enhances immunosuppressive effect of MDSC on CMV-induced adaptive immunity. Oncotarget 2017, 8, 81873. [Google Scholar] [CrossRef][Green Version]
- Youn, J.I.; Gabrilovich, D.I. The biology of myeloid-derived suppressor cells: The blessing and the curse of morphological and functional heterogeneity. Eur. J. Immunol. 2010, 40, 2969–2975. [Google Scholar] [CrossRef]
- Porembka, M.R.; Mitchem, J.B.; Belt, B.A.; Hsieh, C.-S.; Lee, H.-M.; Herndon, J.; Gillanders, W.E.; Linehan, D.C.; Goedegebuure, P. Pancreatic adenocarcinoma induces bone marrow mobilization of myeloid-derived suppressor cells which promote primary tumor growth. Cancer Immunol. Immunother. 2012, 61, 1373–1385. [Google Scholar] [CrossRef]
- Wang, L.; Chang, E.W.; Wong, S.C.; Ong, S.-M.; Chong, D.Q.; Ling, K.L. Increased myeloid-derived suppressor cells in gastric cancer correlate with cancer stage and plasma S100A8/A9 proinflammatory proteins. J. Immunol. 2013, 190, 794–804. [Google Scholar] [CrossRef]
- Diaz-Montero, C.M.; Salem, M.L.; Nishimura, M.I.; Garrett-Mayer, E.; Cole, D.J.; Montero, A.J. Increased circulating myeloid-derived suppressor cells correlate with clinical cancer stage, metastatic tumor burden, and doxorubicin–cyclophosphamide chemotherapy. Cancer Immunol. Immunother. 2009, 58, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.Y.; Spivia, W.R.; Veiras, L.C.; Khan, Z.; Peng, Z.; Jones, A.E.; Bernstein, E.A.; Saito, S.; Okwan-Duodu, D.; Parker, S.J.; et al. ACE overexpression in myeloid cells increases oxidative metabolism and cellular ATP. J. Biol. Chem. 2020, 295, 1369–1384. [Google Scholar] [CrossRef]
- Schulte-Schrepping, J.; Reusch, N.; Paclik, D.; Baßler, K.; Schlickeiser, S.; Zhang, B.; Krämer, B.; Krammer, T.; Brumhard, S.; Bonaguro, L.; et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell 2020, 182, 1419–1440. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.M.; Torrado, E. Protection versus pathology in tuberculosis: Recent insights. Curr. Opin. Immunol. 2012, 24, 431–437. [Google Scholar] [CrossRef]
- Qualls, J.E.; Murray, P.J. Immunometabolism within the tuberculosis granuloma: Amino acids, hypoxia, and cellular respiration. Semin. Immunopathol. 2016, 38, 139–152. [Google Scholar] [CrossRef]
- Ost, M.; Singh, A.; Peschel, A.; Mehling, R.; Rieber, N.; Hartl, D. Myeloid-derived suppressor cells in bacterial infections. Front. Cell. Infect. Microbiol. 2016, 6, 37. [Google Scholar] [CrossRef]
- Janols, H.; Bergenfelz, C.; Allaoui, R.; Larsson, A.M.; Rydén, L.; Björnsson, S.; Janciauskiene, S.; Wullt, M.; Bredberg, A.; Leandersson, K. A high frequency of MDSC in sepsis patients, with the granulocytic subtype dominating in gram-positive cases. J. Leukoc. Biol. 2014, 96, 685–693. [Google Scholar] [CrossRef]
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.-D.; Coopersmith, C.M. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef]
- Periasamy, S.; Avram, D.; McCabe, A.; MacNamara, K.C.; Sellati, T.J.; Harton, J.A. An Immature Myeloid/Myeloid-Suppressor Cell Response Associated with Necrotizing Inflammation Mediates Lethal Pulmonary Tularemia. PLoS Pathog. 2016, 12, e1005517. [Google Scholar] [CrossRef]
- Du Plessis, N.; Loebenberg, L.; Kriel, M.; von Groote-Bidlingmaier, F.; Ribechini, E.; Loxton, A.G.; van Helden, P.D.; Lutz, M.B.; Walzl, G. Increased Frequency of Myeloid-derived Suppressor Cells during Active Tuberculosis and after Recent Mycobacterium tuberculosis Infection Suppresses T-Cell Function. Am. J. Respir. Crit. Care Med. 2013, 188, 724–732. [Google Scholar] [CrossRef]
- Yan, D.; Adeshakin, A.O.; Xu, M.; Afolabi, L.O.; Zhang, G.; Chen, Y.H.; Wan, X. Lipid Metabolic Pathways Confer the Immunosuppressive Function of Myeloid-Derived Suppressor Cells in Tumor. Front. Immunol. 2019, 10, 1399. [Google Scholar] [CrossRef] [PubMed]
- Al-Khami, A.A.; Rodriguez, P.C.; Ochoa, A.C. Metabolic reprogramming of myeloid-derived suppressor cells (MDSC) in cancer. OncoImmunology 2016, 5, e1200771. [Google Scholar] [CrossRef]
- Norata, G.D.; Caligiuri, G.; Chavakis, T.; Matarese, G.; Netea, M.G.; Nicoletti, A.; O’Neill, L.A.; Marelli-Berg, F.M. The cellular and molecular basis of translational immunometabolism. Immunity 2015, 43, 421–434. [Google Scholar] [CrossRef] [PubMed]
- Pearce, E.L.; Pearce, E.J. Metabolic pathways in immune cell activation and quiescence. Immunity 2013, 38, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Filipazzi, P.; Huber, V.; Rivoltini, L. Phenotype, function and clinical implications of myeloid-derived suppressor cells in cancer patients. Cancer Immunol. Immunother. 2012, 61, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Condamine, T.; Ramachandran, I.; Youn, J.-I.; Gabrilovich, D.I. Regulation of tumor metastasis by myeloid-derived suppressor cells. Annu. Rev. Med. 2015, 66, 97–110. [Google Scholar] [CrossRef]
- Wang, W.; Xia, X.; Mao, L.; Wang, S. The CCAAT/enhancer-binding protein family: Its roles in MDSC expansion and function. Front. Immunol. 2019, 10, 1804. [Google Scholar] [CrossRef]
- Peranzoni, E.; Zilio, S.; Marigo, I.; Dolcetti, L.; Zanovello, P.; Mandruzzato, S.; Bronte, V. Myeloid-derived suppressor cell heterogeneity and subset definition. Curr. Opin. Immunol. 2010, 22, 238–244. [Google Scholar] [CrossRef]
- Dumitru, C.A.; Moses, K.; Trellakis, S.; Lang, S.; Brandau, S. Neutrophils and granulocytic myeloid-derived suppressor cells: Immunophenotyping, cell biology and clinical relevance in human oncology. Cancer Immunol. Immunother. 2012, 61, 1155–1167. [Google Scholar] [CrossRef]
- Solito, S.; Marigo, I.; Pinton, L.; Damuzzo, V.; Mandruzzato, S.; Bronte, V. Myeloid-derived suppressor cell heterogeneity in human cancers. Ann. N. Y. Acad. Sci. 2014, 1319, 47–65. [Google Scholar] [CrossRef]
- Mandruzzato, S.; Brandau, S.; Britten, C.M.; Bronte, V.; Damuzzo, V.; Gouttefangeas, C.; Maurer, D.; Ottensmeier, C.; van der Burg, S.H.; Welters, M.J.P.; et al. Toward harmonized phenotyping of human myeloid-derived suppressor cells by flow cytometry: Results from an interim study. Cancer Immunol. Immunother. 2016, 65, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Dorhoi, A.; Du Plessis, N. Monocytic myeloid-derived suppressor cells in chronic infections. Front. Immunol. 2017, 8, 1895. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Hoechst, B.; Duffy, A.; Gamrekelashvili, J.; Fioravanti, S.; Manns, M.P.; Greten, T.F.; Korangy, F. S100A9 a new marker for monocytic human myeloid-derived suppressor cells. Immunology 2012, 136, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Chai, E.; Zhang, L.; Li, C. LOX-1+ PMN-MDSC enhances immune suppression which promotes glioblastoma multiforme progression. Cancer Manag. Res. 2019, 11, 7307. [Google Scholar] [CrossRef] [PubMed]
- Veglia, F.; Tyurin, V.A.; Blasi, M.; De Leo, A.; Kossenkov, A.V.; Donthireddy, L.; To, T.K.J.; Schug, Z.; Basu, S.; Wang, F.; et al. Fatty acid transport protein 2 reprograms neutrophils in cancer. Nature 2019, 569, 73–78. [Google Scholar] [CrossRef]
- Bronte, V.; Brandau, S.; Chen, S.-H.; Colombo, M.P.; Frey, A.B.; Greten, T.F.; Mandruzzato, S.; Murray, P.J.; Ochoa, A.; Ostrand-Rosenberg, S. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun. 2016, 7, 12150. [Google Scholar] [CrossRef]
- Goldmann, O.; Beineke, A.; Medina, E. Identification of a Novel Subset of Myeloid-Derived Suppressor Cells During Chronic Staphylococcal Infection That Resembles Immature Eosinophils. J. Infect. Dis. 2017, 216, 1444–1451. [Google Scholar] [CrossRef]
- Gabrilovich, D.I.; Nagaraj, S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009, 9, 162–174. [Google Scholar] [CrossRef]
- Parker, K.H.; Sinha, P.; Horn, L.A.; Clements, V.K.; Yang, H.; Li, J.; Tracey, K.J.; Ostrand-Rosenberg, S. HMGB1 enhances immune suppression by facilitating the differentiation and suppressive activity of myeloid-derived suppressor cells. Cancer Res. 2014, 74, 5723–5733. [Google Scholar] [CrossRef]
- Chiba, Y.; Mizoguchi, I.; Hasegawa, H.; Ohashi, M.; Orii, N.; Nagai, T.; Sugahara, M.; Miyamoto, Y.; Xu, M.; Owaki, T. Regulation of myelopoiesis by proinflammatory cytokines in infectious diseases. Cell. Mol. Life Sci. 2018, 75, 1363–1376. [Google Scholar] [CrossRef]
- Rajabinejad, M.; Salari, F.; Gorgin Karaji, A.; Rezaiemanesh, A. The role of myeloid-derived suppressor cells in the pathogenesis of rheumatoid arthritis; anti-or pro-inflammatory cells? Artif. Cells Nanomed. Biotechnol. 2019, 47, 4149–4158. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Mao, C.; Wang, X.; Shi, Y.; Tao, Y. Epigenetic crosstalk between hypoxia and tumor driven by HIF regulation. J. Exp. Clin. Cancer Res. 2020, 39, 224. [Google Scholar] [CrossRef] [PubMed]
- Chiu, D.K.-C.; Tse, A.P.-W.; Xu, I.M.-J.; Di Cui, J.; Lai, R.K.-H.; Li, L.L.; Koh, H.-Y.; Tsang, F.H.-C.; Wei, L.L.; Wong, C.-M. Hypoxia inducible factor HIF-1 promotes myeloid-derived suppressor cells accumulation through ENTPD2/CD39L1 in hepatocellular carcinoma. Nat. Commun. 2017, 8, 517. [Google Scholar] [CrossRef] [PubMed]
- Alfaro, C.; Teijeira, A.; Oñate, C.; Pérez, G.; Sanmamed, M.F.; Andueza, M.P.; Alignani, D.; Labiano, S.; Azpilikueta, A.; Rodriguez-Paulete, A. Tumor-produced interleukin-8 attracts human myeloid-derived suppressor cells and elicits extrusion of neutrophil extracellular traps (NETs). Clin. Cancer Res. 2016, 22, 3924–3936. [Google Scholar] [CrossRef]
- Yao, L.; Abe, M.; Kawasaki, K.; Akbar, S.M.F.; Matsuura, B.; Onji, M.; Hiasa, Y. Characterization of liver monocytic myeloid-derived suppressor cells and their role in a murine model of non-alcoholic fatty liver disease. PLoS ONE 2016, 11, e0149948. [Google Scholar] [CrossRef][Green Version]
- Condamine, T.; Gabrilovich, D.I. Molecular mechanisms regulating myeloid-derived suppressor cell differentiation and function. Trends Immunol. 2011, 32, 19–25. [Google Scholar] [CrossRef]
- Fleming, V.; Hu, X.; Weber, R.; Nagibin, V.; Groth, C.; Altevogt, P.; Utikal, J.; Umansky, V. Targeting myeloid-derived suppressor cells to bypass tumor-induced immunosuppression. Front. Immunol. 2018, 9, 398. [Google Scholar] [CrossRef]
- Rodríguez, P.C.; Ochoa, A.C. Arginine regulation by myeloid derived suppressor cells and tolerance in cancer: Mechanisms and therapeutic perspectives. Immunol. Rev. 2008, 222, 180–191. [Google Scholar] [CrossRef]
- Hu, C.; Pang, B.; Lin, G.; Zhen, Y.; Yi, H. Energy metabolism manipulates the fate and function of Tumor myeloid-derived suppressor cells. Br. J. Cancer 2019, 122, 23–29. [Google Scholar] [CrossRef]
- Köstlin, N.; Vogelmann, M.; Spring, B.; Schwarz, J.; Feucht, J.; Härtel, C.; Orlikowsky, T.W.; Poets, C.F.; Gille, C. Granulocytic myeloid-derived suppressor cells from human cord blood modulate T-helper cell response towards an anti-inflammatory phenotype. Immunology 2017, 152, 89–101. [Google Scholar] [CrossRef]
- Derive, M.; Bouazza, Y.; Alauzet, C.; Gibot, S. Myeloid-derived suppressor cells control microbial sepsis. Intensive Care Med. 2012, 38, 1040–1049. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Pan, P.-Y.; Li, Q.; Sato, A.I.; Levy, D.E.; Bromberg, J.; Divino, C.M.; Chen, S.-H. Gr-1+ CD115+ immature myeloid suppressor cells mediate the development of tumor-induced T regulatory cells and T-cell anergy in tumor-bearing host. Cancer Res. 2006, 66, 1123–1131. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Schafer, C.C.; Hough, K.P.; Tousif, S.; Duncan, S.R.; Kearney, J.F.; Ponnazhagan, S.; Hsu, H.-C.; Deshane, J.S. Myeloid-derived suppressor cells impair B cell responses in lung cancer through IL-7 and STAT5. J. Immunol. 2018, 201, 278–295. [Google Scholar] [CrossRef] [PubMed]
- Baumann, T.; Dunkel, A.; Schmid, C.; Schmitt, S.; Hiltensperger, M.; Lohr, K.; Laketa, V.; Donakonda, S.; Ahting, U.; Lorenz-Depiereux, B. Regulatory myeloid cells paralyze T cells through cell–cell transfer of the metabolite methylglyoxal. Nat. Immunol. 2020, 21, 555–566. [Google Scholar] [CrossRef]
- Tsiganov, E.N.; Verbina, E.M.; Radaeva, T.V.; Sosunov, V.V.; Kosmiadi, G.A.; Nikitina, I.Y.; Lyadova, I.V. Gr-1dimCD11b+ Immature Myeloid-Derived Suppressor Cells but Not Neutrophils Are Markers of Lethal Tuberculosis Infection in Mice. J. Immunol. 2014, 192, 4718. [Google Scholar] [CrossRef]
- El Daker, S.; Sacchi, A.; Tempestilli, M.; Carducci, C.; Goletti, D.; Vanini, V.; Colizzi, V.; Lauria, F.N.; Martini, F.; Martino, A. Granulocytic myeloid derived suppressor cells expansion during active pulmonary tuberculosis is associated with high nitric oxide plasma level. PLoS ONE 2015, 10, e0123772. [Google Scholar] [CrossRef]
- Lenaerts, A.; Barry, C.E., III; Dartois, V. Heterogeneity in tuberculosis pathology, microenvironments and therapeutic responses. Immunol. Rev. 2015, 264, 288–307. [Google Scholar] [CrossRef]
- Dorhoi, A.; Kaufmann, S.H. Versatile myeloid cell subsets contribute to tuberculosis-associated inflammation. Eur. J. Immunol. 2015, 45, 2191–2202. [Google Scholar] [CrossRef]
- Rabbani, N.; Thornalley, P.J. Dicarbonyl stress in cell and tissue dysfunction contributing to ageing and disease. Biochem. Biophys. Res. Commun. 2015, 458, 221–226. [Google Scholar] [CrossRef]
- Capece, D.; Verzella, D.; Fischietti, M.; Zazzeroni, F.; Alesse, E. Targeting costimulatory molecules to improve antitumor immunity. BioMed Res. Int. 2012, 2012, 926321. [Google Scholar] [CrossRef]
- Fujimura, T.; Ring, S.; Umansky, V.; Mahnke, K.; Enk, A.H. Regulatory T cells stimulate B7-H1 expression in myeloid-derived suppressor cells in ret melanomas. J. Investig. Dermatol. 2012, 132, 1239–1246. [Google Scholar] [CrossRef] [PubMed]
- Knaul, J.K.; Jörg, S.; Oberbeck-Mueller, D.; Heinemann, E.; Scheuermann, L.; Brinkmann, V.; Mollenkopf, H.-J.; Yeremeev, V.; Kaufmann, S.H.; Dorhoi, A. Lung-residing myeloid-derived suppressors display dual functionality in murine pulmonary tuberculosis. Am. J. Respir. Crit. Care Med. 2014, 190, 1053–1066. [Google Scholar] [CrossRef] [PubMed]
- Obregón-Henao, A.; Henao-Tamayo, M.; Orme, I.M.; Ordway, D.J. Gr1intCD11b+ myeloid-derived suppressor cells in Mycobacterium tuberculosis infection. PLoS ONE 2013, 8, e80669. [Google Scholar] [CrossRef] [PubMed]
- Zahorchak, A.F.; Ezzelarab, M.B.; Lu, L.; Turnquist, H.R.; Thomson, A.W. In Vivo Mobilization and Functional Characterization of Nonhuman Primate Monocytic Myeloid-Derived Suppressor Cells. Am. J. Transpl. 2016, 16, 661–671. [Google Scholar] [CrossRef]
- Dorhoi, A.; Kotzé, L.A.; Berzofsky, J.A.; Sui, Y.; Gabrilovich, D.I.; Garg, A.; Hafner, R.; Khader, S.A.; Schaible, U.E.; Kaufmann, S.H.; et al. Therapies for tuberculosis and AIDS: Myeloid-derived suppressor cells in focus. J. Clin. Investig. 2020, 130, 2789–2799. [Google Scholar] [CrossRef]
- Eisenreich, W.; Rudel, T.; Heesemann, J.; Goebel, W. How viral and intracellular bacterial pathogens reprogram the metabolism of host cells to allow their intracellular replication. Front. Cell. Infect. Microbiol. 2019, 9, 42. [Google Scholar] [CrossRef]
- Ganeshan, K.; Chawla, A. Metabolic regulation of immune responses. Annu. Rev. Immunol. 2014, 32, 609–634. [Google Scholar] [CrossRef]
- Stine, Z.E.; Walton, Z.E.; Altman, B.J.; Hsieh, A.L.; Dang, C.V. MYC, metabolism, and cancer. Cancer Discov. 2015, 5, 1024–1039. [Google Scholar] [CrossRef]
- Cazzaniga, M.; Bonanni, B. Relationship between metabolic reprogramming and mitochondrial activity in cancer cells. Understanding the anticancer effect of metformin and its clinical implications. Anticancer Res. 2015, 35, 5789–5796. [Google Scholar]
- Jellusova, J.; Cato, M.H.; Apgar, J.R.; Ramezani-Rad, P.; Leung, C.R.; Chen, C.; Richardson, A.D.; Conner, E.M.; Benschop, R.J.; Woodgett, J.R. Gsk3 is a metabolic checkpoint regulator in B cells. Nat. Immunol. 2017, 18, 303. [Google Scholar] [CrossRef]
- Escoll, P.; Buchrieser, C. Metabolic reprogramming of host cells upon bacterial infection: Why shift to a Warburg-like metabolism? FEBS J. 2018, 285, 2146–2160. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, L.A.; Kishton, R.J.; Rathmell, J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 2016, 16, 553. [Google Scholar] [CrossRef] [PubMed]
- Domblides, C.; Lartigue, L.; Faustin, B. Metabolic stress in the immune function of T cells, macrophages and dendritic cells. Cells 2018, 7, 68. [Google Scholar] [CrossRef]
- Cohen, S.B.; Gern, B.H.; Delahaye, J.L.; Adams, K.N.; Plumlee, C.R.; Winkler, J.K.; Sherman, D.R.; Gerner, M.Y.; Urdahl, K.B. Alveolar macrophages provide an early Mycobacterium tuberculosis niche and initiate dissemination. Cell Host Microbe 2018, 24, 439–446.e4. [Google Scholar] [CrossRef] [PubMed]
- Kotzé, L.A.; Young, C.; Leukes, V.N.; John, V.; Fang, Z.; Walzl, G.; Lutz, M.B.; du Plessis, N. Mycobacterium tuberculosis and myeloid-derived suppressor cells: Insights into caveolin rich lipid rafts. EBioMedicine 2020, 53, 102670. [Google Scholar] [CrossRef] [PubMed]
- Law, K.; Weiden, M.; Harkin, T.; Tchou-Wong, K.; Chi, C.; Rom, W.N. Increased release of interleukin-1 beta, interleukin-6, and tumor necrosis factor-alpha by bronchoalveolar cells lavaged from involved sites in pulmonary tuberculosis. Am. J. Respir. Crit. Care Med. 1996, 153, 799–804. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Wainwright, H.C.; Locketz, M.; Bekker, L.G.; Walther, G.B.; Dittrich, C.; Visser, A.; Wang, W.; Hsu, F.F.; Wiehart, U. Caseation of human tuberculosis granulomas correlates with elevated host lipid metabolism. EMBO Mol. Med. 2010, 2, 258–274. [Google Scholar] [CrossRef]
- Russell, D.G.; VanderVen, B.C.; Lee, W.; Abramovitch, R.B.; Kim, M.-j.; Homolka, S.; Niemann, S.; Rohde, K.H. Mycobacterium tuberculosis wears what it eats. Cell Host Microbe 2010, 8, 68–76. [Google Scholar] [CrossRef]
- Mayito, J.; Andia, I.; Belay, M.; Jolliffe, D.A.; Kateete, D.P.; Reece, S.T.; Martineau, A.R. Anatomic and Cellular Niches for Mycobacterium tuberculosis in Latent Tuberculosis Infection. J. Infect. Dis. 2018, 219, 685–694. [Google Scholar] [CrossRef]
- Behr, M.A.; Waters, W.R. Is tuberculosis a lymphatic disease with a pulmonary portal? Lancet Infect. Dis. 2014, 14, 250–255. [Google Scholar] [CrossRef]
- Mariotti, S.; Sargentini, V.; Pardini, M.; Giannoni, F.; De Spirito, M.; Gagliardi, M.C.; Greco, E.; Teloni, R.; Fraziano, M.; Nisini, R. Mycobacterium tuberculosis may escape helper T cell recognition by infecting human fibroblasts. Hum. Immunol. 2013, 74, 722–729. [Google Scholar] [CrossRef] [PubMed]
- Neyrolles, O.; Hernández-Pando, R.; Pietri-Rouxel, F.; Fornès, P.; Tailleux, L.; Payán, J.A.B.; Pivert, E.; Bordat, Y.; Aguilar, D.; Prévost, M.-C. Is adipose tissue a place for Mycobacterium tuberculosis persistence? PLoS ONE 2006, 1, e43. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.M.; Ramakrishnan, L. The Role of the Granuloma in Expansion and Dissemination of Early Tuberculous Infection. Cell 2009, 136, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Bénard, A.; Sakwa, I.; Schierloh, P.; Colom, A.; Mercier, I.; Tailleux, L.; Jouneau, L.; Boudinot, P.; Al-Saati, T.; Lang, R. B cells producing type I IFN modulate macrophage polarization in tuberculosis. Am. J. Respir. Crit. Care Med. 2018, 197, 801–813. [Google Scholar] [CrossRef] [PubMed]
- Howard, N.C.; Marin, N.D.; Ahmed, M.; Rosa, B.A.; Martin, J.; Bambouskova, M.; Sergushichev, A.; Loginicheva, E.; Kurepina, N.; Rangel-Moreno, J. Mycobacterium tuberculosis carrying a rifampicin drug resistance mutation reprograms macrophage metabolism through cell wall lipid changes. Nat. Microbiol. 2018, 3, 1099–1108. [Google Scholar] [CrossRef]
- Kelly, B.; O’Neill, L.A.J. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 2015, 25, 771–784. [Google Scholar] [CrossRef]
- Mohamed, E.; Al-Khami, A.A.; Rodriguez, P.C. The cellular metabolic landscape in the tumor milieu regulates the activity of myeloid infiltrates. Cell. Mol. Immunol. 2018, 15, 421–427. [Google Scholar] [CrossRef]
- Moreira, J.D.V.; Hamraz, M.; Abolhassani, M.; Bigan, E.; Pérès, S.; Paulevé, L.; Nogueira, M.L.; Steyaert, J.-M.; Schwartz, L. The redox status of cancer cells supports mechanisms behind the Warburg effect. Metabolites 2016, 6, 33. [Google Scholar] [CrossRef]
- Keibler, M.A.; Wasylenko, T.M.; Kelleher, J.K.; Iliopoulos, O.; Vander Heiden, M.G.; Stephanopoulos, G. Metabolic requirements for cancer cell proliferation. Cancer Metab. 2016, 4, 16. [Google Scholar] [CrossRef]
- Shi, L.; Jiang, Q.; Bushkin, Y.; Subbian, S.; Tyagi, S. Biphasic dynamics of macrophage immunometabolism during Mycobacterium tuberculosis infection. MBio 2019, 10, e02550-18. [Google Scholar] [CrossRef]
- Galván-Peña, S.; O’Neill, L.A. Metabolic reprograming in macrophage polarization. Front. Immunol. 2014, 5, 420. [Google Scholar] [PubMed]
- Ward, P.S.; Thompson, C.B. Metabolic reprogramming: A cancer hallmark even warburg did not anticipate. Cancer Cell 2012, 21, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Salamon, H.; Eugenin, E.A.; Pine, R.; Cooper, A.; Gennaro, M.L. Infection with Mycobacterium tuberculosis induces the Warburg effect in mouse lungs. Sci. Rep. 2015, 5, 18176. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Prados, J.-C.; Través, P.G.; Cuenca, J.; Rico, D.; Aragonés, J.; Martín-Sanz, P.; Cascante, M.; Boscá, L. Substrate fate in activated macrophages: A comparison between innate, classic, and alternative activation. J. Immunol. 2010, 185, 605–614. [Google Scholar] [CrossRef]
- Davies, L.C.; Rice, C.M.; McVicar, D.W.; Weiss, J.M. Diversity and environmental adaptation of phagocytic cell metabolism. J. Leukoc. Biol. 2019, 105, 37–48. [Google Scholar] [CrossRef]
- Everts, B.; Amiel, E.; van der Windt, G.J.; Freitas, T.C.; Chott, R.; Yarasheski, K.E.; Pearce, E.L.; Pearce, E.J. Commitment to glycolysis sustains survival of NO-producing inflammatory dendritic cells. Blood 2012, 120, 1422–1431. [Google Scholar] [CrossRef]
- Lerner, T.R.; Borel, S.; Greenwood, D.J.; Repnik, U.; Russell, M.R.; Herbst, S.; Jones, M.L.; Collinson, L.M.; Griffiths, G.; Gutierrez, M.G. Mycobacterium tuberculosis replicates within necrotic human macrophages. J. Cell Biol. 2017, 216, 583–594. [Google Scholar] [CrossRef]
- Gleeson, L.E.; Sheedy, F.J.; Palsson-McDermott, E.M.; Triglia, D.; O’Leary, S.M.; O’Sullivan, M.P.; O’Neill, L.A.; Keane, J. Cutting edge: Mycobacterium tuberculosis induces aerobic glycolysis in human alveolar macrophages that is required for control of intracellular bacillary replication. J. Immunol. 2016, 196, 2444–2449. [Google Scholar] [CrossRef]
- Cumming, B.M.; Addicott, K.W.; Adamson, J.H.; Steyn, A.J. Mycobacterium tuberculosis induces decelerated bioenergetic metabolism in human macrophages. Elife 2018, 7, e39169. [Google Scholar] [CrossRef]
- Wang, R. Two’s company, three’sa crowd: Can H2S be the third endogenous gaseous transmitter? FASEB J. 2002, 16, 1792–1798. [Google Scholar] [CrossRef]
- Chinta, K.C.; Saini, V.; Glasgow, J.N.; Mazorodze, J.H.; Rahman, M.A.; Reddy, D.; Lancaster, J.R., Jr.; Steyn, A.J. The emerging role of gasotransmitters in the pathogenesis of tuberculosis. Nitric Oxide 2016, 59, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.A.; Cumming, B.M.; Addicott, K.W.; Pacl, H.T.; Russell, S.L.; Nargan, K.; Naidoo, T.; Ramdial, P.K.; Adamson, J.H.; Wang, R. Hydrogen sulfide dysregulates the immune response by suppressing central carbon metabolism to promote tuberculosis. Proc. Natl. Acad. Sci. USA 2020, 117, 6663–6674. [Google Scholar] [CrossRef] [PubMed]
- Schlesinger, L. Entry of Mycobacterium tuberculosis into mononuclear phagocytes. In Tuberculosis; Springer: Berlin/Heidelberg, Germany, 1996; pp. 71–96. [Google Scholar]
- Eum, S.-Y.; Kong, J.-H.; Hong, M.-S.; Lee, Y.-J.; Kim, J.-H.; Hwang, S.-H.; Cho, S.-N.; Via, L.E.; Barry, C.E., III. Neutrophils are the predominant infected phagocytic cells in the airways of patients with active pulmonary TB. Chest 2010, 137, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Haschek, W.M.; Rousseaux, C.; Wallig, M. Chapter 6—Respiratory System. In Fundamentals of Toxicologic Pathology; Elsevier: Amsterdam, The Netherlands, 2010; pp. 93–133. [Google Scholar]
- Lee, W.L.; Downey, G.P. Neutrophil activation and acute lung injury. Curr. Opin. Crit. Care 2001, 7, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Rizo, V.; Martínez-Guzmán, M.A.; Iñiguez-Gutierrez, L.; García-Orozco, A.; Alvarado-Navarro, A.; Fafutis-Morris, M. Neutrophil extracellular traps and its implications in inflammation: An overview. Front. Immunol. 2017, 8, 81. [Google Scholar] [CrossRef]
- Levine, A.P.; Segal, A.W. The NADPH oxidase and microbial killing by neutrophils, with a particular emphasis on the proposed antimicrobial role of myeloperoxidase within the phagocytic vacuole. Microbiol. Spectr. 2017, 4, 599–613. [Google Scholar]
- Fossati, G.; Moulding, D.A.; Spiller, D.G.; Moots, R.J.; White, M.R.; Edwards, S.W. The mitochondrial network of human neutrophils: Role in chemotaxis, phagocytosis, respiratory burst activation, and commitment to apoptosis. J. Immunol. 2003, 170, 1964–1972. [Google Scholar] [CrossRef]
- Van Raam, B.J.; Verhoeven, A.; Kuijpers, T. Mitochondria in neutrophil apoptosis. Int. J. Hematol. 2006, 84, 199–204. [Google Scholar] [CrossRef]
- Furukawa, S.; Saito, H.; Inoue, T.; Matsuda, T.; Fukatsu, K.; Han, I.; Ikeda, S.; Hidemura, A. Supplemental glutamine augments phagocytosis and reactive oxygen intermediate production by neutrophils and monocytes from postoperative patients in vitro. Nutrition 2000, 16, 323–329. [Google Scholar] [CrossRef]
- Boxer, L.A.; Baehner, R.L.; Davis, J. The effect of 2-deoxyglucose on guinea pig polymorphonuclear leukocyte phagocytosis. J. Cell. Physiol. 1977, 91, 89–102. [Google Scholar] [CrossRef]
- Sica, A.; Strauss, L. Energy metabolism drives myeloid-derived suppressor cell differentiation and functions in pathology. J. Leukoc. Biol. 2017, 102, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Groth, C.; Hu, X.; Weber, R.; Fleming, V.; Altevogt, P.; Utikal, J.; Umansky, V. Immunosuppression mediated by myeloid-derived suppressor cells (MDSC) during Tumor progression. Br. J. Cancer 2019, 120, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Hammami, I.; Chen, J.; Murschel, F.; Bronte, V.; De Crescenzo, G.; Jolicoeur, M. Immunosuppressive activity enhances central carbon metabolism and bioenergetics in myeloid-derived suppressor cells in vitro models. BMC Cell Biol. 2012, 13, 18. [Google Scholar] [CrossRef]
- Dibble, C.C.; Manning, B.D. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat. Cell Biol. 2013, 15, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.K. Metabolic reprogramming of immune cells in cancer progression. Immunity 2015, 43, 435–449. [Google Scholar] [CrossRef] [PubMed]
- Al-Khami, A.A.; Zheng, L.; Del Valle, L.; Hossain, F.; Wyczechowska, D.; Zabaleta, J.; Sanchez, M.D.; Dean, M.J.; Rodriguez, P.C.; Ochoa, A.C. Exogenous lipid uptake induces metabolic and functional reprogramming of tumor-associated myeloid-derived suppressor cells. OncoImmunology 2017, 6, e1344804. [Google Scholar] [CrossRef] [PubMed]
- McCaffrey, E.F.; Donato, M.; Keren, L.; Chen, Z.; Fitzpatrick, M.; Jojic, V.; Delmastro, A.; Greenwald, N.F.; Baranski, A.; Graf, W. Multiplexed imaging of human tuberculosis granulomas uncovers immunoregulatory features conserved across tissue and blood. BioRxiv 2020. [Google Scholar] [CrossRef]
- Raychaudhuri, B.; Rayman, P.; Huang, P.; Grabowski, M.; Hambardzumyan, D.; Finke, J.H.; Vogelbaum, M.A. Myeloid derived suppressor cell infiltration of murine and human gliomas is associated with reduction of tumor infiltrating lymphocytes. J. Neuro-Oncol. 2015, 122, 293–301. [Google Scholar] [CrossRef]
- Yu, J.; Du, W.; Yan, F.; Wang, Y.; Li, H.; Cao, S.; Yu, W.; Shen, C.; Liu, J.; Ren, X. Myeloid-derived suppressor cells suppress antitumor immune responses through IDO expression and correlate with lymph node metastasis in patients with breast cancer. J. Immunol. 2013, 190, 3783–3797. [Google Scholar] [CrossRef]
- Korb, V.C.; Chuturgoon, A.A.; Moodley, D. Mycobacterium tuberculosis: Manipulator of protective immunity. Int. J. Mol. Sci. 2016, 17, 131. [Google Scholar] [CrossRef]
- Russell, D.G.; Cardona, P.-J.; Kim, M.-J.; Allain, S.; Altare, F. Foamy macrophages and the progression of the human tuberculosis granuloma. Nat. Immunol. 2009, 10, 943–948. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef] [PubMed]
- Beury, D.W.; Parker, K.H.; Nyandjo, M.; Sinha, P.; Carter, K.A.; Ostrand-Rosenberg, S. Cross-talk among myeloid-derived suppressor cells, macrophages, and tumor cells impacts the inflammatory milieu of solid tumors. J. Leukoc. Biol. 2014, 96, 1109–1118. [Google Scholar] [CrossRef] [PubMed]
- Damaghi, M.; Tafreshi, N.K.; Lloyd, M.C.; Sprung, R.; Estrella, V.; Wojtkowiak, J.W.; Morse, D.L.; Koomen, J.M.; Bui, M.M.; Gatenby, R.A. Chronic acidosis in the Tumor microenvironment selects for overexpression of LAMP2 in the plasma membrane. Nat. Commun. 2015, 6, 8752. [Google Scholar] [CrossRef]
- Renner, K.; Singer, K.; Koehl, G.; Geissler, E.; Peter, K.; Siska, P.; Kreutz, M. Metabolic hallmarks of tumor and immune cells in the tumor microenvironment. Front. Immunol. 2017, 8, 248. [Google Scholar] [CrossRef]
- Yun, J.; Rago, C.; Cheong, I.; Pagliarini, R.; Angenendt, P.; Rajagopalan, H.; Schmidt, K.; Willson, J.K.; Markowitz, S.; Zhou, S. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science 2009, 325, 1555–1559. [Google Scholar] [CrossRef]
- Cai, T.-T.; Ye, S.-B.; Liu, Y.-N.; He, J.; Chen, Q.-Y.; Mai, H.-Q.; Zhang, C.-X.; Cui, J.; Zhang, X.-S.; Busson, P. LMP1-mediated glycolysis induces myeloid-derived suppressor cell expansion in nasopharyngeal carcinoma. PLoS Pathog. 2017, 13, e1006503. [Google Scholar] [CrossRef]
- Liu, G.; Bi, Y.; Shen, B.; Yang, H.; Zhang, Y.; Wang, X.; Liu, H.; Lu, Y.; Liao, J.; Chen, X. SIRT1 limits the function and fate of myeloid-derived suppressor cells in tumors by orchestrating HIF-1α–dependent glycolysis. Cancer Res. 2014, 74, 727–737. [Google Scholar] [CrossRef]
- Ghosh, H.S.; McBurney, M.; Robbins, P.D. SIRT1 negatively regulates the mammalian target of rapamycin. PLoS ONE 2010, 5, e9199. [Google Scholar] [CrossRef]
- Belton, M.; Brilha, S.; Manavaki, R.; Mauri, F.; Nijran, K.; Hong, Y.T.; Patel, N.H.; Dembek, M.; Tezera, L.; Green, J. Hypoxia and tissue destruction in pulmonary TB. Thorax 2016, 71, 1145–1153. [Google Scholar] [CrossRef]
- Tannahill, G.; Curtis, A.; Adamik, J.; Palsson-McDermott, E.; McGettrick, A.; Goel, G.; Frezza, C.; Bernard, N.; Kelly, B.; Foley, N. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 2013, 496, 238. [Google Scholar] [CrossRef] [PubMed]
- LaGory, E.L.; Giaccia, A.J. The ever-expanding role of HIF in Tumor and stromal biology. Nat. Cell Biol. 2016, 18, 356–365. [Google Scholar] [CrossRef] [PubMed]
- Husain, Z.; Huang, Y.; Seth, P.; Sukhatme, V.P. Tumor-derived lactate modifies antitumor immune response: Effect on myeloid-derived suppressor cells and NK cells. J. Immunol. 2013, 191, 1486–1495. [Google Scholar] [CrossRef] [PubMed]
- Hackett, E.E.; Charles-Messance, H.; O’Leary, S.M.; Gleeson, L.E.; Muñoz-Wolf, N.; Case, S.; Wedderburn, A.; Johnston, D.G.W.; Williams, M.A.; Smyth, A.; et al. Mycobacterium tuberculosis Limits Host Glycolysis and IL-1β by Restriction of PFK-M via MicroRNA-21. Cell Rep. 2020, 30, 124–136.e4. [Google Scholar] [CrossRef]
- Veglia, F.; Tyurin, V.; Kagan, V.; Gabrilovich, D. Lipids and Suppressive Functions of MDSC in Cancer; AACR: Philadelphia, PA, USA, 2018. [Google Scholar]
- Peyron, P.; Vaubourgeix, J.; Poquet, Y.; Levillain, F.; Botanch, C.; Bardou, F.; Daffé, M.; Emile, J.-F.; Marchou, B.; Cardona, P.-J. Foamy macrophages from tuberculous patients’ granulomas constitute a nutrient-rich reservoir for M. tuberculosis persistence. PLoS Pathog. 2008, 4, e1000204. [Google Scholar] [CrossRef]
- Veglia, F.; Tyurin, V.; Kagan, V.; Gabrilovich, D. Oxidized Lipids Contribute to the Suppression Function of Myeloid Derived Suppressor Cells in Cancer; AACR: Philadelphia, PA, USA, 2015. [Google Scholar]
- Bailly, C. Regulation of PD-L1 expression on cancer cells with ROS-modulating drugs. Life Sci. 2020, 246, 117403. [Google Scholar] [CrossRef]
- Corzo, C.A.; Cotter, M.J.; Cheng, P.; Cheng, F.; Kusmartsev, S.; Sotomayor, E.; Padhya, T.; McCaffrey, T.V.; McCaffrey, J.C.; Gabrilovich, D.I. Mechanism regulating reactive oxygen species in tumor-induced myeloid-derived suppressor cells. J. Immunol. 2009, 182, 5693–5701. [Google Scholar] [CrossRef]
- Dietlin, T.A.; Hofman, F.M.; Lund, B.T.; Gilmore, W.; Stohlman, S.A.; Van der Veen, R.C. Mycobacteria-induced Gr-1+ subsets from distinct myeloid lineages have opposite effects on T cell expansion. J. Leukoc. Biol. 2007, 81, 1205–1212. [Google Scholar] [CrossRef]
- Nagaraj, S.; Gupta, K.; Pisarev, V.; Kinarsky, L.; Sherman, S.; Kang, L.; Herber, D.L.; Schneck, J.; Gabrilovich, D.I. Altered recognition of antigen is a mechanism of CD8+ T cell tolerance in cancer. Nat. Med. 2007, 13, 828–835. [Google Scholar] [CrossRef]
- Schmielau, J.; Finn, O.J. Activated granulocytes and granulocyte-derived hydrogen peroxide are the underlying mechanism of suppression of t-cell function in advanced cancer patients. Cancer Res. 2001, 61, 4756–4760. [Google Scholar]
- Choi, H.-S.; Rai, P.R.; Chu, H.W.; Cool, C.; Chan, E.D. Analysis of nitric oxide synthase and nitrotyrosine expression in human pulmonary tuberculosis. Am. J. Respir. Crit. Care Med. 2002, 166, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Panday, A.; Sahoo, M.K.; Osorio, D.; Batra, S. NADPH oxidases: An overview from structure to innate immunity-associated pathologies. Cell. Mol. Immunol. 2015, 12, 5–23. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.M.; Segal, B.H.; Frank, A.A.; Holland, S.M.; Orme, I.M. Transient loss of resistance to pulmonary tuberculosis in p47 phox−/− mice. Infect. Immun. 2000, 68, 1231–1234. [Google Scholar] [CrossRef]
- Jian, S.-L.; Chen, W.-W.; Su, Y.-C.; Su, Y.-W.; Chuang, T.-H.; Hsu, S.-C.; Huang, L.-R. Glycolysis regulates the expansion of myeloid-derived suppressor cells in tumor-bearing hosts through prevention of ROS-mediated apoptosis. Cell Death Dis. 2017, 8, e2779. [Google Scholar] [CrossRef] [PubMed]
- Satoh, H.; Moriguchi, T.; Taguchi, K.; Takai, J.; Maher, J.M.; Suzuki, T.; Winnard, P.T., Jr.; Raman, V.; Ebina, M.; Nukiwa, T. Nrf2-deficiency creates a responsive microenvironment for metastasis to the lung. Carcinogenesis 2010, 31, 1833–1843. [Google Scholar] [CrossRef]
- Ohl, K.; Fragoulis, A.; Klemm, P.; Baumeister, J.; Klock, W.; Verjans, E.; Böll, S.; Möllmann, J.; Lehrke, M.; Costa, I. Nrf2 is a central regulator of metabolic reprogramming of myeloid-derived suppressor cells in steady state and sepsis. Front. Immunol. 2018, 9, 1552. [Google Scholar] [CrossRef]
- Ohl, K.; Tenbrock, K. Reactive oxygen species as regulators of MDSC-mediated immune suppression. Front. Immunol. 2018, 9, 2499. [Google Scholar] [CrossRef]
- Corzo, C.A.; Condamine, T.; Lu, L.; Cotter, M.J.; Youn, J.-I.; Cheng, P.; Cho, H.-I.; Celis, E.; Quiceno, D.G.; Padhya, T. HIF-1α regulates function and differentiation of myeloid-derived suppressor cells in the tumor microenvironment. J. Exp. Med. 2010, 207, 2439–2453. [Google Scholar] [CrossRef]
- Gabrilovich, D.I.; Velders, M.P.; Sotomayor, E.M.; Kast, W.M. Mechanism of immune dysfunction in cancer mediated by immature Gr-1+ myeloid cells. J. Immunol. 2001, 166, 5398–5406. [Google Scholar] [CrossRef]
- Drabczyk-Pluta, M.; Werner, T.; Hoffmann, D.; Leng, Q.; Chen, L.; Dittmer, U.; Zelinskyy, G. Granulocytic myeloid-derived suppressor cells suppress virus-specific CD8(+) T cell responses during acute Friend retrovirus infection. Retrovirology 2017, 14, 42. [Google Scholar] [CrossRef]
- Lei, G.-S.; Zhang, C.; Lee, C.-H. Myeloid-derived suppressor cells impair alveolar macrophages through PD-1 receptor ligation during Pneumocystis pneumonia. Infect. Immun. 2015, 83, 572–582. [Google Scholar] [CrossRef] [PubMed]
- Poe, S.L.; Arora, M.; Oriss, T.B.; Yarlagadda, M.; Isse, K.; Khare, A.; Levy, D.E.; Lee, J.S.; Mallampalli, R.K.; Chan, Y.R.; et al. STAT1-regulated lung MDSC-like cells produce IL-10 and efferocytose apoptotic neutrophils with relevance in resolution of bacterial pneumonia. Mucosal Immunol. 2013, 6, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Sammicheli, S.; Kuka, M.; Di Lucia, P.; de Oya, N.J.; De Giovanni, M.; Fioravanti, J.; Cristofani, C.; Maganuco, C.G.; Fallet, B.; Ganzer, L.; et al. Inflammatory monocytes hinder antiviral B cell responses. Sci. Immunol. 2016, 1, eaah6789. [Google Scholar] [CrossRef] [PubMed]
- Fooksman, D.R.; Nussenzweig, M.C.; Dustin, M.L. Myeloid cells limit production of antibody-secreting cells after immunization in the lymph node. J. Immunol. 2014, 192, 1004–1012. [Google Scholar] [CrossRef]
- Lelis, F.J.; Jaufmann, J.; Singh, A.; Fromm, K.; Teschner, A.C.; Pöschel, S.; Schäfer, I.; Beer-Hammer, S.; Rieber, N.; Hartl, D. Myeloid-derived suppressor cells modulate B-cell responses. Immunol. Lett. 2017, 188, 108–115. [Google Scholar] [CrossRef]
- Rastad, J.L.; Green, W.R. Myeloid-derived suppressor cells in murine AIDS inhibit B-cell responses in part via soluble mediators including reactive oxygen and nitrogen species, and TGF-β. Virology 2016, 499, 9–22. [Google Scholar] [CrossRef]
- Park, M.J.; Lee, S.H.; Kim, E.K.; Lee, E.J.; Park, S.H.; Kwok, S.K.; Cho, M.L. Myeloid-derived suppressor cells induce the expansion of regulatory B cells and ameliorate autoimmunity in the sanroque mouse model of systemic lupus erythematosus. Arthritis Rheumatol. 2016, 68, 2717–2727. [Google Scholar] [CrossRef]
- Crook, K.R.; Jin, M.; Weeks, M.F.; Rampersad, R.R.; Baldi, R.M.; Glekas, A.S.; Shen, Y.; Esserman, D.A.; Little, P.; Schwartz, T.A.; et al. Myeloid-derived suppressor cells regulate T cell and B cell responses during autoimmune disease. J. Leukoc. Biol. 2015, 97, 573–582. [Google Scholar] [CrossRef]
- Pålsson-McDermott, E.M.; O’Neill, L.A. Targeting immunometabolism as an anti-inflammatory strategy. Cell Res. 2020, 30, 300–314. [Google Scholar] [CrossRef]
- Du Plessis, N.; Kotze, L.A.; Leukes, V.; Walzl, G. Translational Potential of Therapeutics Targeting Regulatory Myeloid Cells in Tuberculosis. Front. Cell. Infect. Microbiol. 2018, 8, 332. [Google Scholar] [CrossRef]
- Li, L.; Wang, L.; Li, J.; Fan, Z.; Yang, L.; Zhang, Z.; Zhang, C.; Yue, D.; Qin, G.; Zhang, T. Metformin-induced reduction of CD39 and CD73 blocks myeloid-derived suppressor cell activity in patients with ovarian cancer. Cancer Res. 2018, 78, 1779–1791. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Lian, J.; Huang, L.; Zhao, Q.; Liu, S.; Zhang, Z.; Chen, X.; Yue, D.; Li, L.; Li, F. Metformin blocks myeloid-derived suppressor cell accumulation through AMPK-DACH1-CXCL1 axis. Oncoimmunology 2018, 7, e1442167. [Google Scholar] [CrossRef] [PubMed]
- Singhal, A.; Jie, L.; Kumar, P.; Hong, G.S.; Leow, M.K.-S.; Paleja, B.; Tsenova, L.; Kurepina, N.; Chen, J.; Zolezzi, F. Metformin as adjunct antituberculosis therapy. Sci. Transl. Med. 2014, 6, 263ra159. [Google Scholar] [CrossRef] [PubMed]
- Condamine, T.; Dominguez, G.A.; Youn, J.-I.; Kossenkov, A.V.; Mony, S.; Alicea-Torres, K.; Tcyganov, E.; Hashimoto, A.; Nefedova, Y.; Lin, C. Lectin-type oxidized LDL receptor-1 distinguishes population of human polymorphonuclear myeloid-derived suppressor cells in cancer patients. Sci. Immunol. 2016, 1, aaf8943. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Munansangu, B.S.M.; Kenyon, C.; Walzl, G.; Loxton, A.G.; Kotze, L.A.; du Plessis, N. Immunometabolism of Myeloid-Derived Suppressor Cells: Implications for Mycobacterium tuberculosis Infection and Insights from Tumor Biology. Int. J. Mol. Sci. 2022, 23, 3512. https://doi.org/10.3390/ijms23073512
Munansangu BSM, Kenyon C, Walzl G, Loxton AG, Kotze LA, du Plessis N. Immunometabolism of Myeloid-Derived Suppressor Cells: Implications for Mycobacterium tuberculosis Infection and Insights from Tumor Biology. International Journal of Molecular Sciences. 2022; 23(7):3512. https://doi.org/10.3390/ijms23073512
Chicago/Turabian StyleMunansangu, Brian S. M., Colin Kenyon, Gerhard Walzl, André G. Loxton, Leigh A. Kotze, and Nelita du Plessis. 2022. "Immunometabolism of Myeloid-Derived Suppressor Cells: Implications for Mycobacterium tuberculosis Infection and Insights from Tumor Biology" International Journal of Molecular Sciences 23, no. 7: 3512. https://doi.org/10.3390/ijms23073512
APA StyleMunansangu, B. S. M., Kenyon, C., Walzl, G., Loxton, A. G., Kotze, L. A., & du Plessis, N. (2022). Immunometabolism of Myeloid-Derived Suppressor Cells: Implications for Mycobacterium tuberculosis Infection and Insights from Tumor Biology. International Journal of Molecular Sciences, 23(7), 3512. https://doi.org/10.3390/ijms23073512





