Recent Progress on the Salt Tolerance Mechanisms and Application of Tamarisk
Abstract
:1. Introduction
2. The Salt Tolerance Mechanisms of Tamarisk
2.1. Salt Ion Secretion
2.2. Na+ Homeostasis and Osmotic Adjustment
2.3. Efficient ROS Scavenging Mechanism
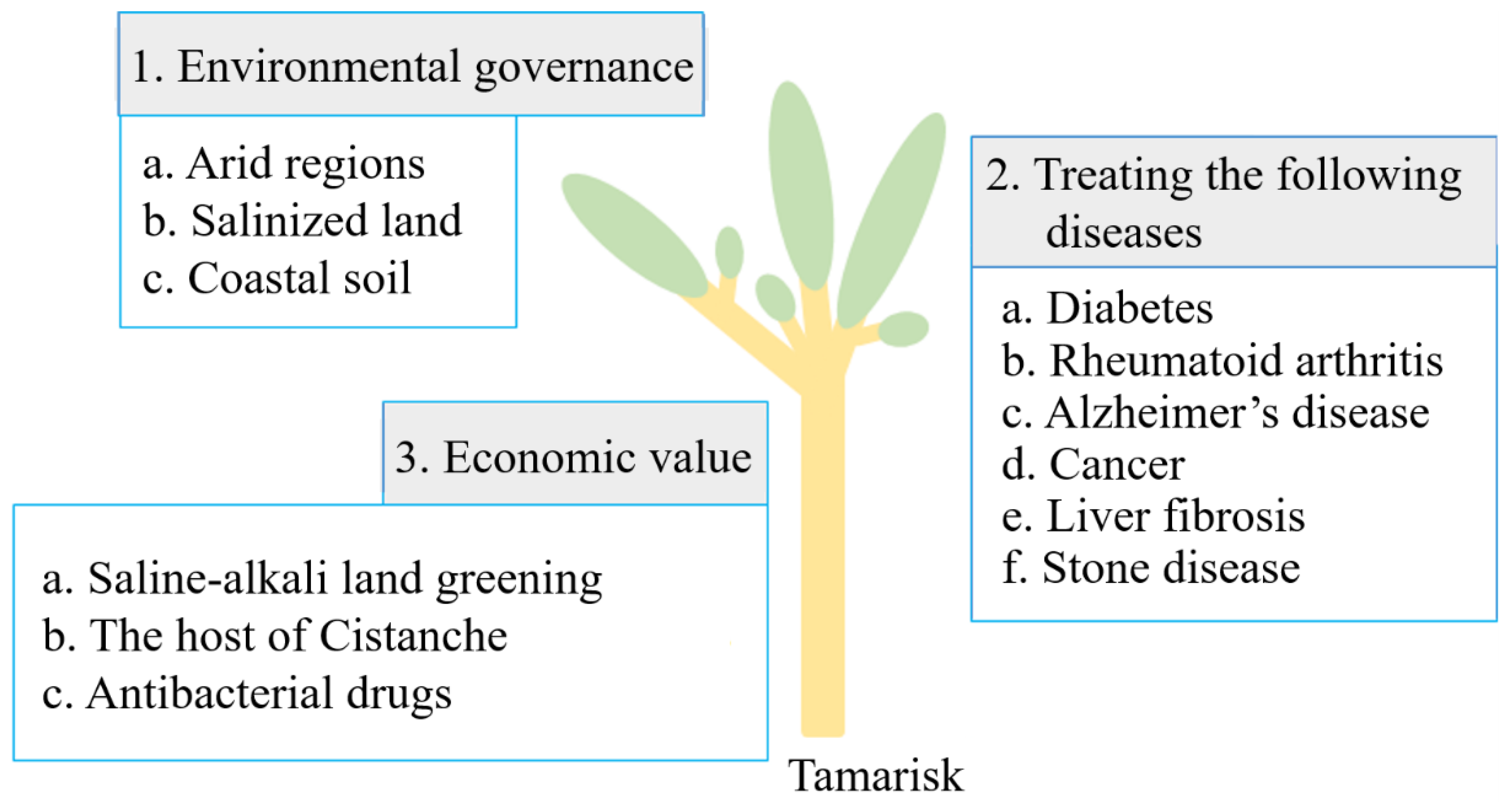
3. Application of Tamarisk
3.1. Significance of Tamarisk to the Environment
3.2. Medicinal Value of Tamarisk
3.2.1. Treatment of Diabetes
3.2.2. Treatment of Rheumatoid Arthritis
3.2.3. Treatment of Alzheimer’s Disease
3.2.4. Treatment of Cancer
3.2.5. Liver Protection
3.2.6. Prevention of Lithiasis
3.3. Economic Value of Tamarisk
4. Outlook and Prospects for Development
Author Contributions
Funding
Conflicts of Interest
References
- Rozema, J.; Flowers, T. Crops for a Salinized World. Science 2008, 322, 1478–1480. [Google Scholar] [CrossRef]
- Morton, M.J.L.; Awlia, M.; Al Tamimi, N.; Saade, S.; Pailles, Y.; Negrão, S.; Tester, M. Salt stress under the scalpel–dissecting the genetics of salt tolerance. Plant J. 2019, 97, 148–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banin, A.; Fish, A. Secondary desertification due to salinization of intensively irrigated lands: The Israeli experience. Environ. Monit. Assess. 1995, 37, 17–37. [Google Scholar] [CrossRef]
- Daliakopoulos, I.; Tsanis, I.; Koutroulis, A.; Kourgialas, N.; Varouchakis, A.; Karatzas, G.P.; Ritsema, C. The threat of soil salinity: A European scale review. Sci. Total Environ. 2016, 573, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Z.; Wang, P.; Jiang, B.; Lei, X.; Wu, J.; Dong, W.; Gao, C. A 2-Cys peroxiredoxin gene from Tamarix hispida improved salt stress tolerance in plants. BMC Plant Biol. 2020, 20, 360. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Liu, J.J.; Banerjee, S.; Zhou, N.; Zhao, Z.Y.; Zhang, K.; Tian, C.Y. Soil pH is equally important as salinity in shaping bacterial communities in saline soils under halophytic vegetation. Sci. Rep. 2018, 8, 4550. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, L.-J.; Huang, R.-D. Cytoskeleton and plant salt stress tolerance. Plant Signal. Behav. 2011, 6, 29–31. [Google Scholar] [CrossRef] [Green Version]
- Pimentel, D. Ethical Issues of Global Corporatization: Agriculture and Beyond. Poult. Sci. 2004, 83, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Ruan, C.J.; da Silva, J.A.T. Metabolomics: Creating new potentials for unraveling the mechanisms in response to salt and drought stress and for the biotechnological improvement of xero-halophytes. Crit. Rev. Biotechnol. 2011, 31, 153–169. [Google Scholar] [CrossRef]
- Bartels, D.; Sunkar, R. Drought and Salt Tolerance in Plants. Crit. Rev. Plant Sci. 2005, 24, 23–58. [Google Scholar] [CrossRef]
- Orlovsky, N.; Japakova, U.; Zhang, H.; Volis, S. Effect of salinity on seed germination, growth and ion content in dimorphic seeds of Salicornia europaea L. (Chenopodiaceae). Plant Divers. 2016, 38, 183–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flowers, T.J.; Colmer, T.D. Salinity tolerance in halophytes. New Phytol. 2008, 179, 945–963. [Google Scholar] [CrossRef] [PubMed]
- Flowers, T.J.; Flowers, S.A. Why does salinity pose such a difficult problem for plant breeders? Agric. Water Manag. 2005, 78, 15–24. [Google Scholar] [CrossRef]
- Yuan, F.; Guo, J.; Shabala, S.; Wang, B. Reproductive Physiology of Halophytes: Current Standing. Front. Plant Sci. 2019, 9, 1954. [Google Scholar] [CrossRef]
- Abd El-Salam, M.M.; Elhakem, A.H. Desertification and its effect on the erosion of vegetation in the south-western region of Saudi Arabia. Environ. Monit. Assess. 2016, 188, 164. [Google Scholar] [CrossRef]
- Campos, J.A.; Aires, U.R.; Da Silva, D.D.; Calijuri, M.L. Environmental fragility and vegetation cover dynamics in the Lapa Grande State Park, MG, Brazil. An. Da Acad. Bras. Ciências 2019, 91, e20170940. [Google Scholar] [CrossRef]
- Feng, Q.; Ma, H.; Jiang, X.; Wang, X.; Cao, S. What Has Caused Desertification in China? Sci. Rep. 2015, 5, 15998. [Google Scholar] [CrossRef] [Green Version]
- Kalir, A.; Poljakoff-Mayber, A. Effect of Salinity on Respiratory Pathways in Root Tips of Tamarix tetragyna. Plant Physiol. 1976, 57, 167–170. [Google Scholar] [CrossRef] [Green Version]
- Han, Z.; Yin, W.; Zhang, J.; Niu, S.; Ren, L. Active Anti-erosion Protection Strategy in Tamarisk (Tamarix aphylla). Sci. Rep. 2013, 3, 3429. [Google Scholar] [CrossRef]
- Mamat, Z.; Halik, U.; Aishan, T.; Aini, A. Ecological effect of the riparian ecosystem in the lower reaches of the Tarim River in northwest China. PLoS ONE 2019, 14, e0208462. [Google Scholar] [CrossRef]
- Zeng, Y.; Zhao, C.; Kundzewicz, Z.; Lv, G. Distribution pattern of Tugai forests species diversity and their relationship to environmental factors in an arid area of China. PLoS ONE 2020, 15, e0232907. [Google Scholar]
- Thomson, W.W.; Liu, L.L. Ultrastructural features of the salt gland of Tamarix aphylla L. Planta 1967, 73, 201–220. [Google Scholar] [CrossRef]
- Thomson, W.W.; Berry, W.L.; Liu, L.L. Localization and secretion of salt by the salt glands of Tamarix aphylla. Proc. Natl. Acad. Sci. USA 1969, 63, 310–317. [Google Scholar] [CrossRef] [Green Version]
- Gao, C.; Wang, Y.; Jiang, B.; Liu, G.; Yu, L.; Wei, Z.; Yang, C. A novel vacuolar membrane H+-ATPase c subunit gene (ThVHAc1) from Tamarix hispida confers tolerance to several abiotic stresses in Saccharomyces cerevisiae. Mol. Biol. Rep. 2010, 38, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Gupta, B.; Huang, B. Mechanism of Salinity Tolerance in Plants: Physiological, Biochemical, and Molecular Characterization. Int. J. Genom. 2014, 2014, 701596. [Google Scholar] [CrossRef]
- Che, B.; Cheng, C.; Fang, J.; Liu, Y.; Jiang, L.; Yu, B. The Recretohalophyte Tamarix TrSOS1 Gene Confers Enhanced Salt Tolerance to Transgenic Hairy Root Composite Cotton Seedlings Exhibiting Virus-Induced Gene Silencing of GhSOS1. Int. J. Mol. Sci. 2019, 20, 2930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, X.; Yan, X.; Yang, Z.; Han, G.; Wang, L.; Yuan, F.; Wang, B. Salt glands of recretohalophyte Tamarix under salinity: Their evolution and adaptation. Ecol. Evol. 2020, 10, 9384–9395. [Google Scholar] [CrossRef]
- Breckle, S.W. Salinity Tolerance of Different Halophyte Types; Springer: Dordrecht, The Netherlands, 1990; pp. 167–175. [Google Scholar]
- Deng, Y.; Feng, Z.; Yuan, F.; Guo, J.; Suo, S.; Wang, B. Identification and Functional Analysis of the Autofluorescent Substance in Limonium bicolor Salt Glands. Plant Physiol. Biochem. 2015, 97, 20–27. [Google Scholar] [CrossRef]
- Ding, F.; Chen, M.; Sui, N.; Wang, B.-S. Ca2+ significantly enhanced development and salt-secretion rate of salt glands of Limonium bicolor under NaCl treatment. S. Afr. J. Bot. 2009, 76, 95–101. [Google Scholar] [CrossRef] [Green Version]
- Flowers, T.J.; Galal, H.K.; Bromham, L. Evolution of halophytes: Multiple origins of salt tolerance in land plants. Funct. Plant Biol. 2010, 37, 604–612. [Google Scholar] [CrossRef]
- Dassanayake, M.; Larkin, J.C. Making Plants Break a Sweat: The Structure, Function, and Evolution of Plant Salt Glands. Front. Plant Sci. 2017, 8, 406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, F.; Leng, B.; Wang, B. Progress in Studying Salt Secretion from the Salt Glands in Recretohalophytes: How Do Plants Secrete Salt? Front. Plant Sci. 2016, 7, 977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Yuan, F.; Liu, Y.; Zhang, M.; Liu, Y.; Zhao, Y.; Wang, B.; Chen, M. Exogenous melatonin enhances salt secretion from salt glands by upregulating the expression of ion transporter and vesicle transport genes in Limonium bicolor. BMC Plant Biol. 2020, 20, 493. [Google Scholar] [CrossRef]
- Lu, C.; Feng, Z.; Yuan, F.; Han, G.; Wang, B. The SNARE protein LbSYP61 participates in salt secretion in Limonium bicolor. Environ. Exp. Bot. 2020, 176, 104076. [Google Scholar] [CrossRef]
- Zhang, M.; Chen, Z.; Yuan, F.; Wang, B.; Chen, M. Integrative transcriptome and proteome analyses provide deep insights into the molecular mechanism of salt tolerance in Limonium bicolor. Plant Mol. Biol. 2022, 108, 127–143. [Google Scholar] [CrossRef]
- Ma, H.; Tian, C.; Feng, G.; Yuan, J. Ability of multicellular salt glands in Tamarix species to secrete Na+ and K+ selectively. Sci. China Life Sci. 2011, 54, 282–289. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Liu, Y.; Zhang, M.; Xu, H.; Ning, K.; Wang, B.; Chen, M. Melatonin increases growth and salt tolerance of Limonium bicolor by improving photosynthetic and antioxidant capacity. BMC Plant Biol. 2022, 22, 16. [Google Scholar] [CrossRef]
- Wu, S.-W.; Kumar, R.; Iswanto, A.B.B.; Kim, J.-Y. Callose balancing at plasmodesmata. J. Exp. Bot. 2018, 69, 5325–5339. [Google Scholar] [CrossRef]
- Lucas, W.J.; Bouché-Pillon, S.; Jackson, D.P.; Nguyen, L.; Baker, L.; Ding, B.; Hake, S. Selective Trafficking of KNOTTED1 Homeodomain Protein and Its mRNA through Plasmodesmata. Science 1995, 270, 1980–1983. [Google Scholar] [CrossRef]
- Miyashima, S.; Roszak, P.; Sevilem, I.; Toyokura, K.; Blob, B.; Heo, J.O.; Mellor, N.; Help-Rinta-Rahko, H.; Otero, S.; Smet, W.; et al. Mobile PEAR transcription factors integrate positional cues to prime cambial growth. Nature 2019, 565, 490–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, F.; Peterson, C.A. Frequencies of plasmodesmata in Allium cepa L. roots: Implications for solute transport pathways. J. Exp. Bot. 2001, 52, 1051–1061. [Google Scholar] [CrossRef] [Green Version]
- Wilson, H.; Mycock, D.; Weiersbye, I.M. The salt glands of Tamarix usneoides E. Mey. ex Bunge (South African Salt Cedar). Int. J. Phytoremediat. 2017, 19, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Ye, J.; Yang, Y.; Lin, H.; Yue, L.; Luo, J.; Long, Y.; Fu, H.; Liu, X.; Zhang, Y.; et al. The SOS2-SCaBP8 Complex Generates and Fine-Tunes an AtANN4-Dependent Calcium Signature under Salt Stress. Dev. Cell 2019, 48, 697–709. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Li, Z.; Lu, M.; Wang, Y. ThNAC13, a NAC Transcription Factor from Tamarix hispida, Confers Salt and Osmotic Stress Tolerance to Transgenic Tamarix and Arabidopsis. Front. Plant Sci. 2017, 8, 635. [Google Scholar] [CrossRef] [Green Version]
- He, Z.; Li, Z.; Lu, H.; Huo, L.; Wang, Z.; Wang, Y.; Ji, X. The NAC Protein from Tamarix hispida, ThNAC7, Confers Salt and Osmotic Stress Tolerance by Increasing Reactive Oxygen Species Scavenging Capability. Plants 2019, 8, 221. [Google Scholar] [CrossRef] [Green Version]
- Zang, D.; Wang, C.; Ji, X.; Wang, Y. Tamarix hispida zinc finger protein ThZFP1 participates in salt and osmotic stress tolerance by increasing proline content and SOD and POD activities. Plant Sci. 2015, 235, 111–121. [Google Scholar] [CrossRef]
- Zang, D.; Wang, L.; Zhang, Y.; Zhao, H.; Wang, Y. ThDof1.4 and ThZFP1 constitute a transcriptional regulatory cascade involved in salt or osmotic stress in Tamarix hispida. Plant Mol. Biol. 2017, 94, 495–507. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.Y.; Wang, Y.C.; Xia, D.A.; Gao, C.Q.; Wang, C.; Yang, C.P. Overexpression of a GST gene (ThGSTZ1) from Tamarix hispida, improves drought and salinity tolerance by enhancing the ability to scavenge reactive oxygen species. Plant Cell Tissue Organ Cult. 2014, 117, 99–112. [Google Scholar] [CrossRef]
- Liu, Z.; Xie, Q.; Tang, F.; Wu, J.; Dong, W.; Wang, C.; Gao, C. The ThSOS3 Gene Improves the Salt Tolerance of Transgenic Tamarix hispida and Arabidopsis thaliana. Front. Plant Sci. 2021, 11, 597480. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, Y.; Wang, C.; Wang, Y.C.; Wang, L.Q. ThNAC12 from Tamarix hispida directly regulates ThPIP2;5 to enhance salt tolerance by modulating reactive oxygen species. Plant Physiol. Biochem. 2021, 163, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Liu, G.; Meng, X.; Liu, Y.; Ji, X.; Li, Y.; Nie, X.; Wang, Y. A WRKY gene from Tamarix hispida, ThWRKY4, mediates abiotic stress responses by modulating reactive oxygen species and expression of stress-responsive genes. Plant Mol. Biol. 2013, 82, 303–320. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.-T.; Wang, C.; Liu, R.; Zhang, Y.; Wang, Y.-C.; Wang, L.-Q. ThHSFA1 Confers Salt Stress Tolerance through Modulation of Reactive Oxygen Species Scavenging by Directly Regulating ThWRKY4. Int. J. Mol. Sci. 2021, 22, 5048. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xia, J.; Fang, Y.; Li, T.; Liu, J. Effects of Salt-Drought Stress on Growth and Physiobiochemical Characteristics of Tamarix chinensis Seedlings. Sci. World J. 2014, 2014, 765840. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Shen, W.; Chan, Z.; Wu, Y. Endogenous Cytokinin Overproduction Modulates ROS Homeostasis and Decreases Salt Stress Resistance in Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 1004. [Google Scholar] [CrossRef] [Green Version]
- Mittler, R. ROS Are Good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef] [Green Version]
- Baral, A. Strawberries under salt stress: ALA and ROS to the rescue. Physiol. Plant. 2019, 167, 2–4. [Google Scholar] [CrossRef] [Green Version]
- Ahanger, M.A.; Singh Tomar, N.; Tittal, M.; Argal, S.; Agarwal, R.M. Plant growth under water/salt stress: ROS production; antioxidants and significance of added potassium under such conditions. Physiol. Mol. Biol. Plants 2017, 23, 731–744. [Google Scholar] [CrossRef]
- Poór, P.; Kovács, J.; Borbély, P.; Takács, Z.; Szepesi, Á.; Tari, I. Salt stress-induced production of reactive oxygen- and nitrogen species and cell death in the ethylene receptor mutant Never ripe and wild type tomato roots. Plant Physiol. Biochem. 2015, 97, 313–322. [Google Scholar] [CrossRef] [Green Version]
- Luan, J.; Dong, J.; Song, X.; Jiang, J.; Li, H. Overexpression of Tamarix hispida ThTrx5 Confers Salt Tolerance to Arabidopsis by Activating Stress Response Signals. Int. J. Mol. Sci. 2020, 21, 1165. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Lei, X.; Wang, P.; Wang, Y.; Lv, J.; Li, X.; Gao, C. Overexpression of ThSAP30BP from Tamarix hispida improves salt tolerance. Plant Physiol. Biochem. 2019, 146, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Torshizi, M.R.; Miri, A.; Shahriari, A.; Dong, Z.; Davidson-Arnott, R. The effectiveness of a multi-row Tamarix windbreak in reducing aeolian erosion and sediment flux, Niatak area, Iran. J. Environ. Manag. 2020, 265, 110486. [Google Scholar] [CrossRef]
- Yang, H.; Xia, J.; Cui, Q.; Liu, J.; Wei, S.; Feng, L.; Dong, K. Effects of different Tamarix chinensis-grass patterns on the soil quality of coastal saline soil in the Yellow River Delta, China. Sci. Total Environ. 2021, 772, 145501. [Google Scholar] [CrossRef] [PubMed]
- Fountoulakis, M.S.; Daskalakis, G.; Papadaki, A.; Kalogerakis, N.; Manios, T. Use of halophytes in pilot-scale horizontal flow constructed wetland treating domestic wastewater. Environ. Sci. Pollut. Res. 2017, 24, 16682–16689. [Google Scholar] [CrossRef] [PubMed]
- Alnuqaydan, A.M.; Rah, B.; Rahb, B. Tamarix articulata (T. articulata)—An Important Halophytic Medicinal Plant with Potential Pharmacological Properties. Curr. Pharm. Biotechnol. 2019, 20, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Bahramsoltani, R.; Farzaei, M.H.; Iranpanah, A.; Hajimahmoudi, M.; Pourjabar, Z.; Daglia, M.; Santarcangelo, C.; Rashidi, K.; Nabavi, S.M.; Rahimi, R. Phytochemical and toxicological evaluation of Tamarix stricta Boiss. Drug Chem. Toxicol. 2019, 45, 223–230. [Google Scholar] [CrossRef]
- Tundis, R.; Loizzo, M.R.; Menichini, F. Natural Products as α-Amylase and α-Glucosidase Inhibitors and their Hypoglycaemic Potential in the Treatment of Diabetes: An Update. Mini-Rev. Med. Chem. 2010, 10, 315–331. [Google Scholar] [CrossRef]
- Cardullo, N.; Muccilli, V.; Pulvirenti, L.; Cornu, A.; Pouységu, L.; Deffieux, D.; Quideau, S.; Tringali, C. C-glucosidic ellagitannins and galloylated glucoses as potential functional food ingredients with anti-diabetic properties: A study of α-glucosidase and α-amylase inhibition. Food Chem. 2019, 313, 126099. [Google Scholar] [CrossRef]
- Malik, M.; Sharif, A.; Hassan, S.U.; Muhammad, F.; Khan, H.M.; Akhtar, B.; Saeed, M. Amelioration of hyperglycaemia and modulation of pro-inflammatory cytokines by Tamarix gallica fractions in alloxan induced diabetic rats. Arch. Physiol. Biochem. 2020. Online ahead of print. [Google Scholar] [CrossRef]
- Hebi, M.; Eddouks, M. Hypolipidemic activity of Tamarix articulata Vahl. in diabetic rats. J. Integr. Med. 2017, 15, 476–482. [Google Scholar] [CrossRef]
- Van Heemst, J.; Jansen, D.T.; Polydorides, S.; Moustakas, A.K.; Bax, M.; Feitsma, A.L.; Bontrop-Elferink, D.G.; Baarse, M.; van der Woude, D.; Wolbink, G.J.; et al. Crossreactivity to vinculin and microbes provides a molecular basis for HLA-based protection against rheumatoid arthritis. Nat. Commun. 2015, 6, 6681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, Y.; Jiang, C.-S.; Sun, N.; Li, W.-Q.; Niu, Y.; Han, H.-Q.; Miao, Z.-H.; Zhao, X.-X.; Zhao, J.; Li, J. Tamaractam, a New Bioactive Lactam from Tamarix ramosissima, Induces Apoptosis in Rheumatoid Arthritis Fibroblast-Like Synoviocytes. Molecules 2017, 22, 96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, Z.; Li, J.; Biswas, S.; Jiang, C.S.; Huang, Y.; Sun, T.; Yao, Y. Ramosissimin, a new flavonol from Tararix ramosissima, induces apoptosis in rheumatoid arthritis fibroblast-like synoviocytes. Pharmazie 2018, 73, 169–173. [Google Scholar] [PubMed]
- Scott, D.L.; Wolfe, F.; Huizinga, T.W. Rheumatoid arthritis. Lancet 2010, 376, 1094–1108. [Google Scholar] [CrossRef]
- Bottini, N.; Firestein, G.S. Duality of fibroblast-like synoviocytes in RA: Passive responders and imprinted aggressors. Nat. Rev. Rheumatol. 2012, 9, 24–33. [Google Scholar] [CrossRef] [Green Version]
- Hodson, R. Alzheimer’s disease. Nature 2018, 559, S1. [Google Scholar] [CrossRef] [Green Version]
- Engelhart, M.J.; Geerlings, M.I.; Ruitenberg, A.; van Swieten, J.C.; Hofman, A.; Witteman, J.C.; Breteler, M.M. Dietary intake of antioxidants and risk of Alzheimer disease. JAMA 2002, 287, 3223–3229. [Google Scholar] [CrossRef] [Green Version]
- Morris, M.C.; Evans, D.A.; Bienias, J.L.; Tangney, C.C.; Bennett, D.A.; Aggarwal, N.; Wilson, R.S.; Scherr, P.A. Dietary intake of antioxidant nutrients and the risk of incident Alzheimer disease in a biracial community study. JAMA 2002, 287, 3230–3237. [Google Scholar] [CrossRef]
- Head, E. Oxidative damage and cognitive dysfunction: Antioxidant treatments to promote healthy brain aging. Neurochem. Res. 2009, 34, 670–678. [Google Scholar] [CrossRef] [Green Version]
- Salissou, M.T.M.; Mahaman, Y.A.R.; Zhu, F.; Huang, F.; Wang, Y.; Xu, Z.; Ke, D.; Wang, Q.; Liu, R.; Wang, J.-Z.; et al. Methanolic extract of Tamarix Gallica attenuates hyperhomocysteinemia induced AD-like pathology and cognitive impairments in rats. Aging 2018, 10, 3229–3248. [Google Scholar] [CrossRef]
- Boulaaba, M.; Tsolmon, S.; Ksouri, R.; Han, J.; Kawada, K.; Smaoui, A.; Isoda, H. Anticancer effect of Tamarix gallica extracts on human colon cancer cells involves Erk1/2 and p38 action on G2/M cell cycle arrest. Cytotechnology 2013, 65, 927–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abaza, M.S.; Al-Attiyah, R.; Bhardwaj, R.; Abbadi, G.; Koyippally, M.; Afzal, M. Syringic acid from Tamarix aucheriana possesses antimitogenic and chemo-sensitizing activities in human colorectal cancer cells. Pharm. Biol. 2013, 51, 1110–1124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abaza, M.S.I.; Afzal, M.; Al-Attiyah, R.J.; Guleri, R. Methylferulate from Tamarix aucheriana inhibits growth and enhances chemosensitivity of human colorectal cancer cells: Possible mechanism of action. BMC Complement. Altern. Med. 2016, 16, 384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altamirano-Barrera, A.; Barranco-Fragoso, B.; Méndez-Sánchez, N. Management Strategies for Liver Fibrosis. Ann. Hepatol. 2017, 16, 48–56. [Google Scholar] [CrossRef]
- Lai, M.; Afdhal, N.H. Liver Fibrosis Determination. Gastroenterol. Clin. N. Am. 2019, 48, 281–289. [Google Scholar] [CrossRef]
- Sun, M.; Kisseleva, T. Reversibility of liver fibrosis. Clin. Res. Hepatol. Gastroenterol. 2015, 39 (Suppl. S1), S60–S63. [Google Scholar] [CrossRef] [Green Version]
- Sekkien, A.; Swilam, N.; Ebada, S.S.; Esmat, A.; El-Khatib, A.H.; Linscheid, M.W.; Singab, A.N. Polyphenols from Tamarix nilotica: LC(-)ESI-MS(n) Profiling and In Vivo Antifibrotic Activity. Molecules 2018, 23, 1411. [Google Scholar] [CrossRef] [Green Version]
- Sehrawat, A.; Sultana, S. Evaluation of possible mechanisms of protective role of Tamarix gallica against DEN initiated and 2-AAF promoted hepatocarcinogenesis in male Wistar rats. Life Sci. 2006, 79, 1456–1465. [Google Scholar] [CrossRef]
- Abouzid, S.; Sleem, A. Hepatoprotective and antioxidant activities of Tamarix nilotica flowers. Pharm. Biol. 2011, 49, 392–395. [Google Scholar] [CrossRef]
- O’Kell, A.L.; Grant, D.C.; Khan, S.R. Pathogenesis of calcium oxalate urinary stone disease: Species comparison of humans, dogs, and cats. Urolithiasis 2017, 45, 329–336. [Google Scholar] [CrossRef]
- Bensatal, A.; Ouahrani, M.R. Inhibition of crystallization of calcium oxalate by the extraction of Tamarix gallica L. Urol. Res. 2008, 36, 283–287. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Lin, H.; Gu, L.; Gao, J.; Tzeng, C.-M. Herba Cistanche (Rou Cong-Rong): One of the Best Pharmaceutical Gifts of Traditional Chinese Medicine. Front. Pharmacol. 2016, 7, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, R.; Yang, Y.; Xu, J.; Pan, Y.; Zhang, W.; Xing, Y.; Ni, H.; Sun, Y.; Hou, Y.; Li, N. Tamarix hohenackeri Bunge exerts anti-inflammatory effects on lipopolysaccharide-activated microglia in vitro. Phytomedicine 2018, 40, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Bao, Y.; Zhu, Y.; Liu, S.; Peng, Z.; Zhang, Y.; Zhou, G. Isorhamnetin, Hispidulin, and Cirsimaritin Identified in Tamarix ramosissima Barks from Southern Xinjiang and Their Antioxidant and Antimicrobial Activities. Molecules 2019, 24, 390. [Google Scholar] [CrossRef] [Green Version]
- Ksouri, R.; Falleh, H.; Megdiche, W.; Trabelsi, N.; Mhamdi, B.; Chaieb, K.; Bakrouf, A.; Magné, C.; Abdelly, C. Antioxidant and antimicrobial activities of the edible medicinal halophyte Tamarix gallica L. and related polyphenolic constituents. Food Chem. Toxicol. 2009, 47, 2083–2091. [Google Scholar] [CrossRef]
- Benmerache, A.; Benteldjoune, M.; Magid, A.A.; Abedini, A.; Berrehal, D.; Kabouche, A.; Gangloff, S.C.; Voutquenne-Nazabadioko, L.; Kabouche, Z. Chemical composition, antioxidant and antibacterial activities of Tamarix balansae J. Gay aerial parts. Nat. Prod. Res. 2017, 31, 2828–2835. [Google Scholar] [CrossRef]
- Khalid, M.; Hassani, D.; Bilal, M.; Butt, Z.A.; Hamayun, M.; Ahmad, A.; Huang, D.; Hussain, A. Identification of oral cavity biofilm forming bacteria and determination of their growth inhibition by Acacia arabica, Tamarix aphylla L. and Melia azedarach L. medicinal plants. Arch. Oral Biol. 2017, 81, 175–185. [Google Scholar] [CrossRef]
- Luziatelli, F.; Crognale, S.; D’Annibale, A.; Moresi, M.; Petruccioli, M.; Ruzzi, M. Screening, isolation, and characterization of glycosyl-hydrolase-producing fungi from desert halophyte plants. Int. Microbiol. 2014, 17, 41–48. [Google Scholar]
- Zhang, P.; Yuan, G.; Zhuang, W.; Xue, S. Ecophysiological responses and adaptation of Tamarix ramosissima to changes in groundwater depth in the Heihe river basin. Acta Ecol. Sin. 2011, 31, 36–46. [Google Scholar]
- Flowers, T.J. Improving crop salt tolerance. J. Exp. Bot. 2004, 55, 307–319. [Google Scholar] [CrossRef]
- Newete, S.W.; Allem, S.M.; Venter, N.; Byrne, M.J. Tamarix efficiency in salt excretion and physiological tolerance to salt-induced stress in South Africa. Int. J. Phytoremediat. 2019, 22, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Chew, M.K. The Monstering of Tamarisk: How Scientists made a Plant into a Problem. J. Hist. Biol. 2009, 42, 231–266. [Google Scholar] [CrossRef] [PubMed]
- Hultine, K.R.; Bean, D.W.; Dudley, T.L.; Gehring, C.A. Species Introductions and Their Cascading Impacts on Biotic Interactions in desert riparian ecosystems. Integr. Comp. Biol. 2015, 55, 587–601. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Gene Resources | Transformed Plants | Contribution to Salt Tolerance | References |
|---|---|---|---|
| TrSOS1 from Tamarix ramosissima | cotton | Na+ homeostasis | Che et al., 2019 [27] |
| Th2CysPrx from Tamarix hispida | T. hispida | Na+ homeostasis | Wang et al., 2020 [6] |
| ThNAC13 from T. hispida | T. hispida | osmotic adjustment | Wang et al., 2017 [46] |
| ThNAC7 from T. hispida | T. hispida | ROS homeostasis, osmotic adjustment | He et al., 2019 [47] |
| ThZFP1 from T. hispida | T. hispida, Arabidopsis | ROS homeostasis, osmotic adjustment | Zang et al., 2015 [48] |
| ThbZIP1 from T. hispida | tobacco | osmotic adjustment | Zang et al., 2017 [49] |
| ThGSTZ1 from T. hispida | T. hispida, Arabidopsis | ROS homeostasis | Yang et al., 2014 [50] |
| ThSOS3 from T. hispida | T. hispida | ROS homeostasis | Liu et al., 2021 [51] |
| ThNAC12 from T. hispida | T. hispida, Arabidopsis | ROS homeostasis | Wang et al., 2021 [52] |
| ThWRKY4 from T. hispida | T. hispida | ROS homeostasis | Zheng et al., 2013 [53] |
| ThHSFA1 from T. hispida | T. hispida | ROS homeostasis | Sun et al., 2021 [54] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duan, Q.; Zhu, Z.; Wang, B.; Chen, M. Recent Progress on the Salt Tolerance Mechanisms and Application of Tamarisk. Int. J. Mol. Sci. 2022, 23, 3325. https://doi.org/10.3390/ijms23063325
Duan Q, Zhu Z, Wang B, Chen M. Recent Progress on the Salt Tolerance Mechanisms and Application of Tamarisk. International Journal of Molecular Sciences. 2022; 23(6):3325. https://doi.org/10.3390/ijms23063325
Chicago/Turabian StyleDuan, Qixin, Zhihui Zhu, Baoshan Wang, and Min Chen. 2022. "Recent Progress on the Salt Tolerance Mechanisms and Application of Tamarisk" International Journal of Molecular Sciences 23, no. 6: 3325. https://doi.org/10.3390/ijms23063325
APA StyleDuan, Q., Zhu, Z., Wang, B., & Chen, M. (2022). Recent Progress on the Salt Tolerance Mechanisms and Application of Tamarisk. International Journal of Molecular Sciences, 23(6), 3325. https://doi.org/10.3390/ijms23063325





