The Prognostic Relevance of PMCA4 Expression in Melanoma: Gender Specificity and Implications for Immune Checkpoint Inhibition
Abstract
:1. Introduction
2. Results
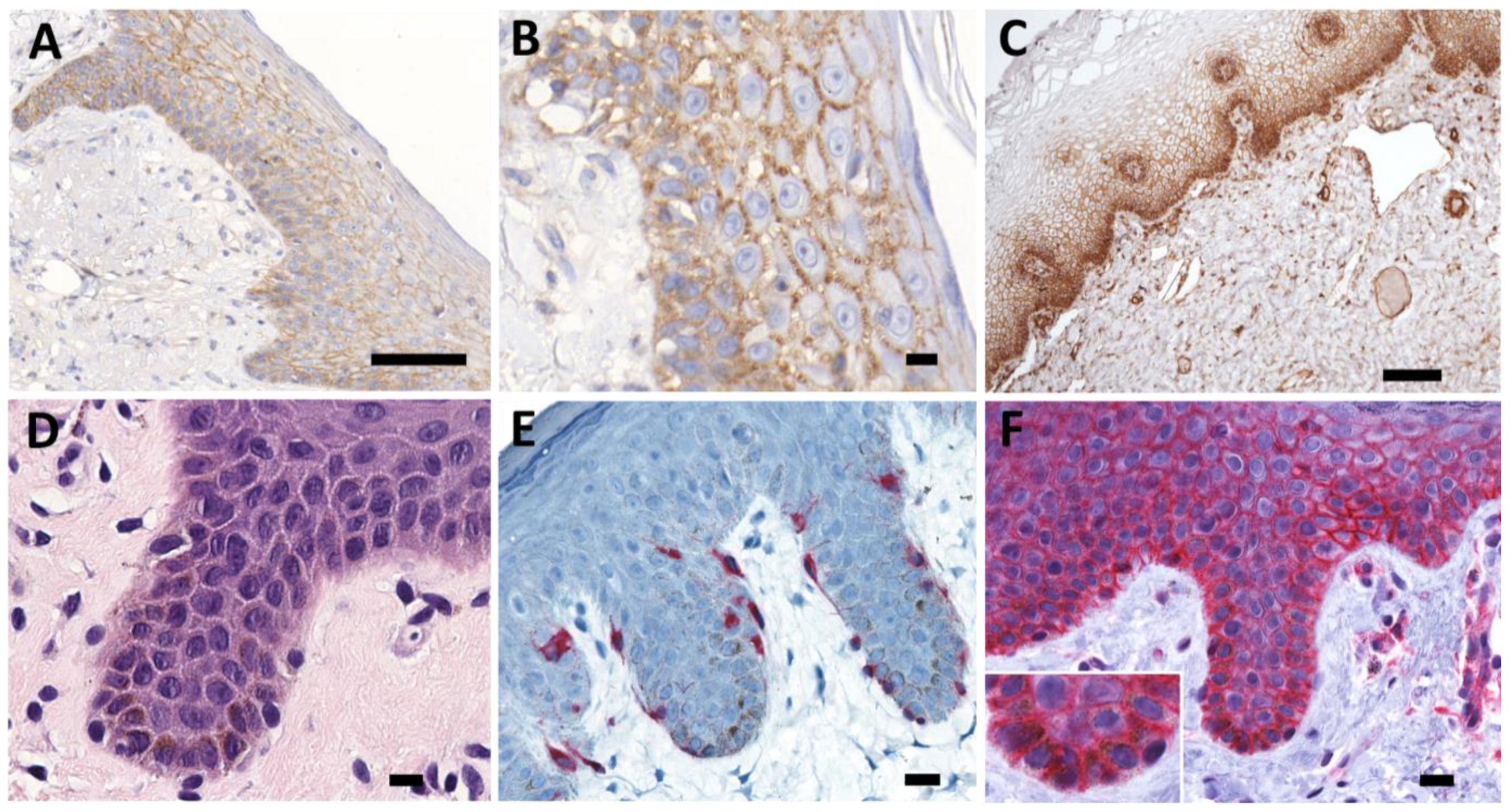
2.1. Differentiation Specific Expression of PMCA4 in Human Epithelia
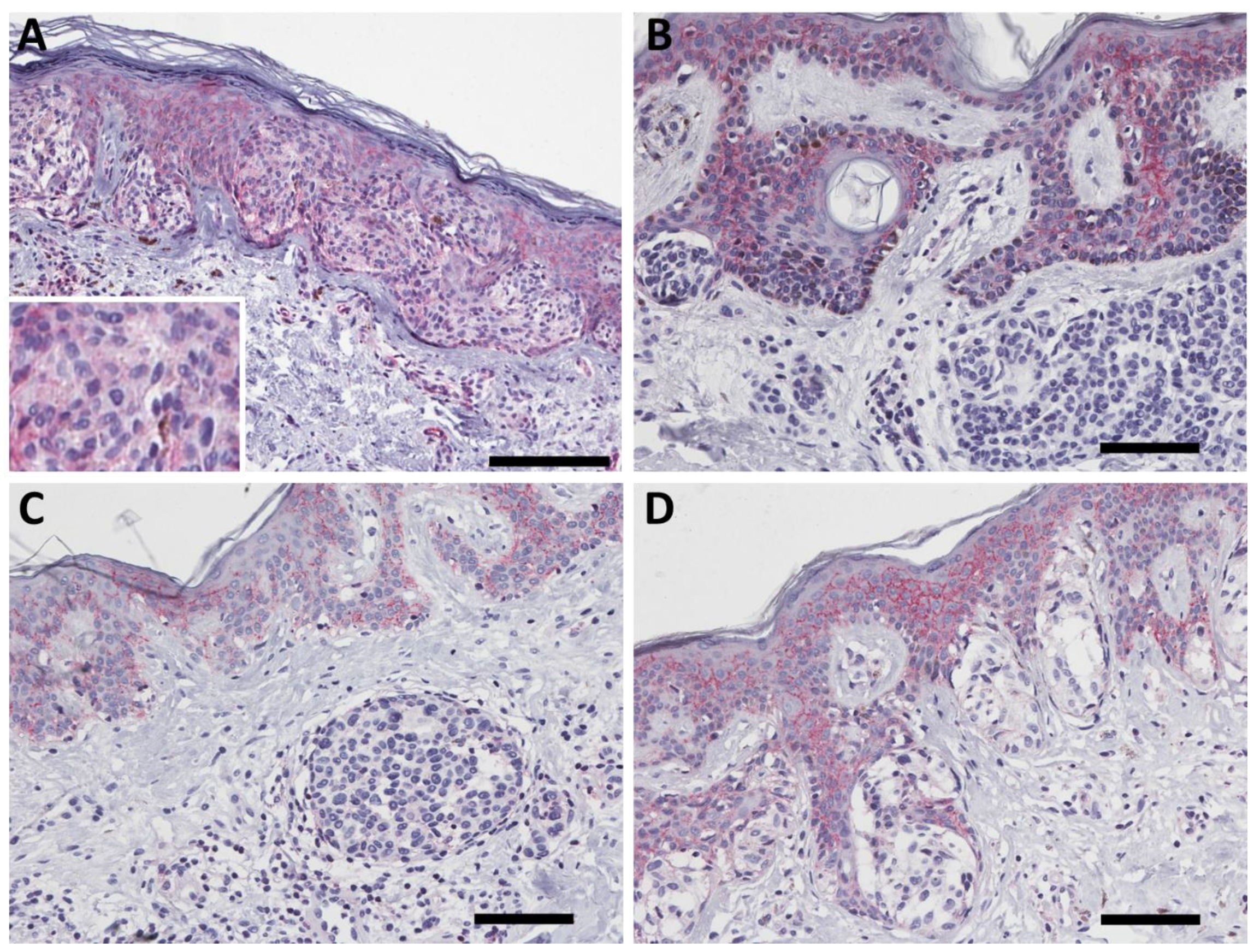
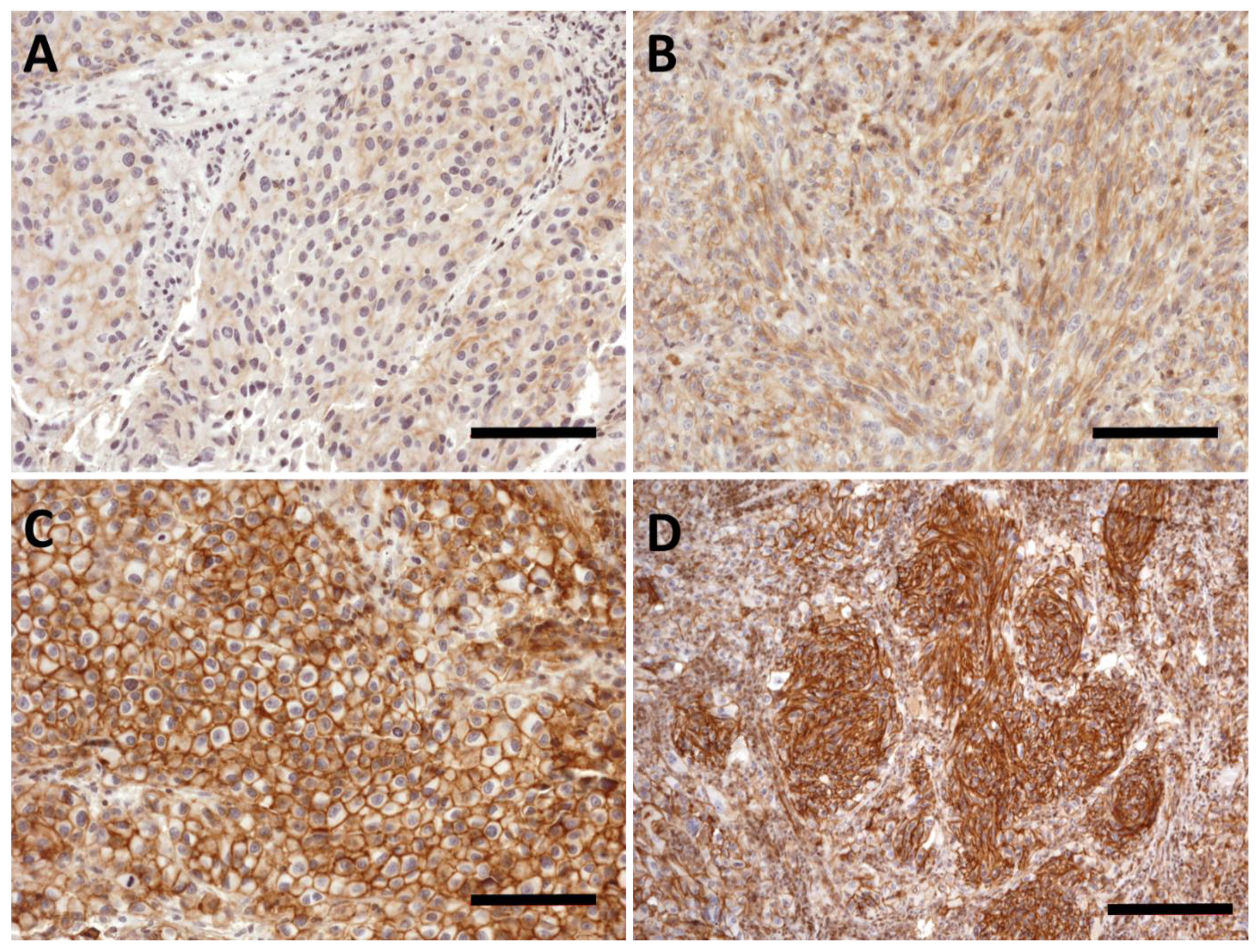
2.2. PMCA4b Expression in Nevus
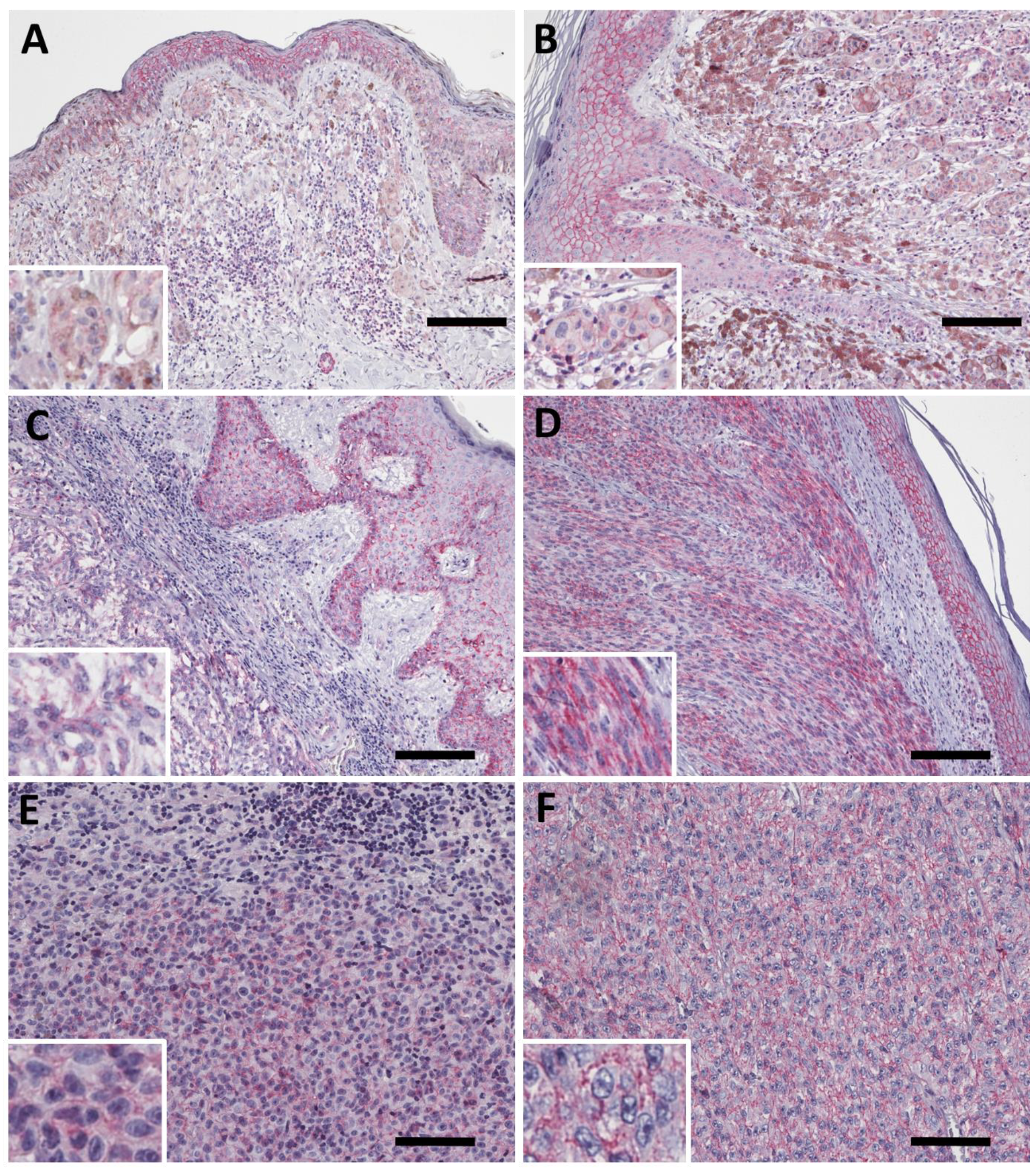
2.3. PMCA4b Expression in Primary Melanoma
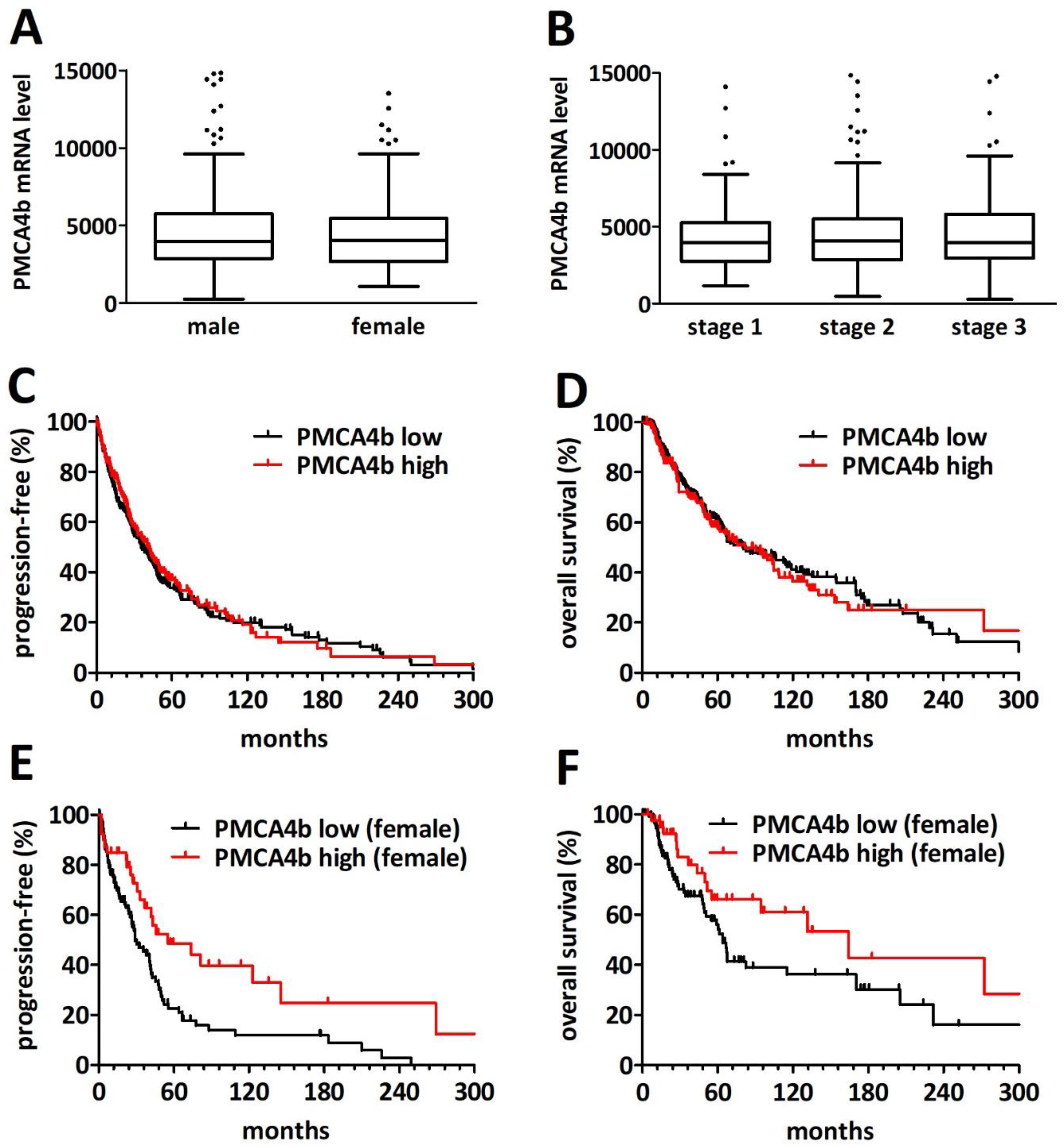
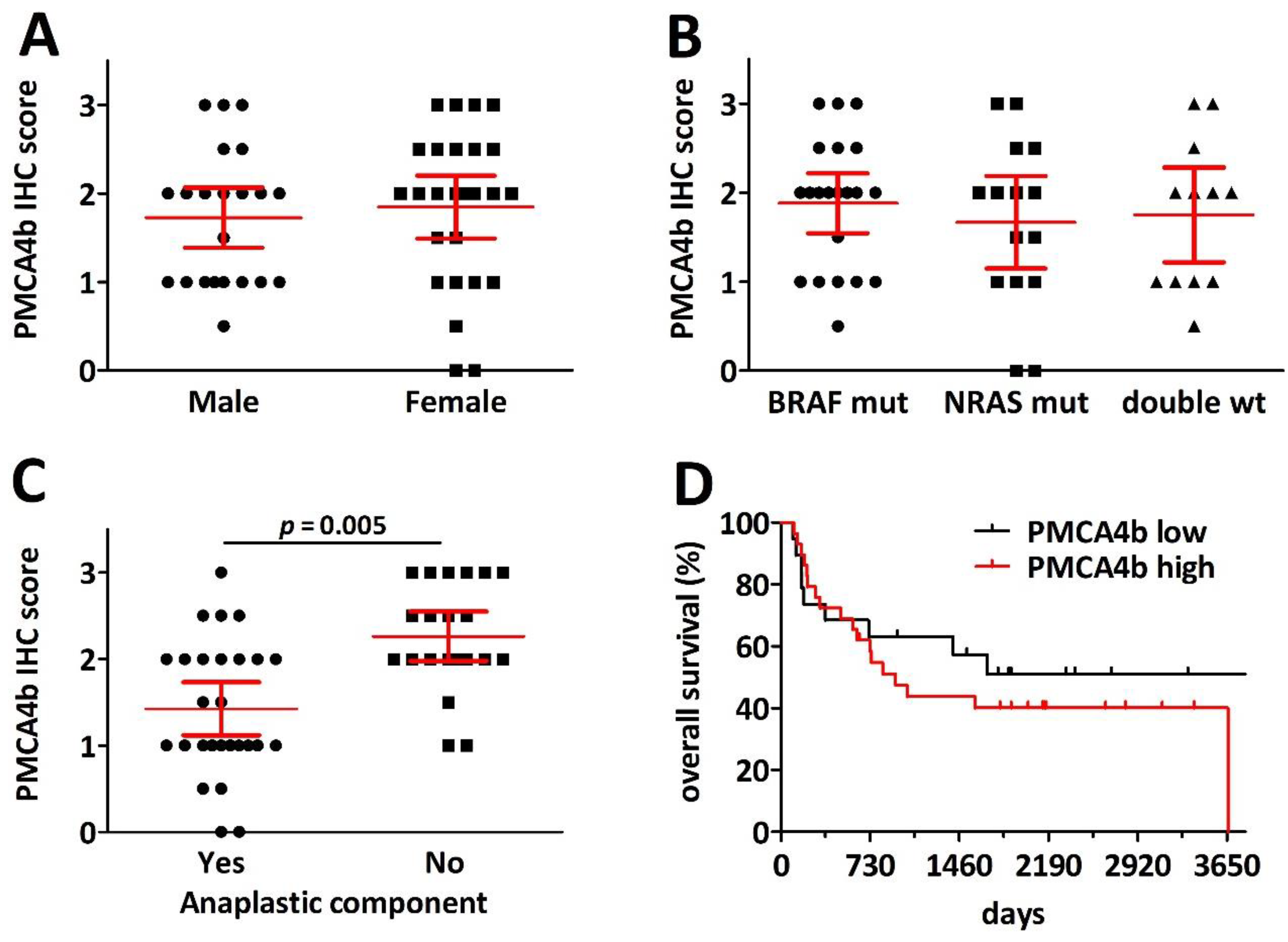
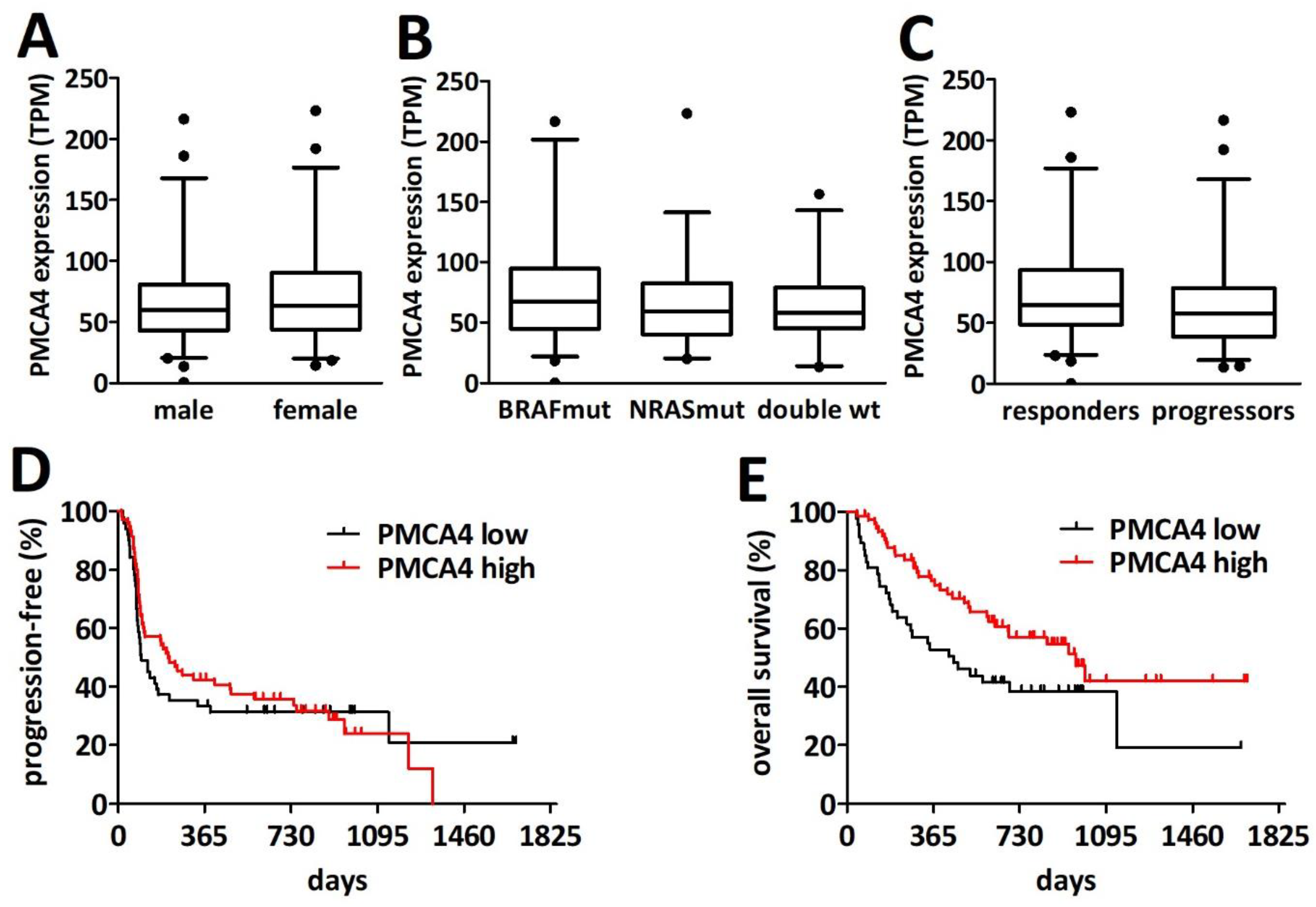
2.4. PMCA4 mRNA Level Is a Gender-Specific Prognosticator in Primary Cutaneous Melanoma
2.5. PMCA4b Expression Is Decreased in Pulmonary Melanoma Metastasis with Anaplasticity but Has No Prognostic Impact after Metastasectomy
2.6. PMCA4 mRNA Levels Are Prognostic for Overall Survival after PD1 Blockade Immune Checkpoint Inhibition Therapy
3. Discussion
4. Methods
4.1. Patient Cohorts
4.2. Immunohistochemistry
4.3. Mutational Analysis
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Eddy, K.; Shah, R.; Chen, S. Decoding Melanoma Development and Progression: Identification of Therapeutic Vulnerabilities. Front. Oncol. 2020, 10, 626129. [Google Scholar] [CrossRef]
- Yeh, I.; von Deimling, A.; Bastian, B.C. Clonal BRAF mutations in melanocytic nevi and initiating role of BRAF in melanocytic neoplasia. J. Natl. Cancer Inst. 2013, 105, 917–919. [Google Scholar] [CrossRef] [Green Version]
- Shain, A.H.; Yeh, I.; Kovalyshyn, I.; Sriharan, A.; Talevich, E.; Gagnon, A.; Dummer, R.; North, J.; Pincus, L.; Ruben, B.; et al. The Genetic Evolution of Melanoma from Precursor Lesions. N. Engl. J. Med. 2015, 373, 1926–1936. [Google Scholar] [CrossRef]
- Curtin, J.A.; Fridlyand, J.; Kageshita, T.; Patel, H.N.; Busam, K.J.; Kutzner, H.; Cho, K.H.; Aiba, S.; Brocker, E.B.; LeBoit, P.E.; et al. Distinct sets of genetic alterations in melanoma. N. Engl. J. Med. 2005, 353, 2135–2147. [Google Scholar] [CrossRef]
- Shain, A.H.; Bastian, B.C. From melanocytes to melanomas. Nat. Rev. Cancer 2016, 16, 345–358. [Google Scholar] [CrossRef]
- Carvajal, R.D.; Schwartz, G.K.; Tezel, T.; Marr, B.; Francis, J.H.; Nathan, P.D. Metastatic disease from uveal melanoma: Treatment options and future prospects. Br. J. Ophthalmol. 2017, 101, 38–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, C.; Merritt, R.; Fu, L.; Pan, Z. Targeting calcium signaling in cancer therapy. Acta Pharm. Sin. B 2017, 7, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Padanyi, R.; Paszty, K.; Hegedus, L.; Varga, K.; Papp, B.; Penniston, J.T.; Enyedi, A. Multifaceted plasma membrane Ca(2+) pumps: From structure to intracellular Ca(2+) handling and cancer. Biochim. Biophys. Acta 2016, 1863, 1351–1363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varga, K.; Hollosi, A.; Paszty, K.; Hegedus, L.; Szakacs, G.; Timar, J.; Papp, B.; Enyedi, A.; Padanyi, R. Expression of calcium pumps is differentially regulated by histone deacetylase inhibitors and estrogen receptor alpha in breast cancer cells. BMC Cancer 2018, 18, 1029. [Google Scholar] [CrossRef] [Green Version]
- Varga, K.; Paszty, K.; Padanyi, R.; Hegedus, L.; Brouland, J.P.; Papp, B.; Enyedi, A. Histone deacetylase inhibitor- and PMA-induced upregulation of PMCA4b enhances Ca2+ clearance from MCF-7 breast cancer cells. Cell Calcium 2014, 55, 78–92. [Google Scholar] [CrossRef] [PubMed]
- Ruschoff, J.H.; Brandenburger, T.; Strehler, E.E.; Filoteo, A.G.; Heinmoller, E.; Aumuller, G.; Wilhelm, B. Plasma membrane calcium ATPase expression in human colon multistep carcinogenesis. Cancer Investig. 2012, 30, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Sritangos, P.; Pena Alarcon, E.; James, A.D.; Sultan, A.; Richardson, D.A.; Bruce, J.I.E. Plasma Membrane Ca(2+) ATPase Isoform 4 (PMCA4) Has an Important Role in Numerous Hallmarks of Pancreatic Cancer. Cancers 2020, 12, 218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hegedus, L.; Garay, T.; Molnar, E.; Varga, K.; Bilecz, A.; Torok, S.; Padanyi, R.; Paszty, K.; Wolf, M.; Grusch, M.; et al. The plasma membrane Ca(2+) pump PMCA4b inhibits the migratory and metastatic activity of BRAF mutant melanoma cells. Int. J. Cancer 2017, 140, 2758–2770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hegedus, L.; Padanyi, R.; Molnar, J.; Paszty, K.; Varga, K.; Kenessey, I.; Sarkozy, E.; Wolf, M.; Grusch, M.; Hegyi, Z.; et al. Histone Deacetylase Inhibitor Treatment Increases the Expression of the Plasma Membrane Ca(2+) Pump PMCA4b and Inhibits the Migration of Melanoma Cells Independent of ERK. Front. Oncol. 2017, 7, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naffa, R.; Vogel, L.; Hegedus, L.; Paszty, K.; Toth, S.; Kelemen, K.; Singh, N.; Remenyi, A.; Kallay, E.; Cserepes, M.; et al. P38 MAPK Promotes Migration and Metastatic Activity of BRAF Mutant Melanoma Cells by Inducing Degradation of PMCA4b. Cells 2020, 9, 1209. [Google Scholar] [CrossRef] [PubMed]
- Naffa, R.; Padanyi, R.; Ignacz, A.; Hegyi, Z.; Jezso, B.; Toth, S.; Varga, K.; Homolya, L.; Hegedus, L.; Schlett, K.; et al. The Plasma Membrane Ca(2+) Pump PMCA4b Regulates Melanoma Cell Migration through Remodeling of the Actin Cytoskeleton. Cancers 2021, 13, 1354. [Google Scholar] [CrossRef] [PubMed]
- Chapman, P.B.; Hauschild, A.; Robert, C.; Haanen, J.B.; Ascierto, P.; Larkin, J.; Dummer, R.; Garbe, C.; Testori, A.; Maio, M.; et al. Improved Survival with Vemurafenib in Melanoma with BRAF V600E Mutation. N. Engl. J. Med. 2011, 364, 2507–2516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robert, C.; Karaszewska, B.; Schachter, J.; Rutkowski, P.; Mackiewicz, A.; Stroiakovski, D.; Lichinitser, M.; Dummer, R.; Grange, F.; Mortier, L.; et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N. Engl. J. Med. 2015, 372, 30–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hodi, F.S.; O’Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C.; et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010, 363, 711–723. [Google Scholar] [CrossRef]
- Robert, C.; Long, G.V.; Brady, B.; Dutriaux, C.; Maio, M.; Mortier, L.; Hassel, J.C.; Rutkowski, P.; McNeil, C.; Kalinka-Warzocha, E.; et al. Nivolumab in previously untreated melanoma without BRAF mutation. N. Engl. J. Med. 2015, 372, 320–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ko, J.S. The Immunology of Melanoma. Clin. Lab. Med. 2017, 37, 449–471. [Google Scholar] [CrossRef] [PubMed]
- Switzer, B.; Puzanov, I.; Skitzki, J.J.; Hamad, L.; Ernstoff, M.S. Managing Metastatic Melanoma in 2022: A Clinical Review. JCO Oncol. Pract. 2022, Op2100686. [Google Scholar] [CrossRef] [PubMed]
- Nebhan, C.A.; Johnson, D.B. Predictive biomarkers of response to immune checkpoint inhibitors in melanoma. Expert Rev. Anticancer Ther. 2020, 20, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Viehof, J.; Livingstone, E.; Loscha, E.; Stockhammer, P.; Bankfalvi, A.; Plones, T.; Mardanzai, K.; Zimmer, L.; Sucker, A.; Schadendorf, D.; et al. Prognostic factors for pulmonary metastasectomy in malignant melanoma: Size matters. Eur. J. Cardiothorac. Surg. 2019, 56, 1104–1109. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Schilling, B.; Liu, D.; Sucker, A.; Livingstone, E.; Jerby-Arnon, L.; Zimmer, L.; Gutzmer, R.; Satzger, I.; Loquai, C.; et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med. 2019, 25, 1916–1927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stafford, N.; Wilson, C.; Oceandy, D.; Neyses, L.; Cartwright, E.J. The Plasma Membrane Calcium ATPases and Their Role as Major New Players in Human Disease. Physiol. Rev. 2017, 97, 1089–1125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Prasad, V.; Hyung, S.W.; Lee, Z.H.; Lee, S.W.; Bhargava, A.; Pearce, D.; Lee, Y.; Kim, H.H. Plasma membrane calcium ATPase regulates bone mass by fine-tuning osteoclast differentiation and survival. J. Cell Biol. 2012, 199, 1145–1158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, R.; Liang, H.; Guo, H.; Wang, Z.; Deng, Q. PMCA4 gene expression is regulated by the androgen receptor in the mouse testis during spermatogenesis. Mol. Med. Rep. 2021, 23, 152. [Google Scholar] [CrossRef] [PubMed]
- Haqq, C.; Nosrati, M.; Sudilovsky, D.; Crothers, J.; Khodabakhsh, D.; Pulliam, B.L.; Federman, S.; Miller, J.R., 3rd; Allen, R.E.; Singer, M.I.; et al. The gene expression signatures of melanoma progression. Proc. Natl. Acad. Sci. USA 2005, 102, 6092–6097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talantov, D.; Mazumder, A.; Yu, J.X.; Briggs, T.; Jiang, Y.; Backus, J.; Atkins, D.; Wang, Y. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin. CancerRes. Off. J. Am. Assoc. Cancer Res. 2005, 11, 7234–7242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betancourt, L.H.; Gil, J.; Sanchez, A.; Doma, V.; Kuras, M.; Murillo, J.R.; Velasquez, E.; Cakir, U.; Kim, Y.; Sugihara, Y.; et al. The Human Melanoma Proteome Atlas-Complementing the melanoma transcriptome. Clin. Transl. Med. 2021, 11, e451. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Li, N.; Jin, L.; Qi, X.; Zhang, C.; Hua, D. The calcium pump PMCA4 prevents epithelial-mesenchymal transition by inhibiting NFATc1-ZEB1 pathway in gastric cancer. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118833. [Google Scholar] [CrossRef] [PubMed]
- Bellenghi, M.; Puglisi, R.; Pontecorvi, G.; De Feo, A.; Care, A.; Mattia, G. Sex and Gender Disparities in Melanoma. Cancers 2020, 12, 1819. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.L.; Flanagan, K.L. Sex differences in immune responses. Nat. Rev. Immunol. 2016, 16, 626–638. [Google Scholar] [CrossRef]
- Conforti, F.; Pala, L.; Bagnardi, V.; De Pas, T.; Martinetti, M.; Viale, G.; Gelber, R.D.; Goldhirsch, A. Cancer immunotherapy efficacy and patients’ sex: A systematic review and meta-analysis. Lancet Oncol. 2018, 19, 737–746. [Google Scholar] [CrossRef]
- Bautista, D.M.; Hoth, M.; Lewis, R.S. Enhancement of calcium signalling dynamics and stability by delayed modulation of the plasma-membrane calcium-ATPase in human T cells. J. Physiol. 2002, 541, 877–894. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, N.; Kollewe, A.; Constantin, C.E.; Henrich, S.; Ritzau-Jost, A.; Bildl, W.; Saalbach, A.; Hallermann, S.; Kulik, A.; Fakler, B.; et al. Neuroplastin and Basigin Are Essential Auxiliary Subunits of Plasma Membrane Ca(2+)-ATPases and Key Regulators of Ca(2+) Clearance. Neuron 2017, 96, 827–838.e829. [Google Scholar] [CrossRef] [Green Version]
- Korthals, M.; Langnaese, K.; Smalla, K.H.; Kahne, T.; Herrera-Molina, R.; Handschuh, J.; Lehmann, A.C.; Mamula, D.; Naumann, M.; Seidenbecher, C.; et al. A complex of Neuroplastin and Plasma Membrane Ca(2+) ATPase controls T cell activation. Sci. Rep. 2017, 7, 8358. [Google Scholar] [CrossRef] [PubMed]
- Supper, V.; Schiller, H.B.; Paster, W.; Forster, F.; Boulegue, C.; Mitulovic, G.; Leksa, V.; Ohradanova-Repic, A.; Machacek, C.; Schatzlmaier, P.; et al. Association of CD147 and Calcium Exporter PMCA4 Uncouples IL-2 Expression from Early TCR Signaling. J. Immunol. 2016, 196, 1387–1399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, P.W.-L.; Pang, S.Y.-Y.; Li, M.; Tse, Z.H.-M.; Kung, M.H.-W.; Sham, P.-C.; Ho, S.-L. PMCA4 (ATP2B4) mutation in familial spastic paraplegia causes delay in intracellular calcium extrusion. Brain Behav. 2015, 5, e00321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2018, 47, D941–D947. [Google Scholar] [CrossRef] [Green Version]
- Jin, X.; Demere, Z.; Nair, K.; Ali, A.; Ferraro, G.B.; Natoli, T.; Deik, A.; Petronio, L.; Tang, A.A.; Zhu, C.; et al. A metastasis map of human cancer cell lines. Nature 2020, 588, 331–336. [Google Scholar] [CrossRef]
- van de Nes, J.; Gessi, M.; Sucker, A.; Möller, I.; Stiller, M.; Horn, S.; Scholz, S.L.; Pischler, C.; Stadtler, N.; Schilling, B.; et al. Targeted next generation sequencing reveals unique mutation profile of primary melanocytic tumors of the central nervous system. J. Neuro-Oncol. 2016, 127, 435–444. [Google Scholar] [CrossRef] [Green Version]
- Lanczky, A.; Gyorffy, B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef]
| Total (n = 32) | ||
|---|---|---|
| Gender | male female | 16 (50%) 16 (50%) |
| Age | <60 years >60 years | 12 (37.5%) 20 (62.5)% |
| Site | trunk extremities head and neck choroidal conjunctival | 8 (25%) 7 (22%) 7 (22%) 7 (22%) 3 (9%) |
| pT stage (NA = 5) | 1 2 3 4 | 7 (28%) 6 (24% 8 (32%) 6 (24%) |
| Morphology | epitheloid spindle cell mixed | 20 (62.5%) 5 (15.5%) 7 (22%) |
| Total (n = 424) | Median PMCA4 mRNA Level | p-Value | ||
|---|---|---|---|---|
| Gender | male female | 262 162 | 3975 4043 | 0.596 |
| Age (NA = 5) | <60 ≥60 | 217 202 | 3991 4035 | 0.785 |
| Site (NA = 41) | trunk extremities head and neck | 156 182 45 | 3985 3962 4070 | 0.861 |
| Stage (NA = 34) | I II III | 80 140 170 | 3967 4068 3986 | 0.874 |
| Mutation | BRAF mutation NRAS mutation Double wildtype | 203 116 105 | 4102 4185 3629 | 0.142 |
| Total (n = 48) | PMCA4 Low (n = 19) | PMCA4 High (n = 29) | p-Value | ||
|---|---|---|---|---|---|
| Gender | male female | 22 (46%) 26 (54%) | 10 (53%) 9 (47%) | 12 (41%) 17 (59%) | 0.557 |
| Age | <60 ≥60 | 24 (50%) 24 (50%) | 12 (63%) 7 (37%) | 12 (41%) 17 (59%) | 0.238 |
| Anaplastic component | yes no | 27 (56%) 21 (44%) | 16 (84%) 3 (16%) | 11 (38%) 18 (62%) | 0.003 |
| Maximum nodule size | <20 mm ≥20 mm | 29 (60%) 19 (40%) | 11 (58%) 8 (42%) | 18 (62%) 11 38%) | 0.773 |
| Mutation | BRAF NRAS double WT | 21 (44%) 15 (31%) 12 (25%) | 7 (37%) 7 (37%) 5 (26%) | 14 (48%) 8 (28%) 7 (24%) | 0.712 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hegedüs, L.; Livingstone, E.; Bánkfalvi, Á.; Viehof, J.; Enyedi, Á.; Bilecz, Á.; Győrffy, B.; Baranyi, M.; Tőkés, A.-M.; Gil, J.; et al. The Prognostic Relevance of PMCA4 Expression in Melanoma: Gender Specificity and Implications for Immune Checkpoint Inhibition. Int. J. Mol. Sci. 2022, 23, 3324. https://doi.org/10.3390/ijms23063324
Hegedüs L, Livingstone E, Bánkfalvi Á, Viehof J, Enyedi Á, Bilecz Á, Győrffy B, Baranyi M, Tőkés A-M, Gil J, et al. The Prognostic Relevance of PMCA4 Expression in Melanoma: Gender Specificity and Implications for Immune Checkpoint Inhibition. International Journal of Molecular Sciences. 2022; 23(6):3324. https://doi.org/10.3390/ijms23063324
Chicago/Turabian StyleHegedüs, Luca, Elisabeth Livingstone, Ágnes Bánkfalvi, Jan Viehof, Ágnes Enyedi, Ágnes Bilecz, Balázs Győrffy, Marcell Baranyi, Anna-Mária Tőkés, Jeovanis Gil, and et al. 2022. "The Prognostic Relevance of PMCA4 Expression in Melanoma: Gender Specificity and Implications for Immune Checkpoint Inhibition" International Journal of Molecular Sciences 23, no. 6: 3324. https://doi.org/10.3390/ijms23063324
APA StyleHegedüs, L., Livingstone, E., Bánkfalvi, Á., Viehof, J., Enyedi, Á., Bilecz, Á., Győrffy, B., Baranyi, M., Tőkés, A.-M., Gil, J., Marko-Varga, G., Griewank, K. G., Zimmer, L., Váraljai, R., Sucker, A., Zaremba, A., Schadendorf, D., Aigner, C., & Hegedüs, B. (2022). The Prognostic Relevance of PMCA4 Expression in Melanoma: Gender Specificity and Implications for Immune Checkpoint Inhibition. International Journal of Molecular Sciences, 23(6), 3324. https://doi.org/10.3390/ijms23063324








